

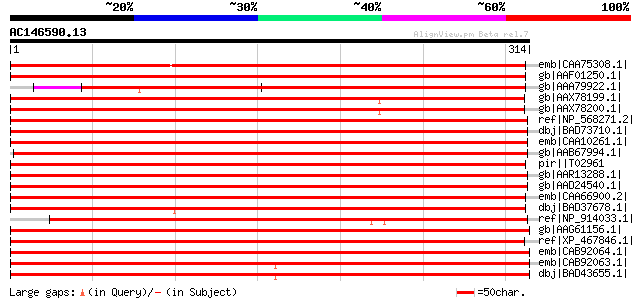
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146590.13 + phase: 0
(314 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA75308.1| annexin [Medicago truncatula] gi|22859608|emb|CA... 425 e-117
gb|AAF01250.1| annexin [Fragaria x ananassa] gi|21264397|sp|P510... 424 e-117
gb|AAA79922.1| annexin 369 e-101
gb|AAX78199.1| putative annexin [Nicotiana tabacum] 355 9e-97
gb|AAX78200.1| putative annexin [Nicotiana tabacum] 355 1e-96
ref|NP_568271.2| annexin, putative [Arabidopsis thaliana] 343 3e-93
dbj|BAD73710.1| putative calcium-binding protein annexin 6 [Oryz... 324 2e-87
emb|CAA10261.1| annexin P38 [Capsicum annuum] 306 4e-82
gb|AAB67994.1| annexin [Gossypium hirsutum] gi|7441511|pir||T108... 302 7e-81
pir||T02961 annexin P33 - maize 302 1e-80
gb|AAR13288.1| Anx1 [Gossypium hirsutum] 301 1e-80
gb|AAD24540.1| vacuole-associated annexin VCaB42 [Nicotiana taba... 301 1e-80
emb|CAA66900.2| annexin p33 [Zea mays] 301 2e-80
dbj|BAD37678.1| putative annexin [Oryza sativa (japonica cultiva... 301 2e-80
ref|NP_914033.1| putative annexin [Oryza sativa (japonica cultiv... 298 1e-79
gb|AAG61156.1| calcium-binding protein annexin 7 [Arabidopsis th... 298 2e-79
ref|XP_467846.1| putative annexin P35 [Oryza sativa (japonica cu... 297 2e-79
emb|CAB92064.1| annexin-like protein [Arabidopsis thaliana] gi|1... 297 2e-79
emb|CAB92063.1| annexin-like protein [Arabidopsis thaliana] gi|1... 297 3e-79
dbj|BAD43655.1| annexin -like protein [Arabidopsis thaliana] gi|... 297 3e-79
>emb|CAA75308.1| annexin [Medicago truncatula] gi|22859608|emb|CAD29698.1| annexin
[Medicago truncatula]
Length = 313
Score = 425 bits (1092), Expect = e-117
Identities = 218/312 (69%), Positives = 260/312 (82%), Gaps = 1/312 (0%)
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATL AP NHSP EDA+ L KA +GWGTDE +I I+G RN+ Q QQIR+AY+ IY EDL
Sbjct: 1 MATLSAPSNHSPNEDAEALRKAFEGWGTDEKTVITILGHRNSNQIQQIRKAYEGIYNEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
IKRLESE+ G+FEKA+YRWIL+PA+R AVLANVAIKS K+Y+VIVEI++VL P+ELL V
Sbjct: 61 IKRLESEIKGDFEKAVYRWILEPAERDAVLANVAIKS-GKNYNVIVEISAVLSPEELLNV 119
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AY RYK+SLEED+AAHTSG+ RQLLVGLV++FRY G EINP LA+ EA ILHE+VK
Sbjct: 120 RRAYVKRYKHSLEEDLAAHTSGHLRQLLVGLVTAFRYVGDEINPKLAQTEAGILHESVKE 179
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
KKG+ EE IRIL TRSKTQL ATFNRYR+ HG SI+KKLL+E SD+F KA++ IR +D
Sbjct: 180 KKGSHEEAIRILTTRSKTQLIATFNRYRETHGTSITKKLLDEGSDEFQKALYTTIRSFND 239
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
H KYYEKV+R A+K++GTDED LTRV+++RA+ DLK I ++YYKRNSV LE VAKE SG
Sbjct: 240 HVKYYEKVVRDAIKKVGTDEDALTRVIVSRAQHDLKVISDVYYKRNSVLLEHVVAKETSG 299
Query: 301 DYKKFLLTLLGK 312
DYKKFLLTLLGK
Sbjct: 300 DYKKFLLTLLGK 311
>gb|AAF01250.1| annexin [Fragaria x ananassa] gi|21264397|sp|P51074|ANX4_FRAAN
Annexin-like protein RJ4
Length = 314
Score = 424 bits (1089), Expect = e-117
Identities = 212/312 (67%), Positives = 259/312 (82%)
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MATL++P N KEDA+ L K+VKGWGT+E AII+I+G RNA QR++IR AY+ +YQEDL
Sbjct: 1 MATLVSPPNFCAKEDAEALRKSVKGWGTNEKAIISILGHRNAGQRKEIRAAYEQLYQEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
+K LESELSG+FEKA+YRW LDPADR AVLANVAIK Y+VI+EI+ + P+ELLAV
Sbjct: 61 LKPLESELSGDFEKAVYRWTLDPADRDAVLANVAIKKSTDVYNVIIEISCIHSPEELLAV 120
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AY RYK+S+EED+AAHT+G R+LLV LV+++RYDG EIN LA EADILH+A+K+
Sbjct: 121 RRAYQLRYKHSVEEDLAAHTTGDIRKLLVALVTAYRYDGHEINAKLANSEADILHDAIKD 180
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
K N EE+IRIL TRSKTQL ATFN+YRDD G SISK LL E ++DF KA+H AIRC++D
Sbjct: 181 KAFNHEEIIRILSTRSKTQLMATFNKYRDDQGISISKNLLEEGANDFQKALHTAIRCLND 240
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
KKY+EKVLR A+KR+GTDED LTRV++TRAE+DL+DIKE+YYK+NSV LE VAK+ SG
Sbjct: 241 PKKYFEKVLRNAIKRVGTDEDALTRVIVTRAERDLRDIKEVYYKKNSVPLEQAVAKDTSG 300
Query: 301 DYKKFLLTLLGK 312
DYK FLLTLLGK
Sbjct: 301 DYKAFLLTLLGK 312
>gb|AAA79922.1| annexin
Length = 271
Score = 369 bits (947), Expect = e-101
Identities = 184/269 (68%), Positives = 225/269 (83%)
Query: 44 QRQQIRQAYQDIYQEDLIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYH 103
+R++IR AY+ +YQEDL+K LESELSG+FEKA+YRW LDPADR AVLANVAIK Y+
Sbjct: 1 ERKEIRAAYEQLYQEDLLKPLESELSGDFEKAVYRWTLDPADRDAVLANVAIKKSTDVYN 60
Query: 104 VIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEIN 163
VI+EI+ + P+ELLAVR AY RYK+S+EED+AAHT+G R+LLV LV+++RYDG EIN
Sbjct: 61 VIIEISCIHSPEELLAVRRAYQLRYKHSVEEDLAAHTTGDIRKLLVALVTAYRYDGHEIN 120
Query: 164 PILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEA 223
LA EADILH+A+K+K N EE+IRIL TRSKTQL ATFN+YRDD G SISK LL E
Sbjct: 121 AKLANSEADILHDAIKDKAFNHEEIIRILSTRSKTQLMATFNKYRDDQGISISKNLLEEG 180
Query: 224 SDDFLKAVHVAIRCIDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYY 283
++DF KA+H AIRC++D KKY+EKVLR A+KR+GTDED LTRV++TRAE+DL+DIKE+YY
Sbjct: 181 ANDFQKALHTAIRCLNDPKKYFEKVLRNAIKRVGTDEDALTRVIVTRAERDLRDIKEVYY 240
Query: 284 KRNSVHLEDTVAKEISGDYKKFLLTLLGK 312
K+NSV LE VAK+ SGDYK FLLTLLGK
Sbjct: 241 KKNSVPLEQAVAKDTSGDYKAFLLTLLGK 269
Score = 55.1 bits (131), Expect = 3e-06
Identities = 36/141 (25%), Positives = 66/141 (46%), Gaps = 3/141 (2%)
Query: 15 DADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGNFEK 74
+AD+L A+K + II I+ R+ Q Y+D + K L E + +F+K
Sbjct: 127 EADILHDAIKDKAFNHEEIIRILSTRSKTQLMATFNKYRDDQGISISKNLLEEGANDFQK 186
Query: 75 AMY---RWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNS 131
A++ R + DP + + AIK + D + + ++L ++ Y+ +
Sbjct: 187 ALHTAIRCLNDPKKYFEKVLRNAIKRVGTDEDALTRVIVTRAERDLRDIKEVYYKKNSVP 246
Query: 132 LEEDVAAHTSGYHRQLLVGLV 152
LE+ VA TSG ++ L+ L+
Sbjct: 247 LEQAVAKDTSGDYKAFLLTLL 267
>gb|AAX78199.1| putative annexin [Nicotiana tabacum]
Length = 317
Score = 355 bits (911), Expect = 9e-97
Identities = 183/314 (58%), Positives = 236/314 (74%), Gaps = 3/314 (0%)
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MAT+ P N SP DA+ + KA +GWGTDE AII+I G RNA Q++ IR+AY+++Y EDL
Sbjct: 1 MATINYPENPSPVADAEAIRKACQGWGTDEKAIISIFGHRNATQKKLIRRAYEELYNEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSIN-KDYHVIVEIASVLQPQELLA 119
+KRLESELSG+FEKA+YRWILDP DR AV+ + AIK DY VI+E + + P+E LA
Sbjct: 61 VKRLESELSGHFEKAVYRWILDPEDRDAVMLHAAIKETPIPDYRVIIEYSCIYSPEEFLA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
V+ AY RYK S+EED+A H++G R+LLV LV +RY G EIN +A EAD LH A+
Sbjct: 121 VKRAYQARYKRSVEEDLAEHSAGDLRKLLVALVGIYRYAGKEINARVANTEADNLHSAIC 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNE--ASDDFLKAVHVAIRC 237
NK+ N EE++RI+ TRS QL AT NRY+DD+G SI+K L ++ A+ ++L A+ IRC
Sbjct: 181 NKEFNHEEIVRIISTRSIPQLIATLNRYKDDYGSSITKHLRDDANAAKEYLVALRTTIRC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
I+D +KY+EKV+R A+ GTDE+ LTRV++TRAEKDLKDIKE+YYKRNSV L+ V+K
Sbjct: 241 INDPQKYHEKVIRYAINESGTDEESLTRVIVTRAEKDLKDIKEIYYKRNSVTLDHAVSKH 300
Query: 298 ISGDYKKFLLTLLG 311
SGDYK FLLTLLG
Sbjct: 301 TSGDYKAFLLTLLG 314
>gb|AAX78200.1| putative annexin [Nicotiana tabacum]
Length = 317
Score = 355 bits (910), Expect = 1e-96
Identities = 183/314 (58%), Positives = 235/314 (74%), Gaps = 3/314 (0%)
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MAT+ P N SP DA+ + KA +GWGTDE AII+I G RNA Q++ IR+AY+++Y EDL
Sbjct: 1 MATINYPENPSPVADAEAIRKACQGWGTDEKAIISIFGHRNATQKKLIRRAYEELYNEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSIN-KDYHVIVEIASVLQPQELLA 119
+KRLESELSG+FEKA+YRWILDP DR AV+ + AIK DY VI+E + + P+E LA
Sbjct: 61 VKRLESELSGHFEKAVYRWILDPEDRDAVMLHAAIKETPIPDYRVIIEYSCIYSPEEFLA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
V+ AY RYK S+EED+A H++G R+LLV LV +RY G EIN +A EAD LH A+
Sbjct: 121 VKRAYQARYKRSVEEDLAEHSAGDLRKLLVALVGIYRYAGKEINARVANTEADNLHSAIC 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNE--ASDDFLKAVHVAIRC 237
NK+ N EE++RI+ TRS QL AT NRY+DD+G SI+K L ++ A+ ++L A+ IRC
Sbjct: 181 NKEFNHEEIVRIISTRSIPQLIATLNRYKDDYGSSITKHLRDDANAAKEYLVALRTTIRC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
I+D +KYYEKV+R A+ GTDE+ LTRV++TRAEKDLKDIKELYYKRNSV L+ ++K
Sbjct: 241 INDPQKYYEKVIRYAINESGTDEESLTRVIVTRAEKDLKDIKELYYKRNSVTLDHALSKH 300
Query: 298 ISGDYKKFLLTLLG 311
SGDYK FLL LLG
Sbjct: 301 TSGDYKAFLLALLG 314
>ref|NP_568271.2| annexin, putative [Arabidopsis thaliana]
Length = 316
Score = 343 bits (881), Expect = 3e-93
Identities = 172/313 (54%), Positives = 235/313 (74%)
Query: 1 MATLIAPMNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
MAT+++P + SP EDA+ + A +GWGT+E+AII+I+G RN QR+ IRQAYQ+IY EDL
Sbjct: 1 MATIVSPPHFSPVEDAENIKAACQGWGTNENAIISILGHRNLFQRKLIRQAYQEIYHEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
I +L+SELSGNFE+A+ W+LDP +R A+LAN+A++ DY V+VEIA + P+++LA
Sbjct: 61 IHQLKSELSGNFERAICLWVLDPPERDALLANLALQKPIPDYKVLVEIACMRSPEDMLAA 120
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
R AY YK+SLEED+A+ T G R+LLV +VS+++YDG EI+ +LA+ EA ILH+ +
Sbjct: 121 RRAYRCLYKHSLEEDLASRTIGDIRRLLVAMVSAYKYDGEEIDEMLAQSEAAILHDEILG 180
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
K + EE IR+L TRS QL A FNRY+D +G SI+K LLN ++++L A+ AIRCI +
Sbjct: 181 KAVDHEETIRVLSTRSSMQLSAIFNRYKDIYGTSITKDLLNHPTNEYLSALRAAIRCIKN 240
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
+YY KVLR ++ +GTDED L RV++TRAEKDL +I LY+KRN+V L+ +AKE SG
Sbjct: 241 PTRYYAKVLRNSINTVGTDEDALNRVIVTRAEKDLTNITGLYFKRNNVSLDQAIAKETSG 300
Query: 301 DYKKFLLTLLGKG 313
DYK FLL LLG G
Sbjct: 301 DYKAFLLALLGHG 313
>dbj|BAD73710.1| putative calcium-binding protein annexin 6 [Oryza sativa (japonica
cultivar-group)] gi|55297623|dbj|BAD68998.1| putative
calcium-binding protein annexin 6 [Oryza sativa
(japonica cultivar-group)]
Length = 316
Score = 324 bits (831), Expect = 2e-87
Identities = 170/315 (53%), Positives = 220/315 (68%), Gaps = 2/315 (0%)
Query: 1 MATLIAP-MNHSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MAT++ P + SP EDAD L KA +GWGTDE A+I ++ R+A QR+QIR Y++ Y E+
Sbjct: 1 MATIVVPPVTPSPAEDADALLKAFQGWGTDEQAVIGVLAHRDATQRKQIRLTYEENYNEN 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
LI+RL+SELSG+ E+AMY W+LDP +R AV+ N A K I++DY VIVEIA ELLA
Sbjct: 61 LIQRLQSELSGDLERAMYHWVLDPVERQAVMVNTATKCIHEDYAVIVEIACTNSSSELLA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
V+ YH YK SLEEDVAA +G R LL+ LVS++RYDG E+N LAK EA ILHE V
Sbjct: 121 VKRTYHVLYKCSLEEDVAARATGNLRSLLLALVSTYRYDGDEVNDALAKSEAKILHETVT 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEAS-DDFLKAVHVAIRCI 238
N + E+IRI+ TRS+ QL ATF+ +RD+ G SI+K L + A + A+ A+RCI
Sbjct: 181 NGDTDHGELIRIVGTRSRAQLNATFSWFRDERGTSITKALQHGADPTGYSHALRTALRCI 240
Query: 239 DDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEI 298
D KY+ KVLR A+ + GT+ED LTRV++ AEKDLK IK+ + KR SV LE + +
Sbjct: 241 SDANKYFVKVLRNAMHKSGTNEDSLTRVIVLHAEKDLKGIKDAFQKRASVALEKAIGNDT 300
Query: 299 SGDYKKFLLTLLGKG 313
SGDYK FL+ LLG G
Sbjct: 301 SGDYKSFLMALLGSG 315
>emb|CAA10261.1| annexin P38 [Capsicum annuum]
Length = 316
Score = 306 bits (785), Expect = 4e-82
Identities = 165/314 (52%), Positives = 210/314 (66%), Gaps = 1/314 (0%)
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P + P EDA+ L KA KGWGT+E II I+ RNA QR+ IR +Y Y ED
Sbjct: 1 MASLKVPASVPDPCEDAEQLKKAFKGWGTNEELIIQILAHRNAAQRKLIRDSYAAAYGED 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+SEL+ +F++ + W L PA+R A LAN A K + VI+EIA EL
Sbjct: 61 LLKDLDSELTSDFQRIVLLWTLSPAERDAYLANEATKRLTASNWVIMEIACTRSSDELFK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AYH RYK S EEDVA HT+G R+LLV L+++FRY+G E+N LA+ EA+ILHE V
Sbjct: 121 ARQAYHTRYKKSFEEDVAYHTTGDFRKLLVPLITAFRYEGEEVNMTLARKEANILHEKVS 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
K N EE+IRI+ TRSKTQL ATFN Y D HG I K L + D++LK + AI C+
Sbjct: 181 GKAYNDEELIRIISTRSKTQLNATFNHYNDQHGHEIIKDLEADDDDEYLKLLRAAIECLK 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+EKVLR A+K +GTDE LTRVV TRAE D++ IKE Y KRNSV L+ + + S
Sbjct: 241 TPEKYFEKVLRVAIKGLGTDEWDLTRVVATRAEVDMERIKEEYNKRNSVTLDRAITGDTS 300
Query: 300 GDYKKFLLTLLGKG 313
GDY++ LL L+G G
Sbjct: 301 GDYERMLLALIGHG 314
>gb|AAB67994.1| annexin [Gossypium hirsutum] gi|7441511|pir||T10807 annexin 2 -
upland cotton (fragment)
Length = 315
Score = 302 bits (774), Expect = 7e-81
Identities = 164/313 (52%), Positives = 214/313 (67%), Gaps = 2/313 (0%)
Query: 3 TLIAPMN-HSPKEDAD-VLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDL 60
TL P++ SP EDA+ L KA +GWGT+E II I+ RNA QR IR+ Y + Y EDL
Sbjct: 1 TLKVPVHVPSPSEDAEWQLRKAFEGWGTNEQLIIDILAHRNAAQRNSIRKVYGEAYGEDL 60
Query: 61 IKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAV 120
+K LE EL+ +FE+A+ + LDPA+R A LAN A K +++EIA ELL V
Sbjct: 61 LKCLEKELTSDFERAVLLFTLDPAERDAHLANEATKKFTSSNWILMEIACSRSSHELLNV 120
Query: 121 RHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKN 180
+ AYH RYK SLEEDVA HT+G +R+LLV LVS+FRY+G E+N LAK EA ILH+ + +
Sbjct: 121 KKAYHARYKKSLEEDVAHHTTGEYRKLLVPLVSAFRYEGEEVNMTLAKSEAKILHDKISD 180
Query: 181 KKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCIDD 240
K EEVIRI+ TRSK QL AT N Y G +I+K L + SD+FLK + I+C+
Sbjct: 181 KHYTDEEVIRIVSTRSKAQLNATLNHYNTSFGNAINKDLKADPSDEFLKLLRAVIKCLTT 240
Query: 241 HKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISG 300
++Y+EKVLR A+ ++G+DE LTRVV TRAE D+ IKE Y +RNS+ LE +AK+ SG
Sbjct: 241 PEQYFEKVLRQAINKLGSDEWALTRVVTTRAEVDMVRIKEAYQRRNSIPLEQAIAKDTSG 300
Query: 301 DYKKFLLTLLGKG 313
DY+KFLL L+G G
Sbjct: 301 DYEKFLLALIGAG 313
>pir||T02961 annexin P33 - maize
Length = 314
Score = 302 bits (773), Expect = 1e-80
Identities = 160/313 (51%), Positives = 211/313 (67%), Gaps = 1/313 (0%)
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P P +D D L KA +GWGT+E+ II+I+G R+A QR+ IR+AY + Y E+
Sbjct: 1 MATLKVPATVPPVADDCDQLRKAFQGWGTNEALIISILGHRDAAQRRAIRRAYAEAYGEE 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L++ + E+SG+FE+A+ W LDPA+R AVLAN A + V+VEIA ++ A
Sbjct: 61 LLRSITDEISGDFERAVILWTLDPAERDAVLANEAARKWKPGNRVLVEIACTRTSAQIFA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AYH R+K SLEED+AAH +G R+LLV LVS++RYDG E+N LA EA +LHE +
Sbjct: 121 TRQAYHERFKRSLEEDIAAHVTGDFRKLLVPLVSTYRYDGPEVNTRLAHSEAKLLHEKIH 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K + +E+IRIL TRSK QL ATFN Y D G I+K L + D++L+ + IRC
Sbjct: 181 HKAYSDDEIIRILTTRSKPQLIATFNHYNDAFGHRINKDLKADPQDEYLRTLRAIIRCFS 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+Y+EKV R A+ +GTDE+ LTRV+ TRAE DLK IKE Y KRNSV LE VA + S
Sbjct: 241 CPDRYFEKVARQAIAGLGTDENSLTRVITTRAEVDLKLIKEAYQKRNSVRLERAVAGDTS 300
Query: 300 GDYKKFLLTLLGK 312
GDY+ LL LLG+
Sbjct: 301 GDYESMLLALLGQ 313
>gb|AAR13288.1| Anx1 [Gossypium hirsutum]
Length = 316
Score = 301 bits (772), Expect = 1e-80
Identities = 164/314 (52%), Positives = 213/314 (67%), Gaps = 1/314 (0%)
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P + +P EDA+ L KA +GWGT+E II I+ RNA QR IR+ Y++ Y ED
Sbjct: 1 MATLKVPAHVPAPSEDAEQLRKAFEGWGTNEQLIIDILAHRNAAQRNLIRKTYREAYGED 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELS +FE+A+ + LDPA+R A LA+ A K + V++EIA EL
Sbjct: 61 LLKSLDEELSSDFERAVVLFTLDPAERDAFLAHEATKRFTSSHWVLMEIACTRSSHELFN 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
VR AYH+ YK SLEEDVA HT G +R+LLV LVS+FRY G E+N LA+ EA IL E +
Sbjct: 121 VRKAYHDLYKKSLEEDVAHHTKGDYRKLLVPLVSAFRYQGEEVNMTLARSEAKILREKIS 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K+ + EEVIRI+ TRSK QL AT N Y G +I+K L + D+FLK + AI+C+
Sbjct: 181 DKQYSDEEVIRIVTTRSKAQLNATLNHYNTAFGNAINKDLKADPEDEFLKLLRAAIKCLT 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+EKVLR A+ ++GTDE LTRVV TRAE D+ IKE Y +RNSV LE +A + S
Sbjct: 241 VPEKYFEKVLRQAINKLGTDEWALTRVVATRAEVDMVRIKEEYQRRNSVTLEKAIAGDTS 300
Query: 300 GDYKKFLLTLLGKG 313
GDY+K LL L+G G
Sbjct: 301 GDYEKMLLALIGAG 314
>gb|AAD24540.1| vacuole-associated annexin VCaB42 [Nicotiana tabacum]
Length = 316
Score = 301 bits (772), Expect = 1e-80
Identities = 161/314 (51%), Positives = 213/314 (67%), Gaps = 1/314 (0%)
Query: 1 MATLIAPMN-HSPKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P + P EDA+ L KA GWGT+E+ II I+ RNA QR+ IR+ Y Y ED
Sbjct: 1 MASLKVPTSVPEPYEDAEQLKKAFAGWGTNEALIIQILAHRNAAQRKLIRETYAAAYGED 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L++EL+ +F++A+ W L PA+R A L N A K + VI+EIA +L
Sbjct: 61 LLKDLDAELTSDFQRAVLLWTLSPAERDAYLVNEATKRLTSSNWVILEIACTRSSDDLFK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AYH RYK SLEEDVA HT+G R+LLV L+++FRY+G E N LA+ EA+ILHE +
Sbjct: 121 ARQAYHARYKKSLEEDVAYHTTGDFRKLLVPLLTAFRYEGEEANMTLARKEANILHEKIS 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K N EE+IRI+ TRSK QL ATFN Y D HG I+K L ++ D++LK + AI C+
Sbjct: 181 DKAYNDEELIRIISTRSKAQLNATFNHYLDQHGSEINKDLETDSDDEYLKLLSAAIECLK 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+K++EKVLR A+K GTDE LTRVV TRAE D++ IKE Y+KRNSV L+ +A + S
Sbjct: 241 TPEKHFEKVLRLAIKGTGTDEWDLTRVVTTRAEVDMERIKEEYHKRNSVPLDRAIAGDTS 300
Query: 300 GDYKKFLLTLLGKG 313
GDY++ LL L+G G
Sbjct: 301 GDYERMLLALIGHG 314
>emb|CAA66900.2| annexin p33 [Zea mays]
Length = 314
Score = 301 bits (771), Expect = 2e-80
Identities = 160/313 (51%), Positives = 211/313 (67%), Gaps = 1/313 (0%)
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P P +D D L KA +GWGT+E+ II+I+G R+A QR+ IR+AY + Y E+
Sbjct: 1 MATLKVPATVPPVADDCDQLRKAFQGWGTNEALIISILGHRDAAQRRAIRRAYAEAYGEE 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L++ + E+SG+FE+A+ W LDPA+R AVLAN A + V+VEIA ++ A
Sbjct: 61 LLRSITDEISGDFERAVILWTLDPAERDAVLANEAARKWKPGNRVLVEIACTRTSAQIFA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AYH R+K SLEED+AAH +G R+LLV LVS++RYDG E+N LA EA +LHE +
Sbjct: 121 TRQAYHERFKRSLEEDIAAHVTGDFRKLLVPLVSTYRYDGPEVNTRLAHSEAKLLHEKIH 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K + +E+IRIL TRSK QL ATFN Y D G I+K L + D++L+ + IRC
Sbjct: 181 HKAYSDDEIIRILTTRSKPQLIATFNHYNDAFGHRINKDLKADPQDEYLRTLRAIIRCFS 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+Y+EKV R A+ +GTDE+ LTRV+ TRAE DLK IKE Y KRNSV LE VA + S
Sbjct: 241 CPDRYFEKVARQAIAGLGTDENSLTRVITTRAEVDLKLIKEAYQKRNSVPLERAVAGDTS 300
Query: 300 GDYKKFLLTLLGK 312
GDY+ LL LLG+
Sbjct: 301 GDYESMLLALLGQ 313
>dbj|BAD37678.1| putative annexin [Oryza sativa (japonica cultivar-group)]
Length = 317
Score = 301 bits (771), Expect = 2e-80
Identities = 162/315 (51%), Positives = 214/315 (67%), Gaps = 3/315 (0%)
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P P +D D L KA +GWGT+E+ II+I+ R+A QR+ IR+AY D Y E+
Sbjct: 1 MATLTVPSAVPPVADDCDQLRKAFQGWGTNEALIISILAHRDAAQRRAIRRAYADTYGEE 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSI--NKDYHVIVEIASVLQPQEL 117
L++ + E+SG+FE+A+ W LDPA+R AVLAN + V+VEIA P +L
Sbjct: 61 LLRSITDEISGDFERAVILWTLDPAERDAVLANEVARKWYPGSGSRVLVEIACARGPAQL 120
Query: 118 LAVRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEA 177
AVR AYH R+K SLEEDVAAH +G R+LLV L+S++RY+G E+N LA EA ILHE
Sbjct: 121 FAVRQAYHERFKRSLEEDVAAHATGDFRKLLVPLISAYRYEGPEVNTKLAHSEAKILHEK 180
Query: 178 VKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRC 237
+++K +E+IRIL TRSK QL ATFNRY D++G I+K L + D+FL + IRC
Sbjct: 181 IQHKAYGDDEIIRILTTRSKAQLIATFNRYNDEYGHPINKDLKADPKDEFLSTLRAIIRC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
+Y+EKV+R A+ +GTDE+ LTR++ TRAE DLK I E Y KRNSV LE VA +
Sbjct: 241 FCCPDRYFEKVIRLAIAGMGTDENSLTRIITTRAEVDLKLITEAYQKRNSVPLERAVAGD 300
Query: 298 ISGDYKKFLLTLLGK 312
SGDY++ LL LLG+
Sbjct: 301 TSGDYERMLLALLGQ 315
>ref|NP_914033.1| putative annexin [Oryza sativa (japonica cultivar-group)]
Length = 323
Score = 298 bits (764), Expect = 1e-79
Identities = 157/299 (52%), Positives = 204/299 (67%), Gaps = 10/299 (3%)
Query: 25 GWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQEDLIKRLESELSGNFEKAMYRWILDPA 84
GWGTDE A+I ++ R+A QR+QIR Y++ Y E+LI+RL+SELSG+ E+AMY W+LDP
Sbjct: 24 GWGTDEQAVIGVLAHRDATQRKQIRLTYEENYNENLIQRLQSELSGDLERAMYHWVLDPV 83
Query: 85 DRYAVLANVAIKSINKDYHVIVEIASVLQPQELLAVRHAYHNRYKNSLEEDVAAHTSGYH 144
+R AV+ N A K I++DY VIVEIA ELLAV+ YH YK SLEEDVAA +G
Sbjct: 84 ERQAVMVNTATKCIHEDYAVIVEIACTNSSSELLAVKRTYHVLYKCSLEEDVAARATGNL 143
Query: 145 RQLLVGLVSSFRYDGVEINPILAKHEADILHEAVKNKKGNIEEVIRILITRSKTQLKATF 204
R LL+ LVS++RYDG E+N LAK EA ILHE V N + E+IRI+ TRS+ QL ATF
Sbjct: 144 RSLLLALVSTYRYDGDEVNDALAKSEAKILHETVTNGDTDHGELIRIVGTRSRAQLNATF 203
Query: 205 NRYRDDHGFSISK--------KLLNEASD--DFLKAVHVAIRCIDDHKKYYEKVLRGALK 254
+ +RD+ G SI+K + L +D + A+ A+RCI D KY+ KVLR A+
Sbjct: 204 SWFRDERGTSITKLHAPRFDHQALQHGADPTGYSHALRTALRCISDANKYFVKVLRNAMH 263
Query: 255 RIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEISGDYKKFLLTLLGKG 313
+ GT+ED LTRV++ AEKDLK IK+ + KR SV LE + + SGDYK FL+ LLG G
Sbjct: 264 KSGTNEDSLTRVIVLHAEKDLKGIKDAFQKRASVALEKAIGNDTSGDYKSFLMALLGSG 322
>gb|AAG61156.1| calcium-binding protein annexin 7 [Arabidopsis thaliana]
Length = 316
Score = 298 bits (762), Expect = 2e-79
Identities = 159/315 (50%), Positives = 213/315 (67%), Gaps = 1/315 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P P+EDA+ L+K+ KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKVPATVPLPEEDAEQLYKSFKGWGTNERMIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+A+ W +PA+RYA LA + K K+ V+VEIA EL
Sbjct: 61 LLKELDRELSGDFERAVMLWTFEPAERYAYLAKESTKMFTKNNWVLVEIACTRSALELFN 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
R AY RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA ILHE +K
Sbjct: 121 ARQAYQARYKTSLEEDVAYHTSGDIRKLLVPLVSTFRYDGDEVNMTLARSEAKILHEKIK 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
K +++IRIL TRSK Q+ AT N Y+++ G S+SK L ++ +++++ + I+C+
Sbjct: 181 EKAYADDDLIRILTTRSKAQISATLNHYKNNFGTSMSKYLKEDSENEYIQLLKAVIKCLT 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+EKVLR A+ ++GTDE GLTRVV TRAE ++ IKE Y +RNSV L+ +AK+
Sbjct: 241 YPEKYFEKVLRQAINKLGTDEWGLTRVVTTRAEFVMERIKEEYIRRNSVPLDRAIAKDTH 300
Query: 300 GDYKKFLLTLLGKGH 314
GDY+ LL LLG H
Sbjct: 301 GDYEDILLALLGHDH 315
>ref|XP_467846.1| putative annexin P35 [Oryza sativa (japonica cultivar-group)]
gi|46805936|dbj|BAD17230.1| putative annexin P35 [Oryza
sativa (japonica cultivar-group)]
gi|46390136|dbj|BAD15571.1| putative annexin P35 [Oryza
sativa (japonica cultivar-group)]
Length = 314
Score = 297 bits (761), Expect = 2e-79
Identities = 160/312 (51%), Positives = 207/312 (66%), Gaps = 1/312 (0%)
Query: 1 MATLIAPMNHSP-KEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MATL P P ED + L KA KGWGT+E II+I+ R+A QR+ IR+AY + Y E+
Sbjct: 1 MATLTVPAAVPPVAEDCEQLRKAFKGWGTNEKLIISILAHRDAAQRRAIRRAYAEAYGEE 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L++ L E+ G FE+A+ +W LDPA+R AVLAN + + +VEIA P +L A
Sbjct: 61 LLRALNDEIHGKFERAVIQWTLDPAERDAVLANEEARKWHPGGRALVEIACTRTPSQLFA 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AYH R+K SLEEDVAAH +G +R+LLV LV+ +RYDG E+N LA EA ILHE +
Sbjct: 121 AKQAYHERFKRSLEEDVAAHITGDYRKLLVPLVTVYRYDGPEVNTSLAHSEAKILHEKIH 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
+K + +E+IRIL TRSK QL ATFN Y D G I+K L + D+FL + IRC
Sbjct: 181 DKAYSDDEIIRILTTRSKAQLLATFNSYNDQFGHPITKDLKADPKDEFLGTLRAIIRCFT 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+Y+EKV+R AL +GTDE+ LTR++ TRAE DLK IKE Y KRNSV LE VAK+ +
Sbjct: 241 CPDRYFEKVIRLALGGMGTDENSLTRIITTRAEVDLKLIKEAYQKRNSVPLERAVAKDTT 300
Query: 300 GDYKKFLLTLLG 311
DY+ LL LLG
Sbjct: 301 RDYEDILLALLG 312
>emb|CAB92064.1| annexin-like protein [Arabidopsis thaliana]
gi|15238105|ref|NP_196585.1| annexin 7 (ANN7)
[Arabidopsis thaliana] gi|11276999|pir||T50027
annexin-like protein - Arabidopsis thaliana
Length = 316
Score = 297 bits (761), Expect = 2e-79
Identities = 159/315 (50%), Positives = 213/315 (67%), Gaps = 1/315 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P P+EDA+ L+KA KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKVPATVPLPEEDAEQLYKAFKGWGTNERMIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+A+ W +PA+R A LA + K K+ V+VEIA EL
Sbjct: 61 LLKELDRELSGDFERAVMLWTFEPAERDAYLAKESTKMFTKNNWVLVEIACTRSALELFN 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDGVEINPILAKHEADILHEAVK 179
+ AY RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA ILHE +K
Sbjct: 121 AKQAYQARYKTSLEEDVAYHTSGDIRKLLVPLVSTFRYDGDEVNMTLARSEAKILHEKIK 180
Query: 180 NKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRCID 239
K +++IRIL TRSK Q+ AT N Y+++ G S+SK L ++ +++++ + I+C+
Sbjct: 181 EKAYADDDLIRILTTRSKAQISATLNHYKNNFGTSMSKYLKEDSENEYIQLLKAVIKCLT 240
Query: 240 DHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKEIS 299
+KY+EKVLR A+ ++GTDE GLTRVV TRAE D++ IKE Y +RNSV L+ +AK+
Sbjct: 241 YPEKYFEKVLRQAINKLGTDEWGLTRVVTTRAEFDMERIKEEYIRRNSVPLDRAIAKDTH 300
Query: 300 GDYKKFLLTLLGKGH 314
GDY+ LL LLG H
Sbjct: 301 GDYEDILLALLGHDH 315
>emb|CAB92063.1| annexin-like protein [Arabidopsis thaliana]
gi|15238094|ref|NP_196584.1| annexin 6 (ANN6)
[Arabidopsis thaliana] gi|11276998|pir||T50026
annexin-like protein - Arabidopsis thaliana
Length = 318
Score = 297 bits (760), Expect = 3e-79
Identities = 163/317 (51%), Positives = 211/317 (66%), Gaps = 3/317 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P N P+ED++ L KA KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKIPANIPLPEEDSEQLHKAFKGWGTNEGMIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+ + W LDP +R A LAN + K K+ V+VEIA E
Sbjct: 61 LLKELDGELSGDFERVVMLWTLDPTERDAYLANESTKLFTKNIWVLVEIACTRPSLEFFK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDG--VEINPILAKHEADILHEA 177
+ AYH RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA LH+
Sbjct: 121 TKQAYHVRYKTSLEEDVAYHTSGNIRKLLVPLVSTFRYDGNADEVNVKLARSEAKTLHKK 180
Query: 178 VKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRC 237
+ K E++IRIL TRSK Q+ AT N ++D G SI+K L +++DD+++ + AI+C
Sbjct: 181 ITEKAYTDEDLIRILTTRSKAQINATLNHFKDKFGSSINKFLKEDSNDDYVQLLKTAIKC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
+ +KY+EKVLR A+ R+GTDE LTRVV TRAE DL+ IKE Y +RNSV L+ +A +
Sbjct: 241 LTYPEKYFEKVLRRAINRMGTDEWALTRVVTTRAEVDLERIKEEYLRRNSVPLDRAIAND 300
Query: 298 ISGDYKKFLLTLLGKGH 314
SGDYK LL LLG H
Sbjct: 301 TSGDYKDMLLALLGHDH 317
>dbj|BAD43655.1| annexin -like protein [Arabidopsis thaliana]
gi|51969424|dbj|BAD43404.1| annexin -like protein
[Arabidopsis thaliana] gi|51969286|dbj|BAD43335.1|
annexin -like protein [Arabidopsis thaliana]
Length = 318
Score = 297 bits (760), Expect = 3e-79
Identities = 163/317 (51%), Positives = 211/317 (66%), Gaps = 3/317 (0%)
Query: 1 MATLIAPMNHS-PKEDADVLWKAVKGWGTDESAIIAIMGQRNAVQRQQIRQAYQDIYQED 59
MA+L P N P+ED++ L KA KGWGT+E II+I+ RNA QR IR Y Y +D
Sbjct: 1 MASLKIPANIPLPEEDSEQLHKAFKGWGTNEGIIISILAHRNATQRSFIRAVYAANYNKD 60
Query: 60 LIKRLESELSGNFEKAMYRWILDPADRYAVLANVAIKSINKDYHVIVEIASVLQPQELLA 119
L+K L+ ELSG+FE+ + W LDP +R A LAN + K K+ V+VEIA E
Sbjct: 61 LLKELDGELSGDFERVVMLWTLDPTERDAYLANESTKLFTKNIWVLVEIACTRPSLEFFK 120
Query: 120 VRHAYHNRYKNSLEEDVAAHTSGYHRQLLVGLVSSFRYDG--VEINPILAKHEADILHEA 177
+ AYH RYK SLEEDVA HTSG R+LLV LVS+FRYDG E+N LA+ EA LH+
Sbjct: 121 TKQAYHVRYKTSLEEDVAYHTSGNIRKLLVPLVSTFRYDGNADEVNVKLARSEAKTLHKK 180
Query: 178 VKNKKGNIEEVIRILITRSKTQLKATFNRYRDDHGFSISKKLLNEASDDFLKAVHVAIRC 237
+ K E++IRIL TRSK Q+ AT N ++D G SI+K L +++DD+++ + AI+C
Sbjct: 181 ITEKAYTDEDLIRILTTRSKAQINATLNHFKDKFGSSINKFLKEDSNDDYVQLLKTAIKC 240
Query: 238 IDDHKKYYEKVLRGALKRIGTDEDGLTRVVITRAEKDLKDIKELYYKRNSVHLEDTVAKE 297
+ +KY+EKVLR A+ R+GTDE LTRVV TRAE DL+ IKE Y +RNSV L+ +A +
Sbjct: 241 LTYPEKYFEKVLRRAINRMGTDEWALTRVVTTRAEVDLERIKEEYLRRNSVPLDRAIAND 300
Query: 298 ISGDYKKFLLTLLGKGH 314
SGDYK LL LLG H
Sbjct: 301 TSGDYKDMLLALLGHDH 317
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 501,869,377
Number of Sequences: 2540612
Number of extensions: 19816284
Number of successful extensions: 60000
Number of sequences better than 10.0: 571
Number of HSP's better than 10.0 without gapping: 517
Number of HSP's successfully gapped in prelim test: 54
Number of HSP's that attempted gapping in prelim test: 55814
Number of HSP's gapped (non-prelim): 1304
length of query: 314
length of database: 863,360,394
effective HSP length: 128
effective length of query: 186
effective length of database: 538,162,058
effective search space: 100098142788
effective search space used: 100098142788
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146590.13


