

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146650.9 - phase: 0 /pseudo
(314 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
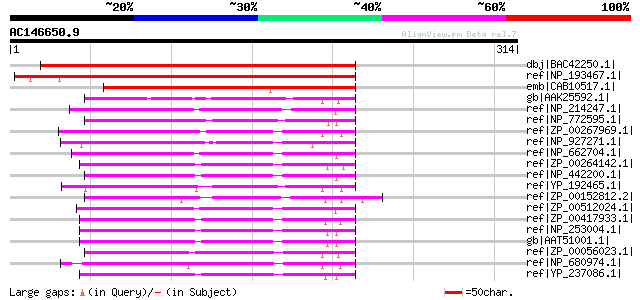
dbj|BAC42250.1| unknown protein [Arabidopsis thaliana] gi|201482... 294 3e-78
ref|NP_193467.1| formyltetrahydrofolate deformylase, putative [A... 291 2e-77
emb|CAB10517.1| formyltransferase purU homolog [Arabidopsis thal... 241 2e-62
gb|AAK25592.1| formyltetrahydrofolate deformylase [Caulobacter c... 109 1e-22
ref|NP_214247.1| formyltetrahydrofolate deformylase [Aquifex aeo... 108 2e-22
ref|NP_772595.1| formyltetrahydrofolate deformylase [Bradyrhizob... 99 1e-19
ref|ZP_00267969.1| COG0788: Formyltetrahydrofolate hydrolase [Rh... 97 5e-19
ref|NP_927271.1| formyltetrahydrofolate deformylase [Gloeobacter... 96 1e-18
ref|NP_662704.1| formyltetrahydrofolate deformylase [Chlorobium ... 96 2e-18
ref|ZP_00264142.1| COG0788: Formyltetrahydrofolate hydrolase [Ps... 96 2e-18
ref|NP_442200.1| phosphoribosylglycinamide formyltransferase [Sy... 95 3e-18
ref|YP_192465.1| Formyltetrahydrofolate deformylase [Gluconobact... 95 3e-18
ref|ZP_00152812.2| COG0788: Formyltetrahydrofolate hydrolase [De... 94 7e-18
ref|ZP_00512024.1| Formyltetrahydrofolate deformylase [Chlorobiu... 93 9e-18
ref|ZP_00417933.1| Formyltetrahydrofolate deformylase [Azotobact... 93 1e-17
ref|NP_253004.1| formyltetrahydrofolate deformylase [Pseudomonas... 93 1e-17
gb|AAT51001.1| PA4314 [synthetic construct] 93 1e-17
ref|ZP_00056023.1| COG0788: Formyltetrahydrofolate hydrolase [Ma... 92 2e-17
ref|NP_680974.1| formyltetrahydrofolate deformylase [Thermosynec... 92 2e-17
ref|YP_237086.1| Formyltetrahydrofolate deformylase [Pseudomonas... 91 4e-17
>dbj|BAC42250.1| unknown protein [Arabidopsis thaliana] gi|20148261|gb|AAM10021.1|
formyltetrahydrofolate deformylase-like [Arabidopsis
thaliana] gi|30695186|ref|NP_851145.1|
formyltetrahydrofolate deformylase, putative
[Arabidopsis thaliana] gi|18422794|ref|NP_568682.1|
formyltetrahydrofolate deformylase, putative
[Arabidopsis thaliana] gi|16648927|gb|AAL24315.1|
formyltetrahydrofolate deformylase-like [Arabidopsis
thaliana]
Length = 323
Score = 294 bits (752), Expect = 3e-78
Identities = 138/195 (70%), Positives = 164/195 (83%)
Query: 20 IRNFSFKSLDLPPLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNK 79
+++ F L SP L G+HVFHC DAVGIVAKLS+CIA++GGNIL DVFVP+N
Sbjct: 19 LKSSRFHGESLDSSVSPVLIPGVHVFHCQDAVGIVAKLSDCIAAKGGNILGYDVFVPENN 78
Query: 80 HVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHC 139
+VFYSRS+F+FDPVKWPR Q++EDF ++Q + A SVVRVP++DPKYKIA+L SKQDHC
Sbjct: 79 NVFYSRSEFIFDPVKWPRSQVDEDFQTIAQRYGALNSVVRVPSIDPKYKIALLLSKQDHC 138
Query: 140 LVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILE 199
LV++LH WQDGKLPVDITCVISNH R SNTHV+RFLERHGIPYH +STT ENKRE +ILE
Sbjct: 139 LVEMLHKWQDGKLPVDITCVISNHERASNTHVMRFLERHGIPYHYVSTTKENKREDDILE 198
Query: 200 LVQNTDFLVLARYMQ 214
LV++TDFLVLARYMQ
Sbjct: 199 LVKDTDFLVLARYMQ 213
>ref|NP_193467.1| formyltetrahydrofolate deformylase, putative [Arabidopsis thaliana]
Length = 328
Score = 291 bits (745), Expect = 2e-77
Identities = 146/218 (66%), Positives = 170/218 (77%), Gaps = 7/218 (3%)
Query: 4 VVRRVSQV--LGLTNSNKIRNFSFKSLD-----LPPLPSPSLSHGIHVFHCPDAVGIVAK 56
++RRVS L T +SFKS L SP L G HVFHCPD VGIVAK
Sbjct: 1 MIRRVSTTSCLSATAFRSFTKWSFKSSQFHGESLDSSVSPLLIPGFHVFHCPDVVGIVAK 60
Query: 57 LSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRS 116
LS+CIA++GGNIL DV VP+NK+VFYSRS+F+FDPVKWPR+QM+EDF ++Q F+A S
Sbjct: 61 LSDCIAAKGGNILGYDVLVPENKNVFYSRSEFIFDPVKWPRRQMDEDFQTIAQKFSALSS 120
Query: 117 VVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRDSNTHVIRFLE 176
VVRVP+LDPKYKIA+L SKQDHCLV++LH WQDGKLPVDITCVISNH R NTHV+RFL+
Sbjct: 121 VVRVPSLDPKYKIALLLSKQDHCLVEMLHKWQDGKLPVDITCVISNHERAPNTHVMRFLQ 180
Query: 177 RHGIPYHCLSTTNENKREGEILELVQNTDFLVLARYMQ 214
RHGI YH L TT++NK E EILELV+ TDFLVLARYMQ
Sbjct: 181 RHGISYHYLPTTDQNKIEEEILELVKGTDFLVLARYMQ 218
>emb|CAB10517.1| formyltransferase purU homolog [Arabidopsis thaliana]
gi|7268488|emb|CAB78739.1| formyltransferase purU
homolog [Arabidopsis thaliana] gi|7433570|pir||H71442
probable formyltransferase purU - Arabidopsis thaliana
Length = 295
Score = 241 bits (616), Expect = 2e-62
Identities = 116/165 (70%), Positives = 137/165 (82%), Gaps = 9/165 (5%)
Query: 59 ECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLKLSQAFNATRSVV 118
+CIA++GGNIL DV VP+NK+VFYSRS+F+FDPVKWPR+QM+EDF ++Q F+A SVV
Sbjct: 21 DCIAAKGGNILGYDVLVPENKNVFYSRSEFIFDPVKWPRRQMDEDFQTIAQKFSALSSVV 80
Query: 119 RVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVI---------SNHHRDSNT 169
RVP+LDPKYKIA+L SKQDHCLV++LH WQDGKLPVDITCVI SNH R NT
Sbjct: 81 RVPSLDPKYKIALLLSKQDHCLVEMLHKWQDGKLPVDITCVISDSGIFGVFSNHERAPNT 140
Query: 170 HVIRFLERHGIPYHCLSTTNENKREGEILELVQNTDFLVLARYMQ 214
HV+RFL+RHGI YH L TT++NK E EILELV+ TDFLVLARYMQ
Sbjct: 141 HVMRFLQRHGISYHYLPTTDQNKIEEEILELVKGTDFLVLARYMQ 185
>gb|AAK25592.1| formyltetrahydrofolate deformylase [Caulobacter crescentus CB15]
gi|16127860|ref|NP_422424.1| formyltetrahydrofolate
deformylase [Caulobacter crescentus CB15]
gi|25286210|pir||D87699 formyltetrahydrofolate
deformylase [imported] - Caulobacter crescentus
Length = 280
Score = 109 bits (272), Expect = 1e-22
Identities = 72/172 (41%), Positives = 99/172 (56%), Gaps = 13/172 (7%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIVAK+S + RG NIL A F Q F+ R VFD R+ + DF
Sbjct: 7 CPDQRGIVAKVSAFLFERGCNILDAQQFDDQETGQFFMR--VVFDADGADREALRGDFGA 64
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L+ F + +R A +Y++ +LASK DHCL DL++ W+ G+LP+DIT V+SNH
Sbjct: 65 LADGFKM-KWTLRNRA--DRYRVLLLASKFDHCLADLVYRWRIGELPMDITGVVSNHPAQ 121
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQ--NTDFLVLARYMQ 214
+ HV + G+ +H L T E K +E E+ +L+Q TD +VLARYMQ
Sbjct: 122 TYAHV----DLSGLDFHHLPVTKETKFEQEAELWKLIQETKTDIVVLARYMQ 169
>ref|NP_214247.1| formyltetrahydrofolate deformylase [Aquifex aeolicus VF5]
gi|2984098|gb|AAC07636.1| formyltetrahydrofolate
deformylase [Aquifex aeolicus VF5]
gi|7433562|pir||G70456 formyltetrahydrofolate
deformylase - Aquifex aeolicus
Length = 283
Score = 108 bits (270), Expect = 2e-22
Identities = 61/181 (33%), Positives = 99/181 (53%), Gaps = 12/181 (6%)
Query: 38 LSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPR 97
+ + I + CPD G+V ++S IA GGNI++ D + + F +R ++ + K PR
Sbjct: 1 MQNAILLVSCPDRKGLVKEISSFIADNGGNIVSFDQHIDEQTKTFLARVEWSLEDFKIPR 60
Query: 98 KQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDIT 157
+++E +F K++Q F+ ++ D K+A+ SKQ+HC DL+H + G+L ++
Sbjct: 61 EKIENEFKKVAQKFSMN---FQISFSDYVKKVAIFVSKQEHCFYDLMHRFYSGELKGEVK 117
Query: 158 CVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILEL----VQNTDFLVLARYM 213
VISNH + T E G+P++ + T ENK E E EL + +VLARYM
Sbjct: 118 LVISNHEKARKT-----AEFFGVPFYHIPKTKENKLEAEKRELELLKEYGVELVVLARYM 172
Query: 214 Q 214
Q
Sbjct: 173 Q 173
>ref|NP_772595.1| formyltetrahydrofolate deformylase [Bradyrhizobium japonicum USDA
110] gi|27354232|dbj|BAC51220.1| formyltetrahydrofolate
deformylase [Bradyrhizobium japonicum USDA 110]
Length = 287
Score = 99.4 bits (246), Expect = 1e-19
Identities = 66/172 (38%), Positives = 92/172 (53%), Gaps = 11/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIV+ +S +A G NIL A F F+ R F + ++ F
Sbjct: 12 CPDRPGIVSAVSTFLAHNGQNILDAQQFDDVETKKFFMRVVFTAADLAVELTALQTGFAA 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
+++ F + A K+ +L SK DHCLVD+L+ W+ G+LP+ T ++SNH R+
Sbjct: 72 IAERFGMEWQMRDRAA---HRKVMLLVSKSDHCLVDILYRWRTGELPMVPTAIVSNHPRE 128
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKREGE--ILELV--QNTDFLVLARYMQ 214
V L+ GIP+H L T E+KRE E IL+LV TD +VLARYMQ
Sbjct: 129 ----VYAGLDFGGIPFHHLPVTKESKREQEAQILDLVAKTGTDLVVLARYMQ 176
>ref|ZP_00267969.1| COG0788: Formyltetrahydrofolate hydrolase [Rhodospirillum rubrum]
Length = 297
Score = 97.4 bits (241), Expect = 5e-19
Identities = 65/189 (34%), Positives = 98/189 (51%), Gaps = 13/189 (6%)
Query: 31 PPLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVF 90
PP P+ I CPD G+VA +S + S+G I+ A + + F+ R+ F
Sbjct: 7 PPPPTDPRKAIILTITCPDGFGLVAAVSGFLNSQGAFIIEAAYYSDPDTGRFFMRTVFRS 66
Query: 91 DPVKWPRK-QMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQD 149
D P + E F +++ F T +V + + K K+ + S+ HCL DLLH WQ
Sbjct: 67 DTAGLPSLGALRECFAPVAERFEMTWDLV---SAERKPKVVIAVSRFGHCLYDLLHRWQA 123
Query: 150 GKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQNT--D 205
G+L V+I ++SNH + R E HGIP+H L T K +E IL+++ ++ D
Sbjct: 124 GQLHVEIPAIVSNH-----KDLARLAEWHGIPFHHLPVTTGGKEAQEEAILKVIDDSSAD 178
Query: 206 FLVLARYMQ 214
+VLARYMQ
Sbjct: 179 LVVLARYMQ 187
>ref|NP_927271.1| formyltetrahydrofolate deformylase [Gloeobacter violaceus PCC 7421]
gi|35214900|dbj|BAC92266.1| formyltetrahydrofolate
deformylase [Gloeobacter violaceus PCC 7421]
Length = 300
Score = 95.9 bits (237), Expect = 1e-18
Identities = 65/189 (34%), Positives = 100/189 (52%), Gaps = 14/189 (7%)
Query: 32 PLPSPSLSHGIH--VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKH-VFYSRSDF 88
P P ++S + CPD GIVA +S+C+ GGNIL +D +F+ R +F
Sbjct: 10 PAPDTAMSPATRTLLISCPDRRGIVAAVSQCVLEAGGNILRSDQHTTDPLGGIFFMRLEF 69
Query: 89 VFDPVKWPRKQMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQ 148
++ F +++ F +VR +PK ++A+ S+ DHC VDLL Q
Sbjct: 70 LYANAPADDDGFARCFAPVAERFAMDWRLVRTG--EPK-RMALFVSRLDHCFVDLLWRRQ 126
Query: 149 DGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLS--TTNENKREGEILELVQ-NTD 205
G+LPV I V+SNH + ++G+PYH L+ TN+ RE ++L L++ D
Sbjct: 127 SGELPVKIPLVVSNH-----PDLEPVAAQYGLPYHYLAIDKTNQPAREAQMLNLLEGEVD 181
Query: 206 FLVLARYMQ 214
F+VLARYM+
Sbjct: 182 FIVLARYMR 190
>ref|NP_662704.1| formyltetrahydrofolate deformylase [Chlorobium tepidum TLS]
gi|21647842|gb|AAM73046.1| formyltetrahydrofolate
deformylase [Chlorobium tepidum TLS]
Length = 289
Score = 95.5 bits (236), Expect = 2e-18
Identities = 60/180 (33%), Positives = 90/180 (49%), Gaps = 12/180 (6%)
Query: 39 SHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRK 98
S I + CPD VG+VA+++ I RGGNIL + V ++ F+ R + D P +
Sbjct: 8 SKAILLLSCPDRVGLVARIANFIYERGGNILDLNEHVDVDERQFFLRVSWSLDHFSIPAE 67
Query: 99 QMEEDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITC 158
+E F L + F A ++ + ++AV SK DHCL ++L G+ +D+
Sbjct: 68 DLESAFAPLGREFRAN---WQIRLSGKRSRMAVFVSKYDHCLREILWRHSLGEFDIDLPL 124
Query: 159 VISNHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARYMQ 214
VISNH + +E HGIP+H + T E K E ++ D +VLARYMQ
Sbjct: 125 VISNH-----PDLAPLVEAHGIPFHVIPVTPEAKAAAEQRQMALCDEHGIDTIVLARYMQ 179
>ref|ZP_00264142.1| COG0788: Formyltetrahydrofolate hydrolase [Pseudomonas fluorescens
PfO-1]
Length = 294
Score = 95.5 bits (236), Expect = 2e-18
Identities = 59/175 (33%), Positives = 93/175 (52%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A+ G I A F+ R + D + + + + E
Sbjct: 19 VISCPDRVGIVAKVSNFLAAHNGWITEASHHSDNQVGWFFMRHEIRADSLPFGIEVLREK 78
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ + K ++ ++AS++ HCL DLLH W +L +I+CVISNH
Sbjct: 79 FAPIAEEFSMDW---RITDTEQKKRVVLMASRESHCLADLLHRWHSDELDCEISCVISNH 135
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKREG--EILELVQNTD--FLVLARYMQ 214
+ +E HGIPY+ + ++K+ E+ LV+ D +VLARYMQ
Sbjct: 136 -----DDLRSMVEWHGIPYYHVPVNPQDKQPAFDEVSRLVKQHDAEVVVLARYMQ 185
>ref|NP_442200.1| phosphoribosylglycinamide formyltransferase [Synechocystis sp. PCC
6803] gi|1001129|dbj|BAA10270.1|
phosphoribosylglycinamide formyltransferase
[Synechocystis sp. PCC 6803]
gi|2500008|sp|Q55135|PURU_SYNY3 Formyltetrahydrofolate
deformylase (Formyl-FH(4) hydrolase)
Length = 284
Score = 94.7 bits (234), Expect = 3e-18
Identities = 57/172 (33%), Positives = 92/172 (53%), Gaps = 12/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD GIVA++++ I GNI+ AD + +F +R ++ D + R ++ + +
Sbjct: 12 CPDQPGIVAQIAQFIYQNQGNIIHADQHTDFSSGLFLNRVEWQLDNFRLSRPELLSAWSQ 71
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
L++ AT ++ D ++A+ SKQDHCL+D+L W+ G+L +I +ISNH
Sbjct: 72 LAEQLQATW---QIHFSDQLPRLALWVSKQDHCLLDILWRWRSGELRCEIPLIISNH--- 125
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENKREGEILELV----QNTDFLVLARYMQ 214
+ ++ GI +HCL T ENK E EL D +VLA+Y+Q
Sbjct: 126 --PDLKSIADQFGIDFHCLPITKENKLAQETAELALLKQYQIDLVVLAKYLQ 175
>ref|YP_192465.1| Formyltetrahydrofolate deformylase [Gluconobacter oxydans 621H]
gi|58002915|gb|AAW61809.1| Formyltetrahydrofolate
deformylase [Gluconobacter oxydans 621H]
Length = 292
Score = 94.7 bits (234), Expect = 3e-18
Identities = 65/193 (33%), Positives = 102/193 (52%), Gaps = 23/193 (11%)
Query: 33 LPSPSLSHGIHVF--HCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVF 90
+ +PS+ +V CP+ GIVA +S +A NI A F ++ VF+ R F
Sbjct: 1 MTAPSVPQTTYVLTLSCPNRPGIVAAISGRLAELNANITEAQQFDDRDSTVFFMRIVFEI 60
Query: 91 DPVKWPRKQMEEDFLKLSQAFNAT-----RSVVRVPALDPKYKIAVLASKQDHCLVDLLH 145
+ +Q+ E L + F+ RSV K K+ ++ S+ DHCLVDLL+
Sbjct: 61 TDGQTSMQQLREALAVLGETFSMDWALHDRSV--------KPKVLLMVSRFDHCLVDLLY 112
Query: 146 GWQDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQN 203
W+ G+LP++ ++SNH R+ V L+ +GIP+H L T + K +E +IL+L
Sbjct: 113 RWRIGELPIEPVGIVSNHPRE----VFADLDFYGIPFHYLPVTKDTKPAQEAQILDLFAA 168
Query: 204 T--DFLVLARYMQ 214
T + ++LARYMQ
Sbjct: 169 TGAELVILARYMQ 181
>ref|ZP_00152812.2| COG0788: Formyltetrahydrofolate hydrolase [Dechloromonas aromatica
RCB]
Length = 289
Score = 93.6 bits (231), Expect = 7e-18
Identities = 69/197 (35%), Positives = 96/197 (48%), Gaps = 24/197 (12%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFL- 105
CPD VGI+A++S IA GG IL + ++ R + D + + + E F
Sbjct: 15 CPDQVGIIARVSGFIAGNGGWILESSFHSDVLTGRYFMRIEIKADSLPFLLAEFRERFRI 74
Query: 106 ----KLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVIS 161
LS + S V K ++ VL SKQ+HCL DLL WQ +L ++I CVIS
Sbjct: 75 EVAEPLSMTWQINDSAV-------KKRVVVLVSKQEHCLYDLLARWQAKELDIEIPCVIS 127
Query: 162 NHHRDSNTHVIRFLERHGIPYHCLSTTNENKRE--GEILELVQNT--DFLVLARYMQATG 217
NH F+E HGIP+H + T +NK EI + ++ D +VLARYMQ
Sbjct: 128 NHDTFRG-----FVEWHGIPFHHVPVTADNKAAAYAEIQRIFEDVRGDSMVLARYMQVLS 182
Query: 218 ---MT*LTFTMVCCHHS 231
LT ++ HHS
Sbjct: 183 PELCDALTGKIINIHHS 199
>ref|ZP_00512024.1| Formyltetrahydrofolate deformylase [Chlorobium limicola DSM 245]
gi|67783899|gb|EAM43280.1| Formyltetrahydrofolate
deformylase [Chlorobium limicola DSM 245]
Length = 287
Score = 93.2 bits (230), Expect = 9e-18
Identities = 60/177 (33%), Positives = 93/177 (51%), Gaps = 12/177 (6%)
Query: 42 IHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQME 101
I + CPD G+V+++S I RGGNIL D V + +F+ R + D P +++
Sbjct: 9 ILLLSCPDRAGLVSRISHFIYERGGNILDLDEHVDTVEKMFFIRVSWSTDHFSIPPTELD 68
Query: 102 EDFLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVIS 161
E F L++ F A+ ++ D K ++A+ S+ DHCL +LL G+ +DI ++S
Sbjct: 69 EAFSPLAKEFGAS---WKIRLGDRKMRVALFVSRYDHCLQELLWRHSIGEFRIDIPLIVS 125
Query: 162 NHHRDSNTHVIRFLERHGIPYHCLSTTNENKREGEILEL----VQNTDFLVLARYMQ 214
NH + R+GIP+H T +K+E E EL + D +VLARYMQ
Sbjct: 126 NH-----PDLEPLALRYGIPFHVFPVTAASKQEIEQQELGLLRDHDIDTVVLARYMQ 177
>ref|ZP_00417933.1| Formyltetrahydrofolate deformylase [Azotobacter vinelandii AvOP]
gi|67086099|gb|EAM05569.1| Formyltetrahydrofolate
deformylase [Azotobacter vinelandii AvOP]
Length = 283
Score = 92.8 bits (229), Expect = 1e-17
Identities = 59/175 (33%), Positives = 89/175 (50%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A+ G I A F+ R + D + + ++
Sbjct: 7 VIACPDRVGIVAKVSNFLATYNGWITEASHHSDTQSGWFFMRHEIRADSLPFDLEEFRRA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ RV ++ ++AS++ HCL DLLH W G+LP +I CVISNH
Sbjct: 67 FAPIAREFSMKW---RVSDSSELKRVVLMASRESHCLADLLHRWHSGELPCEIPCVISNH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKRE--GEILELVQN--TDFLVLARYMQ 214
+ +E HGIPY + ++K E+ L++ D +VLARYMQ
Sbjct: 124 -----DELRSMVEWHGIPYCHVPVDPQDKEPAFAEVSRLIREHAADTVVLARYMQ 173
>ref|NP_253004.1| formyltetrahydrofolate deformylase [Pseudomonas aeruginosa PAO1]
gi|9950538|gb|AAG07702.1| formyltetrahydrofolate
deformylase [Pseudomonas aeruginosa PAO1]
gi|11348559|pir||C83105 formyltetrahydrofolate
deformylase PA4314 [imported] - Pseudomonas aeruginosa
(strain PAO1) gi|46164795|ref|ZP_00137792.2| COG0788:
Formyltetrahydrofolate hydrolase [Pseudomonas aeruginosa
UCBPP-PA14]
Length = 283
Score = 92.8 bits (229), Expect = 1e-17
Identities = 58/175 (33%), Positives = 90/175 (51%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A+ G I A + F+ R + D + + +
Sbjct: 7 VIACPDGVGIVAKVSNFLATYNGWITEASHHSDNDNGWFFMRHEIRADSLPFDLDGFRQA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ + K ++ ++ASK+ HCL DLLH W G+L +I CVI+NH
Sbjct: 67 FAPIAREFSMEW---RITDSEVKKRVVLMASKESHCLADLLHRWHSGELDCEIPCVIANH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKREG--EILELV--QNTDFLVLARYMQ 214
+ +E HGIPY + ++K+ E+ L+ D +VLARYMQ
Sbjct: 124 -----DDLRSMVEWHGIPYFHVPVDPQDKQPAFDEVSRLIDEHGADCIVLARYMQ 173
>gb|AAT51001.1| PA4314 [synthetic construct]
Length = 284
Score = 92.8 bits (229), Expect = 1e-17
Identities = 58/175 (33%), Positives = 90/175 (51%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +A+ G I A + F+ R + D + + +
Sbjct: 7 VIACPDGVGIVAKVSNFLATYNGWITEASHHSDNDNGWFFMRHEIRADSLPFDLDGFRQA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ + K ++ ++ASK+ HCL DLLH W G+L +I CVI+NH
Sbjct: 67 FAPIAREFSMEW---RITDSEVKKRVVLMASKESHCLADLLHRWHSGELDCEIPCVIANH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKREG--EILELV--QNTDFLVLARYMQ 214
+ +E HGIPY + ++K+ E+ L+ D +VLARYMQ
Sbjct: 124 -----DDLRSMVEWHGIPYFHVPVDPQDKQPAFDEVSRLIDEHGADCIVLARYMQ 173
>ref|ZP_00056023.1| COG0788: Formyltetrahydrofolate hydrolase [Magnetospirillum
magnetotacticum MS-1]
Length = 286
Score = 92.0 bits (227), Expect = 2e-17
Identities = 59/172 (34%), Positives = 93/172 (53%), Gaps = 12/172 (6%)
Query: 47 CPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEEDFLK 106
CPD VGIVA +S +++ I A F F+ R F + P ++E+ F
Sbjct: 13 CPDTVGIVAAVSGFLSTHDCFITEAAQFGDPLSSRFFMRIVFGAGAMTPPMAELEKLFTG 72
Query: 107 LSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNHHRD 166
+++ F + + ++ K ++ +LASK HCL DLLH + G LP++I VISNH
Sbjct: 73 VAERF---QMIWKLHDCRKKARVVILASKFGHCLNDLLHRYHTGSLPIEIPAVISNHQ-- 127
Query: 167 SNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQ--NTDFLVLARYMQ 214
+ +E HGIPYH L+ +K +E ++E+++ + D +VLARYMQ
Sbjct: 128 ---DMRSIVEWHGIPYHYLAVDKHDKLTQENRVMEVIERADADLVVLARYMQ 176
>ref|NP_680974.1| formyltetrahydrofolate deformylase [Thermosynechococcus elongatus
BP-1] gi|22293904|dbj|BAC07736.1| formyltetrahydrofolate
deformylase [Thermosynechococcus elongatus BP-1]
Length = 291
Score = 92.0 bits (227), Expect = 2e-17
Identities = 65/191 (34%), Positives = 103/191 (53%), Gaps = 18/191 (9%)
Query: 32 PLPSPSLSHGIHVFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFD 91
P+P+ +LS CPD G+VAKL++ + GNI+ AD +F SR ++ +
Sbjct: 2 PMPTMTLS-----ISCPDQRGLVAKLAQFVYRYNGNIVHADHHTDAVAGIFLSRLEWELE 56
Query: 92 PVKWPRKQMEEDFLKLSQ---AFNATRSV-VRVPALDPKYKIAVLASKQDHCLVDLLHGW 147
+ PR Q+ F+ +Q F + + V ++ A D Y++A+ S+QDHCL DLL
Sbjct: 57 GFEIPRDQIATTFINYAQREKIFTSWQGVRWQLRASDIPYRLAIWVSRQDHCLWDLLLRQ 116
Query: 148 QDGKLPVDITCVISNHHRDSNTHVIRFLERHGIPYHCLSTTNENK--REGEILELVQN-- 203
+ G L +I +ISNH H+ E+ GI +H + T E K E + L+L+++
Sbjct: 117 RAGDLFAEIPLIISNHE-----HLRPIAEQFGIDFHYIPVTPETKPLAEAKQLQLLKDYR 171
Query: 204 TDFLVLARYMQ 214
D +VLA+YMQ
Sbjct: 172 IDLVVLAKYMQ 182
>ref|YP_237086.1| Formyltetrahydrofolate deformylase [Pseudomonas syringae pv.
syringae B728a] gi|63257952|gb|AAY39048.1|
Formyltetrahydrofolate deformylase [Pseudomonas syringae
pv. syringae B728a]
Length = 283
Score = 90.9 bits (224), Expect = 4e-17
Identities = 60/175 (34%), Positives = 86/175 (48%), Gaps = 12/175 (6%)
Query: 44 VFHCPDAVGIVAKLSECIASRGGNILAADVFVPQNKHVFYSRSDFVFDPVKWPRKQMEED 103
V CPD VGIVAK+S +AS G I A F+ R + D + + E
Sbjct: 7 VIACPDRVGIVAKVSNFLASHNGWITEASHHSDNLSGWFFMRHEIRADTLPFDLDGFREA 66
Query: 104 FLKLSQAFNATRSVVRVPALDPKYKIAVLASKQDHCLVDLLHGWQDGKLPVDITCVISNH 163
F +++ F+ R+ K ++ ++AS++ HCL DLLH W +L DI CVISNH
Sbjct: 67 FTPIAEEFSMDW---RITDSAQKKRVVLMASRESHCLADLLHRWHSDELDCDIACVISNH 123
Query: 164 HRDSNTHVIRFLERHGIPYHCLSTTNENKRE--GEILELV--QNTDFLVLARYMQ 214
+ +E H IPY+ + ++K E+ LV D +VLARYMQ
Sbjct: 124 Q-----DLRSMVEWHDIPYYHVPVDPKDKEPAFAEVSRLVGHHQADVVVLARYMQ 173
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.333 0.144 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,696,676
Number of Sequences: 2540612
Number of extensions: 18918115
Number of successful extensions: 74579
Number of sequences better than 10.0: 190
Number of HSP's better than 10.0 without gapping: 172
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 74098
Number of HSP's gapped (non-prelim): 195
length of query: 314
length of database: 863,360,394
effective HSP length: 128
effective length of query: 186
effective length of database: 538,162,058
effective search space: 100098142788
effective search space used: 100098142788
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146650.9


