

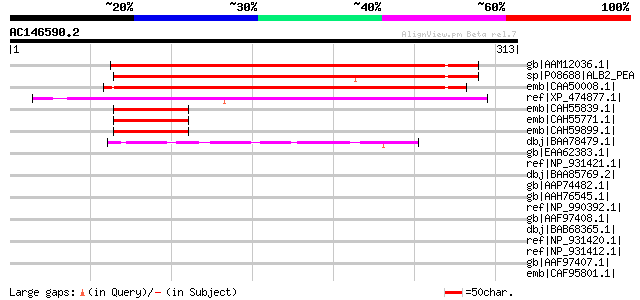
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.2 + phase: 0
(313 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM12036.1| anther-specific protein [Pisum sativum] 270 3e-71
sp|P08688|ALB2_PEA Albumin 2 (PA2) gi|169033|gb|AAA33641.1| majo... 261 2e-68
emb|CAA50008.1| mung bean seed albumin [Vigna radiata var. radia... 230 4e-59
ref|XP_474877.1| OSJNBa0021F22.4 [Oryza sativa (japonica cultiva... 217 3e-55
emb|CAH55839.1| seed albumin 2 [Pisum sativum] 65 3e-09
emb|CAH55771.1| seed albumin 2 [Pisum sativum] 65 3e-09
emb|CAH59899.1| seed albumin 2 [Pisum sativum] 63 1e-08
dbj|BAA78479.1| matrix metalloproteinase-2 [Oncorhynchus mykiss] 49 2e-04
gb|EAA62383.1| predicted protein [Aspergillus nidulans FGSC A4] ... 45 0.002
ref|NP_931421.1| photopexin B [Photorhabdus luminescens subsp. l... 42 0.018
dbj|BAA85769.2| gelatinase A [Oryzias latipes] 42 0.018
gb|AAP74482.1| matrix metalloproteinase 2 [Danio rerio] gi|37620... 41 0.039
gb|AAH76545.1| Matrix metalloproteinase 2 [Danio rerio] 41 0.039
ref|NP_990392.1| vitronectin [Gallus gallus] gi|1922282|emb|CAA7... 40 0.067
gb|AAF97408.1| photopexin B [Photorhabdus luminescens] 40 0.067
dbj|BAB68365.1| gelatinase [Paralichthys olivaceus] 40 0.067
ref|NP_931420.1| photopexin A [Photorhabdus luminescens subsp. l... 39 0.15
ref|NP_931412.1| hypothetical protein plu4231 [Photorhabdus lumi... 39 0.20
gb|AAF97407.1| photopexin A [Photorhabdus luminescens] 39 0.20
emb|CAF95801.1| unnamed protein product [Tetraodon nigroviridis] 39 0.20
>gb|AAM12036.1| anther-specific protein [Pisum sativum]
Length = 230
Score = 270 bits (691), Expect = 3e-71
Identities = 133/228 (58%), Positives = 169/228 (73%), Gaps = 2/228 (0%)
Query: 63 PIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEH 122
P YINAAFR+S N E YLF++++YVL++YA G+ +D ++NGPL + G+ SL T FG +
Sbjct: 4 PGYINAAFRSSFNGERYLFIDDKYVLVDYAPGTRDDKLLNGPLPLPAGFKSLDGTVFGTY 63
Query: 123 GIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDA 182
G+DCAFDTD +A+IF N ALI+YAP T NDKI+SGP KI+DMFPFFK TVFE G+DA
Sbjct: 64 GVDCAFDTDNDEAFIFYENFTALINYAPHTYNDKIISGPKKISDMFPFFKGTVFENGIDA 123
Query: 183 AFRAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEAAFASHGT 241
AFR+ E YLF+G YA I+Y L+ I+ I GFP GTVFENG++AAFASH T
Sbjct: 124 AFRSTKEKEVYLFKGDLYARIDYGKNYLVQSIKNISTGFPCFTGTVFENGVDAAFASHRT 183
Query: 242 KEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKILPRNN 289
EAY FKG+YYA + +PG DDY+IGG VK IL NWPSL+ I+P+ +
Sbjct: 184 NEAYFFKGDYYALVKISPGGIDDYIIGG-VKPILENWPSLRGIIPQKS 230
>sp|P08688|ALB2_PEA Albumin 2 (PA2) gi|169033|gb|AAA33641.1| major seed albumin
gi|169029|gb|AAA02981.1| albumin 2
gi|225836|prf||1314296A albumin
Length = 231
Score = 261 bits (666), Expect = 2e-68
Identities = 122/227 (53%), Positives = 166/227 (72%), Gaps = 3/227 (1%)
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGI 124
YINAAFR+S NNEAYLF+N++YVL++YA G++ND ++ GP + DG+ SL +T FG +G+
Sbjct: 6 YINAAFRSSQNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRDGFKSLNQTVFGSYGV 65
Query: 125 DCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAF 184
DC+FDTD +A+IF CALIDYAP + DKI+ GP KI DMFPFF+ TVFE G+DAA+
Sbjct: 66 DCSFDTDNDEAFIFYEKFCALIDYAPHSNKDKIILGPKKIADMFPFFEGTVFENGIDAAY 125
Query: 185 RAHSSNEAYLFRGGHYALINYSSKRLIH--IETIRHGFPSLIGTVFENGLEAAFASHGTK 242
R+ E YLF+G YA I+Y + +++ I++IR+GFP T+FE+G +AAFASH T
Sbjct: 126 RSTRGKEVYLFKGDQYARIDYETNSMVNKEIKSIRNGFPCFRNTIFESGTDAAFASHKTN 185
Query: 243 EAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKILPRNN 289
E Y FKG+YYA + PG+TDD ++ G V+ L WPSL+ I+P N
Sbjct: 186 EVYFFKGDYYARVTVTPGATDDQIMDG-VRKTLDYWPSLRGIIPLEN 231
>emb|CAA50008.1| mung bean seed albumin [Vigna radiata var. radiata]
gi|2129908|pir||S58127 seed albumin - mung bean
Length = 272
Score = 230 bits (587), Expect = 4e-59
Identities = 120/226 (53%), Positives = 152/226 (67%), Gaps = 4/226 (1%)
Query: 59 VENIPIYINAAFRASANN-EAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLART 117
+ N+P YINAAFR S+ + E Y F N+YV + Y G T D I+ I G+PSLA T
Sbjct: 1 MSNLP-YINAAFRFSSRDYEVYFFAKNKYVRLQYTPGKTEDKILTNLRLISSGFPSLAGT 59
Query: 118 PFGEHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFE 177
PF E GID AF T+ ++AY+FSAN A IDYAPGTTNDKIL+GP I +MFP ++TVF
Sbjct: 60 PFAEPGIDSAFHTEASEAYVFSANNRAYIDYAPGTTNDKILAGPTTIAEMFPVLRNTVFA 119
Query: 178 RGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLI-HIETIRHGFPSLIGTVFENGLEAAF 236
+D+AFR+ E YLF+G Y I+Y SK+L+ I I GFP L GT FE+G++A+F
Sbjct: 120 DSIDSAFRSTKGKEVYLFKGNKYVRIDYDSKQLVGSIRNISDGFPVLNGTGFESGIDASF 179
Query: 237 ASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLK 282
ASH EAYLFKG+ Y I++ PG TDD L+G V+ IL WP LK
Sbjct: 180 ASHKEPEAYLFKGDKYVRIHFTPGKTDDTLVGD-VRPILDGWPVLK 224
>ref|XP_474877.1| OSJNBa0021F22.4 [Oryza sativa (japonica cultivar-group)]
gi|38346471|emb|CAE03710.2| OSJNBa0021F22.4 [Oryza
sativa (japonica cultivar-group)]
Length = 295
Score = 217 bits (553), Expect = 3e-55
Identities = 122/290 (42%), Positives = 173/290 (59%), Gaps = 17/290 (5%)
Query: 15 IDDNGIRRELPEETYTPHERDQVQKRDVSSSQDNEEVELHSYGAVENIPIYINAAFRASA 74
+D+N R++L + V +R +S ++EV + V ++ YI++AFR++
Sbjct: 1 MDENNQRKKLVIQ--------DVPERQLSPDDADDEVNEYYTEGVTDVDNYIDSAFRSTR 52
Query: 75 NNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAFDTD--- 131
N+AY+F+ E V++NY + +D II+G YI + SL T F EHGID AF
Sbjct: 53 KNKAYIFIREENVVMNYGPATRDDKIISGLRYIGNTLQSLVGTAFAEHGIDAAFACHDNH 112
Query: 132 -----KTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFRA 186
+++A IFSANLCA I++AP TT D+I+ GP I+ MFPFFK T FE+G+DAAF +
Sbjct: 113 GFLCARSEAMIFSANLCARINFAPRTTRDRIIQGPKTISQMFPFFKGTSFEKGIDAAFES 172
Query: 187 HSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGT-VFENGLEAAFASHGTKEAY 245
+ EAYLF+G +ALINYS LI I I F +F + AA ASH +K+ Y
Sbjct: 173 TVTGEAYLFKGAEFALINYSRPILIEIRPIVDVFKCFRDCYLFATDIGAALASHVSKDVY 232
Query: 246 LFKGEYYANIYYAPGSTDDYLIGGRVKLILSNWPSLKKILPRNNGRLDLH 295
LFK Y + PG T+ Y+IGG +++ NWPSLK ILPR N LD++
Sbjct: 233 LFKENDYLLFHLTPGETNHYIIGGPKEIVPRNWPSLKGILPRKNKALDIY 282
>emb|CAH55839.1| seed albumin 2 [Pisum sativum]
Length = 51
Score = 64.7 bits (156), Expect = 3e-09
Identities = 28/46 (60%), Positives = 39/46 (83%)
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDG 110
YINAAFR+S NNEAYLF+N++YVL++YA G++ND ++ GP + DG
Sbjct: 6 YINAAFRSSQNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRDG 51
Score = 43.5 bits (101), Expect = 0.008
Identities = 20/44 (45%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query: 124 IDCAF-DTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITD 166
I+ AF + +AY+F + L+DYAPGT+NDK+L GP + D
Sbjct: 7 INAAFRSSQNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRD 50
Score = 35.0 bits (79), Expect = 2.8
Identities = 16/38 (42%), Positives = 24/38 (63%)
Query: 232 LEAAFASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGG 269
+ AAF S EAYLF + Y + YAPG+++D ++ G
Sbjct: 7 INAAFRSSQNNEAYLFINDKYVLLDYAPGTSNDKVLYG 44
>emb|CAH55771.1| seed albumin 2 [Pisum sativum]
Length = 51
Score = 64.7 bits (156), Expect = 3e-09
Identities = 28/46 (60%), Positives = 39/46 (83%)
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDG 110
YINAAFR+S NNEAYLF+N++YVL++YA G++ND ++ GP + DG
Sbjct: 6 YINAAFRSSRNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRDG 51
Score = 43.5 bits (101), Expect = 0.008
Identities = 20/44 (45%), Positives = 30/44 (67%), Gaps = 1/44 (2%)
Query: 124 IDCAFDTDKT-QAYIFSANLCALIDYAPGTTNDKILSGPMKITD 166
I+ AF + + +AY+F + L+DYAPGT+NDK+L GP + D
Sbjct: 7 INAAFRSSRNNEAYLFINDKYVLLDYAPGTSNDKVLYGPTPVRD 50
Score = 35.0 bits (79), Expect = 2.8
Identities = 16/38 (42%), Positives = 24/38 (63%)
Query: 232 LEAAFASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGG 269
+ AAF S EAYLF + Y + YAPG+++D ++ G
Sbjct: 7 INAAFRSSRNNEAYLFINDKYVLLDYAPGTSNDKVLYG 44
>emb|CAH59899.1| seed albumin 2 [Pisum sativum]
Length = 51
Score = 63.2 bits (152), Expect = 1e-08
Identities = 27/46 (58%), Positives = 38/46 (81%)
Query: 65 YINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDG 110
YINAAFR+S NNE YLF+N++YVL++YA G++ND ++ GP + DG
Sbjct: 6 YINAAFRSSRNNETYLFINDKYVLLDYAPGTSNDKVLYGPTPVRDG 51
Score = 42.0 bits (97), Expect = 0.023
Identities = 19/44 (43%), Positives = 29/44 (65%), Gaps = 1/44 (2%)
Query: 124 IDCAFDTDKT-QAYIFSANLCALIDYAPGTTNDKILSGPMKITD 166
I+ AF + + + Y+F + L+DYAPGT+NDK+L GP + D
Sbjct: 7 INAAFRSSRNNETYLFINDKYVLLDYAPGTSNDKVLYGPTPVRD 50
Score = 33.5 bits (75), Expect = 8.2
Identities = 15/38 (39%), Positives = 23/38 (60%)
Query: 232 LEAAFASHGTKEAYLFKGEYYANIYYAPGSTDDYLIGG 269
+ AAF S E YLF + Y + YAPG+++D ++ G
Sbjct: 7 INAAFRSSRNNETYLFINDKYVLLDYAPGTSNDKVLYG 44
>dbj|BAA78479.1| matrix metalloproteinase-2 [Oncorhynchus mykiss]
Length = 655
Score = 48.9 bits (115), Expect = 2e-04
Identities = 51/195 (26%), Positives = 79/195 (40%), Gaps = 30/195 (15%)
Query: 61 NIPIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFG 120
N P+ +A A E + F + RG + GP+ + +P L T
Sbjct: 464 NEPVVFDAV--AQIRGETFFFKDRFIFRSTNFRGKPS-----GPMLVATYWPELPVT--- 513
Query: 121 EHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGL 180
ID A++ + +F A I A D+I G K D +G+
Sbjct: 514 ---IDAAYENPLEEKTVFFAGNEMWIFNA-----DQIEKGYPKRISSIGLPTDL---KGI 562
Query: 181 DAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGLEAAFA 237
DAAF S + YLF G + N + K++ GFP LI + +G+++AF+
Sbjct: 563 DAAFSFGKSQKTYLFAGDQFWRYNEAKKKM------DLGFPKLIADSWNGIPDGIDSAFS 616
Query: 238 SHGTKEAYLFKGEYY 252
+G Y FKG +Y
Sbjct: 617 LNGIDYTYFFKGAHY 631
Score = 42.4 bits (98), Expect = 0.018
Identities = 41/159 (25%), Positives = 66/159 (40%), Gaps = 25/159 (15%)
Query: 62 IPIYINAAFRASANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYP---SLARTP 118
+P+ I+AA+ + F NE + N + I GYP S P
Sbjct: 510 LPVTIDAAYENPLEEKTVFFAGNEMWIFNADQ-------------IEKGYPKRISSIGLP 556
Query: 119 FGEHGIDCAFDTDKTQ-AYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFE 177
GID AF K+Q Y+F+ + + A + L P I D + D
Sbjct: 557 TDLKGIDAAFSFGKSQKTYLFAGDQFWRYNEAKKKMD---LGFPKLIADSWNGIPD---- 609
Query: 178 RGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETI 216
G+D+AF + + Y F+G HY ++ SS +++ + I
Sbjct: 610 -GIDSAFSLNGIDYTYFFKGAHYFKMDDSSLKIVKLGEI 647
>gb|EAA62383.1| predicted protein [Aspergillus nidulans FGSC A4]
gi|67538064|ref|XP_662806.1| hypothetical protein
AN5202_2 [Aspergillus nidulans FGSC A4]
gi|49095756|ref|XP_409339.1| predicted protein
[Aspergillus nidulans FGSC A4]
Length = 168
Score = 45.4 bits (106), Expect = 0.002
Identities = 34/120 (28%), Positives = 47/120 (38%), Gaps = 2/120 (1%)
Query: 66 INAAFRASANNEAYLFMNN-EYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGI 124
++A F +AY F Y I++ GS I G I D +PSL FG
Sbjct: 2 VDAVFYHKGIKQAYFFGGRGRYARIDFVPGSAGGKITFGLAAIADHWPSLKSIGFGTVDA 61
Query: 125 DCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAF 184
D + + Y FS A I P + +D + GP IT F+ +DA F
Sbjct: 62 ILPIDGSQDEGYYFSGAHFARIKLVPSSDDDTFVDGPWVITQKLASLNKAGFDT-IDAPF 120
Score = 38.1 bits (87), Expect = 0.33
Identities = 30/97 (30%), Positives = 43/97 (43%), Gaps = 8/97 (8%)
Query: 180 LDAAFRAHSSNEAYLFRG-GHYALINY-----SSKRLIHIETIRHGFPSLIGTVFENGLE 233
+DA F +AY F G G YA I++ K + I +PSL F ++
Sbjct: 2 VDAVFYHKGIKQAYFFGGRGRYARIDFVPGSAGGKITFGLAAIADHWPSLKSIGFGT-VD 60
Query: 234 AAFASHGTK-EAYLFKGEYYANIYYAPGSTDDYLIGG 269
A G++ E Y F G ++A I P S DD + G
Sbjct: 61 AILPIDGSQDEGYYFSGAHFARIKLVPSSDDDTFVDG 97
>ref|NP_931421.1| photopexin B [Photorhabdus luminescens subsp. laumondii TTO1]
gi|36787513|emb|CAE16615.1| photopexin B [Photorhabdus
luminescens subsp. laumondii TTO1]
Length = 340
Score = 42.4 bits (98), Expect = 0.018
Identities = 50/210 (23%), Positives = 84/210 (39%), Gaps = 23/210 (10%)
Query: 75 NNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAFDTDKTQ 134
N+ YLF+N+E + N + + + P I + +PSL + ID + + +
Sbjct: 2 NSNTYLFLNSENIRYNDSEDKADT---SYPQTISNDWPSLPIE--FQKDIDDVINLNGS- 55
Query: 135 AYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFR-AHSSNEAY 193
Y F + D A ++ GP I + +P K T FE G+DAA + +
Sbjct: 56 LYFFKGSQYLKFDIAKAL----VIDGPKPIIEGWPGLKGTGFENGIDAATEWVDTKQDVV 111
Query: 194 LFRGGHYALINYSSKRLIHIETIRHGFPSLIGTV-----FENGLEAAFASHGTKEAYLFK 248
F G + S I+ +TI + GT+ F L A T A ++
Sbjct: 112 CFFKGRDCIDYTVSSHSINKKTISDRW----GTIGKYAGFSEDLNAVILWKNTAGATIY- 166
Query: 249 GEYYANIYYAPGSTDDYLIGGRVKLILSNW 278
++ + YY +T + I G I + W
Sbjct: 167 --FFKDSYYIQYNTKSHAIDGEPSFIQAYW 194
>dbj|BAA85769.2| gelatinase A [Oryzias latipes]
Length = 657
Score = 42.4 bits (98), Expect = 0.018
Identities = 42/154 (27%), Positives = 71/154 (45%), Gaps = 25/154 (16%)
Query: 103 GPLYICDGYPSLARTPFGEHGIDCAFDTDKTQAYIFSANLCALIDYAPGTTNDKILSG-P 161
GPL + +P L ID A++ + +F A + A D++ SG P
Sbjct: 502 GPLLVATYWPDLPAK------IDAAYENPAEEKTVFFAGNQFWVYRA-----DELQSGYP 550
Query: 162 MKITDMFPFFKDTVFERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFP 221
+++++ D V + +DAAF + + YLF G + Y R +T+ GFP
Sbjct: 551 RRLSNLD--LPDDV--QRIDAAFNFRRNRKTYLFSGDKFW--RYDEDR----KTMDPGFP 600
Query: 222 SLIGTVFE---NGLEAAFASHGTKEAYLFKGEYY 252
LI + + L++AF+ +G +Y FKG +Y
Sbjct: 601 KLIADSWNGIPDDLDSAFSLNGIDYSYFFKGNHY 634
>gb|AAP74482.1| matrix metalloproteinase 2 [Danio rerio]
gi|37620187|ref|NP_932333.1| matrix metalloproteinase 2
[Danio rerio]
Length = 657
Score = 41.2 bits (95), Expect = 0.039
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 9/77 (11%)
Query: 179 GLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIG---TVFENGLEAA 235
G+DAA+ H + + Y+F G + N + K++ GFP +I T + L+ A
Sbjct: 564 GIDAAYSFHKTKKTYIFAGNKFWRYNEAKKKM------DPGFPKIIADSWTAVPDDLDGA 617
Query: 236 FASHGTKEAYLFKGEYY 252
+ +G +Y FK +Y
Sbjct: 618 LSLNGDGHSYFFKDSHY 634
Score = 33.5 bits (75), Expect = 8.2
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 9/101 (8%)
Query: 118 PFGEHGIDCAFDTDKTQ-AYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVF 176
P HGID A+ KT+ YIF+ N + A + P I D + D
Sbjct: 559 PSDLHGIDAAYSFHKTKKTYIFAGNKFWRYNEAKKKMDPGF---PKIIADSWTAVPDD-- 613
Query: 177 ERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIR 217
LD A + +Y F+ HY ++ S+ ++I + ++
Sbjct: 614 ---LDGALSLNGDGHSYFFKDSHYLKMDDSTLKIIKVGEVK 651
>gb|AAH76545.1| Matrix metalloproteinase 2 [Danio rerio]
Length = 657
Score = 41.2 bits (95), Expect = 0.039
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 9/77 (11%)
Query: 179 GLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIG---TVFENGLEAA 235
G+DAA+ H + + Y+F G + N + K++ GFP +I T + L+ A
Sbjct: 564 GIDAAYSFHKTKKTYIFAGNKFWRYNEAKKKM------DPGFPKIIADSWTAVPDDLDGA 617
Query: 236 FASHGTKEAYLFKGEYY 252
+ +G +Y FK +Y
Sbjct: 618 LSLNGDGHSYFFKDSHY 634
Score = 33.5 bits (75), Expect = 8.2
Identities = 26/101 (25%), Positives = 43/101 (41%), Gaps = 9/101 (8%)
Query: 118 PFGEHGIDCAFDTDKTQ-AYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVF 176
P HGID A+ KT+ YIF+ N + A + P I D + D
Sbjct: 559 PSDLHGIDAAYSFHKTKKTYIFAGNKFWRYNEAKKKMDPGF---PKIIADSWTAVPDD-- 613
Query: 177 ERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIR 217
LD A + +Y F+ HY ++ S+ ++I + ++
Sbjct: 614 ---LDGALSLNGDGHSYFFKDSHYLKMDDSTLKIIKVGEVK 651
>ref|NP_990392.1| vitronectin [Gallus gallus] gi|1922282|emb|CAA71914.1| vitronectin
[Gallus gallus]
Length = 453
Score = 40.4 bits (93), Expect = 0.067
Identities = 30/91 (32%), Positives = 44/91 (47%), Gaps = 17/91 (18%)
Query: 177 ERGLDAAF-RAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGL 232
E +DAAF R + + YLF+G Y + + + G+P I FE N +
Sbjct: 201 EGPIDAAFTRINCQGKTYLFKGSQYWRFDDGA--------LDPGYPRDISEGFEGIPNDI 252
Query: 233 EAAFAS-----HGTKEAYLFKGEYYANIYYA 258
+AAFA HG + Y FKG+YY + +A
Sbjct: 253 DAAFALPAHSYHGNERVYFFKGKYYWSYDFA 283
>gb|AAF97408.1| photopexin B [Photorhabdus luminescens]
Length = 340
Score = 40.4 bits (93), Expect = 0.067
Identities = 50/213 (23%), Positives = 81/213 (37%), Gaps = 29/213 (13%)
Query: 75 NNEAYLFMNNEYVLINYARGSTN-DYIINGPLYICDGYPSLARTPFGEHGIDCAFDTDKT 133
N++ Y F+N+E + N + + DY P +I +P L I+ D D
Sbjct: 2 NSKTYFFLNSENIRYNDSEDKADTDY----PQFISHDWPGLP--------IEFQKDIDDV 49
Query: 134 -----QAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFR-AH 187
Y F + D A ++ GP I D +P K T FE G+DAA
Sbjct: 50 INLSGSLYFFKGSQYLKFDIAKAL----VIDGPKPIIDGWPGLKGTGFENGIDAATEWMD 105
Query: 188 SSNEAYLFRGGHYALINYSSKRLIHIETI--RHGFPSLIGTVFENGLEAAFASHGTKEAY 245
+ + F G + S I+ +TI R G F L+A +
Sbjct: 106 TKQDVVCFFKGRDCIDYTVSSHTINKKTISERWGATGKYAG-FSEDLDAVILWKNITGSI 164
Query: 246 LFKGEYYANIYYAPGSTDDYLIGGRVKLILSNW 278
++ ++ + YY +T ++I G I + W
Sbjct: 165 IY---FFKDSYYIQYNTKSHVIDGEPSFIQAYW 194
>dbj|BAB68365.1| gelatinase [Paralichthys olivaceus]
Length = 658
Score = 40.4 bits (93), Expect = 0.067
Identities = 24/76 (31%), Positives = 39/76 (50%), Gaps = 9/76 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGLEAAF 236
+DAAF + + YLF G + + +K + GFP LI + +GL++AF
Sbjct: 566 IDAAFNFRKNKKTYLFAGDKFWRYDEETK------IMDAGFPRLIADSWNGIPDGLDSAF 619
Query: 237 ASHGTKEAYLFKGEYY 252
+ +G +Y FKG +Y
Sbjct: 620 SLNGIDYSYFFKGNHY 635
>ref|NP_931420.1| photopexin A [Photorhabdus luminescens subsp. laumondii TTO1]
gi|36787512|emb|CAE16614.1| photopexin A [Photorhabdus
luminescens subsp. laumondii TTO1]
Length = 564
Score = 39.3 bits (90), Expect = 0.15
Identities = 30/101 (29%), Positives = 47/101 (45%), Gaps = 5/101 (4%)
Query: 176 FERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEA 234
F+R +D + S Y F+G Y + + ++I + I G+P L GT FENG++A
Sbjct: 265 FQRHIDDVINLNGS--LYFFKGSQYLKFDIAKAQVIDGPKPIIEGWPGLAGTEFENGIDA 322
Query: 235 AFASHGTKE--AYLFKGEYYANIYYAPGSTDDYLIGGRVKL 273
A KE FKG+ N + + D I R ++
Sbjct: 323 ATEWIDVKEDIVCFFKGKECINYTVSSHTIDKKNIAERWRI 363
Score = 36.2 bits (82), Expect = 1.3
Identities = 55/211 (26%), Positives = 85/211 (40%), Gaps = 24/211 (11%)
Query: 74 ANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAFDTDKT 133
+N Y F+N+E + N N+ P I + +P+L F H ID + + +
Sbjct: 224 SNTNTYFFLNSENIKYN---DPLNNIDTGYPQPIRNNWPNLP-IEFQRH-IDDVINLNGS 278
Query: 134 QAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFRAHSSNE-- 191
Y F + D A +++ GP I + +P T FE G+DAA E
Sbjct: 279 -LYFFKGSQYLKFDIAKA----QVIDGPKPIIEGWPGLAGTEFENGIDAATEWIDVKEDI 333
Query: 192 AYLFRGGHYALINYS-SKRLIHIETIRHGFPSLIG--TVFENGLEAAFASHGTKE-AYLF 247
F+G INY+ S I + I + + G F L+AA + YLF
Sbjct: 334 VCFFKGKE--CINYTVSSHTIDKKNIAERW-RITGEYAKFSANLDAAILWRNSGYFIYLF 390
Query: 248 KGEYYANIYYAPGSTDDYLIGGRVKLILSNW 278
KG Y ++ S + G +LI + W
Sbjct: 391 KGNEYLRLHLMLNS-----MYGEPELIQTKW 416
>ref|NP_931412.1| hypothetical protein plu4231 [Photorhabdus luminescens subsp.
laumondii TTO1] gi|36787504|emb|CAE16603.1| unnamed
protein product [Photorhabdus luminescens subsp.
laumondii TTO1]
Length = 333
Score = 38.9 bits (89), Expect = 0.20
Identities = 40/138 (28%), Positives = 61/138 (43%), Gaps = 14/138 (10%)
Query: 75 NNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAFDTDKTQ 134
N+ YLF+N + N ++ I P I + +P L F H ID + +
Sbjct: 2 NSHVYLFLNADNARYNEISKKSD---IAYPQPISNDWPDLP-IEFQRH-IDDVINLNG-Y 55
Query: 135 AYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFRAHSSNEAYL 194
Y F + D A K++ GP I D +P K T FE G+DAA ++N
Sbjct: 56 LYFFKGSQYVKFDIAKA----KVIDGPKFIADGWPGLKGTEFENGIDAA-TEFTTNIVCF 110
Query: 195 FRGG---HYALINYSSKR 209
F+G YA+ +++ KR
Sbjct: 111 FKGSDCIDYAVNSHTIKR 128
Score = 35.4 bits (80), Expect = 2.2
Identities = 24/75 (32%), Positives = 36/75 (48%), Gaps = 4/75 (5%)
Query: 176 FERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEA 234
F+R +D + Y F+G Y + + ++I + I G+P L GT FENG++A
Sbjct: 42 FQRHIDDVINLNGY--LYFFKGSQYVKFDIAKAKVIDGPKFIADGWPGLKGTEFENGIDA 99
Query: 235 AFASHGTKEAYLFKG 249
A T FKG
Sbjct: 100 A-TEFTTNIVCFFKG 113
>gb|AAF97407.1| photopexin A [Photorhabdus luminescens]
Length = 564
Score = 38.9 bits (89), Expect = 0.20
Identities = 29/101 (28%), Positives = 47/101 (45%), Gaps = 5/101 (4%)
Query: 176 FERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEA 234
F+R +D + Y F+G Y + + ++I ++I G+P L GT FENG++A
Sbjct: 265 FQRHIDDVINLNGC--LYFFKGSQYLKFDIAKAQVIDGPKSIIEGWPGLAGTEFENGIDA 322
Query: 235 AFASHGTKE--AYLFKGEYYANIYYAPGSTDDYLIGGRVKL 273
A KE FKG+ N + + D I R ++
Sbjct: 323 ATEWIDAKEDIVCFFKGKECINYTVSSHAIDKKTIAERWRI 363
Score = 38.1 bits (87), Expect = 0.33
Identities = 55/210 (26%), Positives = 85/210 (40%), Gaps = 22/210 (10%)
Query: 74 ANNEAYLFMNNEYVLINYARGSTNDYIINGPLYICDGYPSLARTPFGEHGIDCAFDTDKT 133
+N Y F+N+E + N N+ P I + +P+L F H ID + +
Sbjct: 224 SNTNTYFFLNSENIKYN---DPLNNIDTGYPQPIRNNWPNLP-IEFQRH-IDDVINLNGC 278
Query: 134 QAYIFSANLCALIDYAPGTTNDKILSGPMKITDMFPFFKDTVFERGLDAAFRAHSSNE-- 191
Y F + D A +++ GP I + +P T FE G+DAA + E
Sbjct: 279 -LYFFKGSQYLKFDIAKA----QVIDGPKSIIEGWPGLAGTEFENGIDAATEWIDAKEDI 333
Query: 192 AYLFRGGHYALINYS-SKRLIHIETIRHGFP-SLIGTVFENGLEAAFASHGTKE-AYLFK 248
F+G INY+ S I +TI + + F L+AA + YLFK
Sbjct: 334 VCFFKGKE--CINYTVSSHAIDKKTIAERWRITEEYAKFSANLDAAILWRNSGYFIYLFK 391
Query: 249 GEYYANIYYAPGSTDDYLIGGRVKLILSNW 278
G Y ++ S + G +LI + W
Sbjct: 392 GNEYLRLHMMLNS-----LYGEPELIQTKW 416
Score = 35.0 bits (79), Expect = 2.8
Identities = 20/61 (32%), Positives = 32/61 (51%), Gaps = 3/61 (4%)
Query: 176 FERGLDAAFRAHSSNEAYLFRGGHYALINYSSKRLIH-IETIRHGFPSLIGTVFENGLEA 234
F+R +D + Y F+G Y + + ++I + I G+P L GT FENG++A
Sbjct: 42 FQRHIDDVINLNGF--LYFFKGSQYLKFDIAKAKVIDGPKPIIEGWPGLAGTEFENGIDA 99
Query: 235 A 235
A
Sbjct: 100 A 100
>emb|CAF95801.1| unnamed protein product [Tetraodon nigroviridis]
Length = 629
Score = 38.9 bits (89), Expect = 0.20
Identities = 24/76 (31%), Positives = 39/76 (50%), Gaps = 9/76 (11%)
Query: 180 LDAAFRAHSSNEAYLFRGGHYALINYSSKRLIHIETIRHGFPSLIGTVFE---NGLEAAF 236
+DAAF + + YLF G + Y R +T+ FP LIG + + +++AF
Sbjct: 537 IDAAFNFRKNKKTYLFAGDKFW--RYDEDR----KTMDANFPRLIGESWNGIPDDIDSAF 590
Query: 237 ASHGTKEAYLFKGEYY 252
+ +G +Y FKG +Y
Sbjct: 591 SLNGIDYSYFFKGNHY 606
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.139 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,012,358
Number of Sequences: 2540612
Number of extensions: 26640336
Number of successful extensions: 51820
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 51638
Number of HSP's gapped (non-prelim): 125
length of query: 313
length of database: 863,360,394
effective HSP length: 128
effective length of query: 185
effective length of database: 538,162,058
effective search space: 99559980730
effective search space used: 99559980730
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146590.2


