

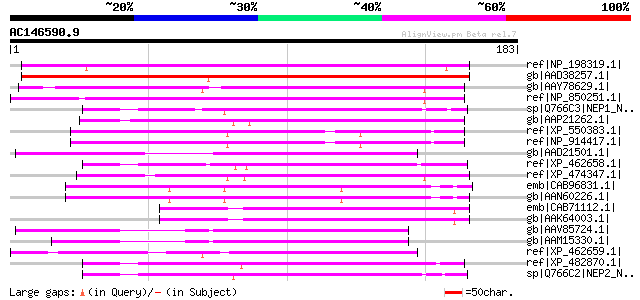
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.9 - phase: 0 /pseudo
(183 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198319.1| aspartyl protease family protein [Arabidopsis t... 154 1e-36
gb|AAD38257.1| Hypothetical Protein [Arabidopsis thaliana] gi|15... 153 2e-36
gb|AAY78629.1| aspartyl protease family protein [Arabidopsis tha... 128 7e-29
ref|NP_850251.1| aspartyl protease family protein [Arabidopsis t... 128 7e-29
sp|Q766C3|NEP1_NEPGR Aspartic proteinase nepenthesin-1 precursor... 105 6e-22
gb|AAP21262.1| At2g03200 [Arabidopsis thaliana] gi|7487145|pir||... 102 4e-21
ref|XP_550383.1| putative CDR1 [Oryza sativa (japonica cultivar-... 100 1e-20
ref|NP_914417.1| P0509B06.7 [Oryza sativa (japonica cultivar-gro... 100 1e-20
gb|AAD21501.1| putative chloroplast nucleoid DNA binding protein... 99 7e-20
ref|XP_462658.1| OSJNBa0064H22.10 [Oryza sativa (japonica cultiv... 98 1e-19
ref|XP_474347.1| OSJNBa0064G10.12 [Oryza sativa (japonica cultiv... 97 2e-19
emb|CAB96831.1| nucleoid DNA-binding protein cnd41-like protein ... 95 8e-19
gb|AAN60226.1| unknown [Arabidopsis thaliana] 95 1e-18
emb|CAB71112.1| putative protein [Arabidopsis thaliana] gi|15228... 95 1e-18
gb|AAK64003.1| AT3g61820/F15G16_210 [Arabidopsis thaliana] 95 1e-18
gb|AAV85724.1| At2g28040 [Arabidopsis thaliana] gi|28392898|gb|A... 94 2e-18
gb|AAM15330.1| putative chloroplast nucleoid DNA binding protein... 93 3e-18
ref|XP_462659.1| OSJNBa0064H22.11 [Oryza sativa (japonica cultiv... 92 5e-18
ref|XP_482870.1| putative nucleoid DNA-binding protein [Oryza sa... 91 1e-17
sp|Q766C2|NEP2_NEPGR Aspartic proteinase nepenthesin-2 precursor... 90 3e-17
>ref|NP_198319.1| aspartyl protease family protein [Arabidopsis thaliana]
gi|37935737|gb|AAP72988.1| CDR1 [Arabidopsis thaliana]
Length = 437
Score = 154 bits (389), Expect = 1e-36
Identities = 78/168 (46%), Positives = 99/168 (58%), Gaps = 6/168 (3%)
Query: 5 SLLILFYFSLCFIISLSHALNN-----GFSVELIHRDSSKSPLYQPTQNKYQHIVNAARR 59
SL SLC + SL + N GF+ +LIHRDS KSP Y P + Q + NA R
Sbjct: 3 SLFSSVLLSLCLLSSLFLSNANAKPKLGFTADLIHRDSPKSPFYNPMETSSQRLRNAIHR 62
Query: 60 SINRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCK 119
S+NR HF + T PQ + + GEYLM S+GTPPF + IADTGSD++W QC PC
Sbjct: 63 SVNRVFHFTEKDNTPQPQIDLTSNSGEYLMNVSIGTPPFPIMAIADTGSDLLWTQCAPCD 122
Query: 120 ECYNQTTPKFKPSKSSTYKNIPCSSDLCKSGQQGAVC-TDQNFCEYSI 166
+CY Q P F P SSTYK++ CSS C + + A C T+ N C YS+
Sbjct: 123 DCYTQVDPLFDPKTSSTYKDVSCSSSQCTALENQASCSTNDNTCSYSL 170
>gb|AAD38257.1| Hypothetical Protein [Arabidopsis thaliana]
gi|15217764|ref|NP_176663.1| aspartyl protease family
protein [Arabidopsis thaliana] gi|25404498|pir||E96671
hypothetical protein F13O11.13 [imported] - Arabidopsis
thaliana
Length = 431
Score = 153 bits (387), Expect = 2e-36
Identities = 71/163 (43%), Positives = 101/163 (61%), Gaps = 1/163 (0%)
Query: 5 SLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRA 64
SL+ SL + +++ +GF+++LIHRDS KSP Y + Q + NA RRS
Sbjct: 3 SLIFATLLSLLLLSNVNAYPKDGFTIDLIHRDSPKSPFYNSAETSSQRMRNAIRRSARST 62
Query: 65 NHFYKT-ALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYN 123
F A N+PQS + + GEYLM S+GTPP + IADTGSD++W QC PC++CY
Sbjct: 63 LQFSNDDASPNSPQSFITSNRGEYLMNISIGTPPVPILAIADTGSDLIWTQCNPCEDCYQ 122
Query: 124 QTTPKFKPSKSSTYKNIPCSSDLCKSGQQGAVCTDQNFCEYSI 166
QT+P F P +SSTY+ + CSS C++ + + TD+N C Y+I
Sbjct: 123 QTSPLFDPKESSTYRKVSCSSSQCRALEDASCSTDENTCSYTI 165
>gb|AAY78629.1| aspartyl protease family protein [Arabidopsis thaliana]
gi|15222357|ref|NP_174430.1| aspartyl protease family
protein [Arabidopsis thaliana] gi|25513600|pir||E86440
probable chloroplast nucleoid DNA binding protein
T8E3.12 - Arabidopsis thaliana
gi|12322538|gb|AAG51267.1| chloroplast nucleoid DNA
binding protein, putative [Arabidopsis thaliana]
Length = 445
Score = 128 bits (322), Expect = 7e-29
Identities = 72/164 (43%), Positives = 95/164 (57%), Gaps = 11/164 (6%)
Query: 4 CSLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINR 63
CSLL + +F S S A +VELIHRDS SPLY P + A RSI+R
Sbjct: 9 CSLLAISFF----FASNSSANRENLTVELIHRDSPHSPLYNPHHTVSDRLNAAFLRSISR 64
Query: 64 ANHFY-KTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECY 122
+ F KT L QS +I + GEY M+ S+GTPP K++ IADTGSD+ W+QC+PC++CY
Sbjct: 65 SRRFTTKTDL----QSGLISNGGEYFMSISIGTPPSKVFAIADTGSDLTWVQCKPCQQCY 120
Query: 123 NQTTPKFKPSKSSTYKNIPCSSDLCK--SGQQGAVCTDQNFCEY 164
Q +P F KSSTYK C S C+ S + ++ C+Y
Sbjct: 121 KQNSPLFDKKKSSTYKTESCDSKTCQALSEHEEGCDESKDICKY 164
>ref|NP_850251.1| aspartyl protease family protein [Arabidopsis thaliana]
Length = 447
Score = 128 bits (322), Expect = 7e-29
Identities = 69/166 (41%), Positives = 92/166 (54%), Gaps = 4/166 (2%)
Query: 1 MNTCSLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRS 60
M T LL F F + S H N FSVELIHRDS SP+Y P + A RS
Sbjct: 1 MATQILLCFFLFFSVTLSSSGHPKN--FSVELIHRDSPLSPIYNPQITVTDRLNAAFLRS 58
Query: 61 INRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKE 120
++R+ F QS +I GE+ M+ ++GTPP K++ IADTGSD+ W+QC+PC++
Sbjct: 59 VSRSRRFNHQLSQTDLQSGLIGADGEFFMSITIGTPPIKVFAIADTGSDLTWVQCKPCQQ 118
Query: 121 CYNQTTPKFKPSKSSTYKNIPCSSDLCK--SGQQGAVCTDQNFCEY 164
CY + P F KSSTYK+ PC S C+ S + N C+Y
Sbjct: 119 CYKENGPIFDKKKSSTYKSEPCDSRNCQALSSTERGCDESNNICKY 164
>sp|Q766C3|NEP1_NEPGR Aspartic proteinase nepenthesin-1 precursor (Nepenthesin-I)
gi|41016421|dbj|BAD07474.1| aspartic proteinase
nepenthesin I [Nepenthes gracilis]
Length = 437
Score = 105 bits (262), Expect = 6e-22
Identities = 56/142 (39%), Positives = 80/142 (55%), Gaps = 13/142 (9%)
Query: 27 GFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTP---QSTVIPD 83
GF + L H DS K+ K+Q + A R R A+ N P +++V
Sbjct: 40 GFQIMLEHVDSGKN------LTKFQLLERAIERGSRRLQRL--EAMLNGPSGVETSVYAG 91
Query: 84 HGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCS 143
GEYLM S+GTP I DTGSD++W QC+PC +C+NQ+TP F P SS++ +PCS
Sbjct: 92 DGEYLMNLSIGTPAQPFSAIMDTGSDLIWTQCQPCTQCFNQSTPIFNPQGSSSFSTLPCS 151
Query: 144 SDLCKSGQQGAVCTDQNFCEYS 165
S LC++ C++ NFC+Y+
Sbjct: 152 SQLCQA-LSSPTCSN-NFCQYT 171
>gb|AAP21262.1| At2g03200 [Arabidopsis thaliana] gi|7487145|pir||T02706
hypothetical protein At2g03200 [imported] - Arabidopsis
thaliana gi|30678047|ref|NP_565298.2| aspartyl protease
family protein [Arabidopsis thaliana]
Length = 461
Score = 102 bits (255), Expect = 4e-21
Identities = 56/145 (38%), Positives = 76/145 (51%), Gaps = 9/145 (6%)
Query: 26 NGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQST---VIP 82
+GF + L H DS K+ K Q +N +NR A+ + P T P
Sbjct: 43 SGFRLSLRHVDSGKN---LTKIQKIQRGINRGFHRLNRLGAVAVLAVASKPDDTNNIKAP 99
Query: 83 DHG---EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKN 139
HG E+LM S+G P K I DTGSD++W QC+PC EC++Q TP F P KSS+Y
Sbjct: 100 THGGSGEFLMELSIGNPAVKYSAIVDTGSDLIWTQCKPCTECFDQPTPIFDPEKSSSYSK 159
Query: 140 IPCSSDLCKSGQQGAVCTDQNFCEY 164
+ CSS LC + + D++ CEY
Sbjct: 160 VGCSSGLCNALPRSNCNEDKDACEY 184
>ref|XP_550383.1| putative CDR1 [Oryza sativa (japonica cultivar-group)]
gi|55296252|dbj|BAD67993.1| putative CDR1 [Oryza sativa
(japonica cultivar-group)] gi|55296112|dbj|BAD67831.1|
putative CDR1 [Oryza sativa (japonica cultivar-group)]
Length = 454
Score = 100 bits (250), Expect = 1e-20
Identities = 61/156 (39%), Positives = 86/156 (55%), Gaps = 18/156 (11%)
Query: 23 ALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQ----- 77
A GFSVE IHRDS +SP + P + + AARRS+ RA +A ++
Sbjct: 29 ASGGGFSVEFIHRDSPRSPFHDPAFTAHGRALAAARRSVARAAAIAGSASSSASGGGAAD 88
Query: 78 ---STVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQT------TPK 128
S V+ EYLMT ++G+PP + IADTGSD+VW++ CK+ N T T +
Sbjct: 89 DVVSKVVSRSFEYLMTVNLGSPPRSMLAIADTGSDLVWVK---CKKGNNDTSSAAAPTTQ 145
Query: 129 FKPSKSSTYKNIPCSSDLCKSGQQGAVCTDQNFCEY 164
F PS+SSTY + C +D C++ + A C D + C Y
Sbjct: 146 FDPSRSSTYGRVSCQTDACEALGR-ATCDDGSNCAY 180
>ref|NP_914417.1| P0509B06.7 [Oryza sativa (japonica cultivar-group)]
Length = 451
Score = 100 bits (250), Expect = 1e-20
Identities = 61/156 (39%), Positives = 86/156 (55%), Gaps = 18/156 (11%)
Query: 23 ALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQ----- 77
A GFSVE IHRDS +SP + P + + AARRS+ RA +A ++
Sbjct: 26 ASGGGFSVEFIHRDSPRSPFHDPAFTAHGRALAAARRSVARAAAIAGSASSSASGGGAAD 85
Query: 78 ---STVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQT------TPK 128
S V+ EYLMT ++G+PP + IADTGSD+VW++ CK+ N T T +
Sbjct: 86 DVVSKVVSRSFEYLMTVNLGSPPRSMLAIADTGSDLVWVK---CKKGNNDTSSAAAPTTQ 142
Query: 129 FKPSKSSTYKNIPCSSDLCKSGQQGAVCTDQNFCEY 164
F PS+SSTY + C +D C++ + A C D + C Y
Sbjct: 143 FDPSRSSTYGRVSCQTDACEALGR-ATCDDGSNCAY 177
>gb|AAD21501.1| putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana] gi|25407941|pir||F84679 hypothetical protein
At2g28010 [imported] - Arabidopsis thaliana
gi|15226315|ref|NP_180368.1| aspartyl protease family
protein [Arabidopsis thaliana]
Length = 396
Score = 98.6 bits (244), Expect = 7e-20
Identities = 51/145 (35%), Positives = 73/145 (50%), Gaps = 24/145 (16%)
Query: 3 TCSLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSIN 62
T +++ SLCF+ + + + +GF+++LIHR S+ S TQ+
Sbjct: 5 TTIIVLFLQISLCFLFTTTASPPHGFTMDLIHRRSNASSRVSNTQSG------------- 51
Query: 63 RANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECY 122
++P + + D+ YLM VGTPPF++ I DTGS+I W QC PC CY
Sbjct: 52 -----------SSPYANTVFDNSVYLMKLQVGTPPFEIQAIIDTGSEITWTQCLPCVHCY 100
Query: 123 NQTTPKFKPSKSSTYKNIPCSSDLC 147
Q P F PSKSST+K C C
Sbjct: 101 EQNAPIFDPSKSSTFKEKRCDGHSC 125
>ref|XP_462658.1| OSJNBa0064H22.10 [Oryza sativa (japonica cultivar-group)]
gi|38344829|emb|CAD40873.2| OSJNBa0064H22.10 [Oryza
sativa (japonica cultivar-group)]
Length = 444
Score = 97.8 bits (242), Expect = 1e-19
Identities = 58/151 (38%), Positives = 80/151 (52%), Gaps = 20/151 (13%)
Query: 27 GFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQSTV------ 80
G V L H D+ + +++Q + AARRS +R + A T P ++
Sbjct: 30 GLRVHLTHVDAHGN------YSRHQLLRRAARRSHHRMSRLVARA-TGVPMTSSKAAGGG 82
Query: 81 ---IPDH---GEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKS 134
+P H GE+LM S+GTP I DTGSD+VW QC+PC +C+ Q+TP F PS S
Sbjct: 83 DLQVPVHAGNGEFLMDVSIGTPALAYSAIVDTGSDLVWTQCKPCVDCFKQSTPVFDPSSS 142
Query: 135 STYKNIPCSSDLCKSGQQGAVCTDQNFCEYS 165
STY +PCSS C S + CT + C Y+
Sbjct: 143 STYATVPCSSASC-SDLPTSKCTSASKCGYT 172
>ref|XP_474347.1| OSJNBa0064G10.12 [Oryza sativa (japonica cultivar-group)]
gi|38344196|emb|CAE05761.2| OSJNBa0064G10.12 [Oryza
sativa (japonica cultivar-group)]
Length = 451
Score = 97.1 bits (240), Expect = 2e-19
Identities = 52/155 (33%), Positives = 79/155 (50%), Gaps = 16/155 (10%)
Query: 25 NNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQ------- 77
NN S+ L+HRD+ Y +++ +V R R H K + +T
Sbjct: 60 NNNPSLSLVHRDAISGATYPSRRHQ---VVGLVARDNARVEHLEKRLVASTSPYLPEDLV 116
Query: 78 STVIPD----HGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSK 133
S V+P GEY + VG+PP Y + D+GSD++W+QC PC++CY QT P F P+
Sbjct: 117 SEVVPGVDDGSGEYFVRVGVGSPPTDQYLVVDSGSDVIWVQCRPCEQCYAQTDPLFDPAA 176
Query: 134 SSTYKNIPCSSDLCK--SGQQGAVCTDQNFCEYSI 166
SS++ + C S +C+ SG D C+YS+
Sbjct: 177 SSSFSGVSCGSAICRTLSGTGCGGGGDAGKCDYSV 211
>emb|CAB96831.1| nucleoid DNA-binding protein cnd41-like protein [Arabidopsis
thaliana] gi|22136986|gb|AAM91722.1| putative nucleoid
DNA-binding protein cnd41 [Arabidopsis thaliana]
gi|18176136|gb|AAL59990.1| putative nucleoid DNA-binding
protein cnd41 [Arabidopsis thaliana]
gi|15238250|ref|NP_196637.1| aspartyl protease family
protein [Arabidopsis thaliana] gi|11358587|pir||T50785
nucleoid DNA-binding protein cnd41-like protein -
Arabidopsis thaliana
Length = 464
Score = 95.1 bits (235), Expect = 8e-19
Identities = 56/158 (35%), Positives = 81/158 (50%), Gaps = 15/158 (9%)
Query: 21 SHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNA---------ARRSINRANHFYKTA 71
S A N S+ ++H + S L + + I+ ++ S N AN +
Sbjct: 56 SKASNTKSSLRVVHMHGACSHLSSDARVDHDEIIRRDQARVESIYSKLSKNSANEVSEAK 115
Query: 72 LTNTP-QSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPC-KECYNQTTPKF 129
T P +S + G Y++T +GTP L + DTGSD+ W QCEPC CY+Q PKF
Sbjct: 116 STELPAKSGITLGSGNYIVTIGIGTPKHDLSLVFDTGSDLTWTQCEPCLGSCYSQKEPKF 175
Query: 130 KPSKSSTYKNIPCSSDLCKSGQQGAVCTDQNFCEYSIL 167
PS SSTY+N+ CSS +C+ + C+ N C YSI+
Sbjct: 176 NPSSSSTYQNVSCSSPMCEDAES---CSASN-CVYSIV 209
>gb|AAN60226.1| unknown [Arabidopsis thaliana]
Length = 464
Score = 94.7 bits (234), Expect = 1e-18
Identities = 56/157 (35%), Positives = 80/157 (50%), Gaps = 15/157 (9%)
Query: 21 SHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNA---------ARRSINRANHFYKTA 71
S A N S+ ++H + S L + + I+ ++ S N AN +
Sbjct: 56 SKASNTKSSLRVVHMHGACSHLSSDARVDHDEIIRRDQARVESIYSKLSKNSANEVSEAK 115
Query: 72 LTNTP-QSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPC-KECYNQTTPKF 129
T P +S + G Y++T +GTP L + DTGSD+ W QCEPC CY+Q PKF
Sbjct: 116 STELPAKSGITLGSGNYIVTIGIGTPKHDLSLVFDTGSDLTWTQCEPCLGSCYSQKEPKF 175
Query: 130 KPSKSSTYKNIPCSSDLCKSGQQGAVCTDQNFCEYSI 166
PS SSTY+N+ CSS +C+ + C+ N C YSI
Sbjct: 176 NPSSSSTYQNVSCSSPMCEDAES---CSASN-CVYSI 208
>emb|CAB71112.1| putative protein [Arabidopsis thaliana]
gi|15228618|ref|NP_191741.1| aspartyl protease family
protein [Arabidopsis thaliana] gi|11357465|pir||T47974
hypothetical protein F15G16.210 - Arabidopsis thaliana
Length = 483
Score = 94.7 bits (234), Expect = 1e-18
Identities = 46/114 (40%), Positives = 60/114 (52%), Gaps = 7/114 (6%)
Query: 55 NAARRSINRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQ 114
NA +R+ A F ++ Q + GEY M VGTP +Y + DTGSD+VWLQ
Sbjct: 108 NATKRTPRTAGGFSGAVISGLSQGS-----GEYFMRLGVGTPATNVYMVLDTGSDVVWLQ 162
Query: 115 CEPCKECYNQTTPKFKPSKSSTYKNIPCSSDLCKSGQQGAVCTDQ--NFCEYSI 166
C PCK CYNQT F P KS T+ +PC S LC+ + C + C Y +
Sbjct: 163 CSPCKACYNQTDAIFDPKKSKTFATVPCGSRLCRRLDDSSECVTRRSKTCLYQV 216
>gb|AAK64003.1| AT3g61820/F15G16_210 [Arabidopsis thaliana]
Length = 362
Score = 94.7 bits (234), Expect = 1e-18
Identities = 46/114 (40%), Positives = 60/114 (52%), Gaps = 7/114 (6%)
Query: 55 NAARRSINRANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQ 114
NA +R+ A F ++ Q + GEY M VGTP +Y + DTGSD+VWLQ
Sbjct: 108 NATKRTPRTAGGFSGAVISGLSQGS-----GEYFMRLGVGTPATNVYMVLDTGSDVVWLQ 162
Query: 115 CEPCKECYNQTTPKFKPSKSSTYKNIPCSSDLCKSGQQGAVCTDQ--NFCEYSI 166
C PCK CYNQT F P KS T+ +PC S LC+ + C + C Y +
Sbjct: 163 CSPCKACYNQTDAIFDPKKSKTFATVPCGSRLCRRLDDSSECVTRRSKTCLYQV 216
>gb|AAV85724.1| At2g28040 [Arabidopsis thaliana] gi|28392898|gb|AAO41885.1|
putative chloroplast nucleoid DNA binding protein
[Arabidopsis thaliana] gi|30683732|ref|NP_180371.2|
aspartyl protease family protein [Arabidopsis thaliana]
Length = 395
Score = 94.0 bits (232), Expect = 2e-18
Identities = 50/142 (35%), Positives = 73/142 (51%), Gaps = 24/142 (16%)
Query: 3 TCSLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSIN 62
T + I F+I+ + + GF+++LIHR S+ S
Sbjct: 5 TTMIAIFLQIITYFLITTTASSPQGFTIDLIHRRSNAS---------------------- 42
Query: 63 RANHFYKTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECY 122
++ + T L +P + + D EYLM +GTPPF++ + DTGS+ +W QC PC CY
Sbjct: 43 -SSRVFNTQL-GSPYADTVFDTYEYLMKLQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCY 100
Query: 123 NQTTPKFKPSKSSTYKNIPCSS 144
NQT P F PSKSST+K I C +
Sbjct: 101 NQTAPIFDPSKSSTFKEIRCDT 122
>gb|AAM15330.1| putative chloroplast nucleoid DNA binding protein [Arabidopsis
thaliana] gi|4063754|gb|AAC98462.1| putative chloroplast
nucleoid DNA binding protein [Arabidopsis thaliana]
gi|25407946|pir||A84680 hypothetical protein At2g28040
[imported] - Arabidopsis thaliana
Length = 389
Score = 93.2 bits (230), Expect = 3e-18
Identities = 48/129 (37%), Positives = 70/129 (54%), Gaps = 24/129 (18%)
Query: 16 FIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNT 75
F+I+ + + GF+++LIHR S+ S ++ + T L +
Sbjct: 12 FLITTTASSPQGFTIDLIHRRSNAS-----------------------SSRVFNTQL-GS 47
Query: 76 PQSTVIPDHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSS 135
P + + D EYLM +GTPPF++ + DTGS+ +W QC PC CYNQT P F PSKSS
Sbjct: 48 PYADTVFDTYEYLMKLQIGTPPFEIEAVLDTGSEHIWTQCLPCVHCYNQTAPIFDPSKSS 107
Query: 136 TYKNIPCSS 144
T+K I C +
Sbjct: 108 TFKEIRCDT 116
>ref|XP_462659.1| OSJNBa0064H22.11 [Oryza sativa (japonica cultivar-group)]
gi|38344830|emb|CAD40872.2| OSJNBa0064H22.11 [Oryza
sativa (japonica cultivar-group)]
Length = 464
Score = 92.4 bits (228), Expect = 5e-18
Identities = 55/161 (34%), Positives = 78/161 (48%), Gaps = 26/161 (16%)
Query: 1 MNTCSLLILFYFSLCFIISLSHALNNGFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRS 60
MN LL+L + + S A F +EL D+S + T+++ RR+
Sbjct: 1 MNAAVLLLLLALAA---LPASCAPPRSFRLELASVDASAADAANLTEHEL------LRRA 51
Query: 61 INRANHFY--------------KTALTNTPQSTVIPDHGEYLMTYSVGTPPFKLYGIADT 106
I R+ + K + TP ++P GEYL+ +GTPP+K DT
Sbjct: 52 IQRSRYRLAGIGMARGEAASARKAVVAETP---IMPAGGEYLVKLGIGTPPYKFTAAIDT 108
Query: 107 GSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSSDLC 147
SD++W QC+PC CY+Q P F P SSTY +PCSSD C
Sbjct: 109 ASDLIWTQCQPCTGCYHQVDPMFNPRVSSTYAALPCSSDTC 149
>ref|XP_482870.1| putative nucleoid DNA-binding protein [Oryza sativa (japonica
cultivar-group)] gi|42407407|dbj|BAD09565.1| putative
nucleoid DNA-binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 448
Score = 91.3 bits (225), Expect = 1e-17
Identities = 51/149 (34%), Positives = 74/149 (49%), Gaps = 18/149 (12%)
Query: 27 GFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQSTVIP---- 82
GF ++L H D+ S K + + A RRS R A + V+
Sbjct: 27 GFQLKLRHVDAHGS------YTKLELVTRAIRRSRARVAALQAVAAAAATVAPVVDPITA 80
Query: 83 -------DHGEYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSS 135
GEYLM ++GTPP + + DTGSD++W QC PC C +Q TP F+P++S+
Sbjct: 81 ARILVAASQGEYLMDLAIGTPPLRYTAMVDTGSDLIWTQCAPCVLCADQPTPYFRPARSA 140
Query: 136 TYKNIPCSSDLCKSGQQGAVCTDQNFCEY 164
TY+ +PC S LC + A C ++ C Y
Sbjct: 141 TYRLVPCRSPLCAALPYPA-CFQRSVCVY 168
>sp|Q766C2|NEP2_NEPGR Aspartic proteinase nepenthesin-2 precursor (Nepenthesin-II)
gi|41016423|dbj|BAD07475.1| aspartic proteinase
nepenthesin II [Nepenthes gracilis]
Length = 438
Score = 90.1 bits (222), Expect = 3e-17
Identities = 50/140 (35%), Positives = 71/140 (50%), Gaps = 9/140 (6%)
Query: 27 GFSVELIHRDSSKSPLYQPTQNKYQHIVNAARRSINRANHFYKTALTNTPQST-VIPDHG 85
G V+L DS K+ KY+ I A +R R +++ T V G
Sbjct: 41 GLRVDLEQVDSGKN------LTKYELIKRAIKRGERRMRSINAMLQSSSGIETPVYAGDG 94
Query: 86 EYLMTYSVGTPPFKLYGIADTGSDIVWLQCEPCKECYNQTTPKFKPSKSSTYKNIPCSSD 145
EYLM ++GTP I DTGSD++W QCEPC +C++Q TP F P SS++ +PC S
Sbjct: 95 EYLMNVAIGTPDSSFSAIMDTGSDLIWTQCEPCTQCFSQPTPIFNPQDSSSFSTLPCESQ 154
Query: 146 LCKSGQQGAVCTDQNFCEYS 165
C+ C + N C+Y+
Sbjct: 155 YCQD-LPSETC-NNNECQYT 172
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.136 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 319,762,934
Number of Sequences: 2540612
Number of extensions: 13218692
Number of successful extensions: 29915
Number of sequences better than 10.0: 676
Number of HSP's better than 10.0 without gapping: 492
Number of HSP's successfully gapped in prelim test: 192
Number of HSP's that attempted gapping in prelim test: 29278
Number of HSP's gapped (non-prelim): 698
length of query: 183
length of database: 863,360,394
effective HSP length: 120
effective length of query: 63
effective length of database: 558,486,954
effective search space: 35184678102
effective search space used: 35184678102
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146590.9


