

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.6 - phase: 0 /pseudo
(670 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
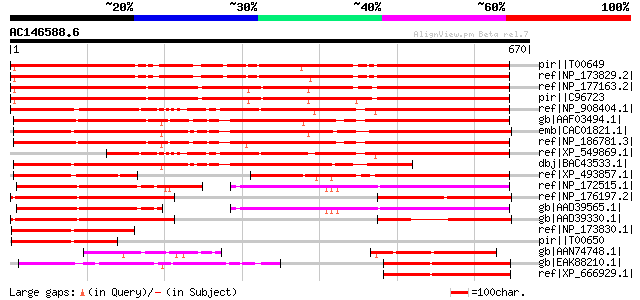
Score E
Sequences producing significant alignments: (bits) Value
pir||T00649 hypothetical protein F3I6.12 - Arabidopsis thaliana ... 677 0.0
ref|NP_173829.2| paired amphipathic helix repeat-containing prot... 672 0.0
ref|NP_177163.2| paired amphipathic helix repeat-containing prot... 667 0.0
pir||C96723 hypothetical protein F20P5.21 [imported] - Arabidops... 667 0.0
ref|NP_908404.1| putative co-repressor protein [Oryza sativa (ja... 642 0.0
gb|AAF03494.1| unknown protein [Arabidopsis thaliana] 598 e-169
emb|CAC01821.1| transcriptional regulatory-like protein [Arabido... 595 e-168
ref|NP_186781.3| paired amphipathic helix repeat-containing prot... 593 e-168
ref|XP_549869.1| transcriptional co-repressor -like [Oryza sativ... 470 e-131
dbj|BAC43533.1| putative transcriptional regulatory protein [Ara... 441 e-122
ref|XP_493857.1| Similar to Arabidopsis chromosome I BAC genomic... 303 2e-80
ref|NP_172515.1| paired amphipathic helix repeat-containing prot... 282 2e-74
ref|NP_176197.2| paired amphipathic helix repeat-containing prot... 279 2e-73
gb|AAD39565.1| T10O24.5 [Arabidopsis thaliana] gi|25402600|pir||... 275 5e-72
gb|AAD39330.1| Hypothetical protein [Arabidopsis thaliana] gi|25... 235 4e-60
ref|NP_173830.1| paired amphipathic helix repeat-containing prot... 209 3e-52
pir||T00650 hypothetical protein F3I6.13 - Arabidopsis thaliana ... 179 3e-43
gb|AAN74748.1| hypothetical protein [Marchantia polymorpha] 172 4e-41
gb|EAK88210.1| Sin3 like paired amphipathic helix containing pro... 169 3e-40
ref|XP_666929.1| transcriptional regulatory-like protein [Crypto... 169 3e-40
>pir||T00649 hypothetical protein F3I6.12 - Arabidopsis thaliana
gi|2829870|gb|AAC00578.1| Hypothetical protein
[Arabidopsis thaliana]
Length = 1263
Score = 677 bits (1748), Expect = 0.0
Identities = 364/662 (54%), Positives = 471/662 (70%), Gaps = 51/662 (7%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ + KYDEFLEVMK+FK+QR+DTAGVI RVKEL
Sbjct: 1 MVGGGSAQKLTTNDALAYLKAVKDKFQDQRGKYDEFLEVMKNFKSQRVDTAGVITRVKEL 60
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYK 117
FKGH++LILGFNTFLPKG+ ITL +D QP KK VEFEEAI+FV KIK RFQG+DRVYK
Sbjct: 61 FKGHQELILGFNTFLPKGFEITLQPEDGQPPLKKRVEFEEAISFVNKIKTRFQGDDRVYK 120
Query: 118 TFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLL 177
+FLDILNMYR++ K IT VYQEV+ LF+DH DLL EFTHFLPDTS A A S + +
Sbjct: 121 SFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEFTHFLPDTS--ATASIPSVKTSV- 177
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRR 237
R+R ++ DK+++ T H D D +H D + +R + + +KE R +++
Sbjct: 178 RERGVSLA-------DKKDRIITPHPDHDYGTEHIDQDRERPIKKENKEHMRGTNKENEH 230
Query: 238 EERDRRERERDDRDYDNDGNLERLPHKKKSVH-RATDPGTAEPLHDADEKLDLLPNSSTC 296
RD RD++ E+ +KK+ +H R DP + + + +P+SST
Sbjct: 231 ---------RDARDFEPHSKKEQFLNKKQKLHIRGDDPAE---ISNQSKLSGAVPSSSTY 278
Query: 297 EDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGK 356
++K ++KS S LA +++VKEKL N +YQEFL+CL+++S+EII+R EL +LVG+L+G
Sbjct: 279 DEKGAMKSY-SQDLAIVDRVKEKL-NASEYQEFLRCLNLFSKEIISRPELQSLVGNLIGV 336
Query: 357 YADIMEGFDDFVTQCEKNE-------------GFLAGVMNKKSLWNEGHGQKPLKVEEKD 403
Y D+M+ F +F+ QCEKNE G L+G++ KKSLW+EG +P ++D
Sbjct: 337 YPDLMDSFIEFLVQCEKNEKRQICNLLNLLAEGLLSGILTKKSLWSEGKYPQPSLDNDRD 396
Query: 404 RDRGRDDGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNV 463
++ RDDG+++RD E +K+ I ++ LS E ++ L N
Sbjct: 397 QEHKRDDGLRDRDHEKERLEKAAANLKWAKPISELDLSNC-----EQCTPSYRLLPKN-- 449
Query: 464 LPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDM 523
P +++ QKTE+G VLNDHWVSVTSGSEDYSF HMRKNQYEESLF+CEDDRFELDM
Sbjct: 450 YPISIAS---QKTEIGKLVLNDHWVSVTSGSEDYSFSHMRKNQYEESLFKCEDDRFELDM 506
Query: 524 LLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLK 583
LLES+N+T+K VEE++ K+N + + + PIR+E+HL+ALNLRCIERLYGDHGLDVM+VLK
Sbjct: 507 LLESVNSTTKHVEELLTKINSNELKTNSPIRVEDHLTALNLRCIERLYGDHGLDVMDVLK 566
Query: 584 KNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLS 643
KN SLALPVILTRLKQKQEEWARCR DF+KVWAEIYAKNY+KSLDHRSFYFKQQD+K LS
Sbjct: 567 KNVSLALPVILTRLKQKQEEWARCRSDFDKVWAEIYAKNYYKSLDHRSFYFKQQDSKKLS 626
Query: 644 TK 645
K
Sbjct: 627 MK 628
>ref|NP_173829.2| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 1353
Score = 672 bits (1734), Expect = 0.0
Identities = 361/653 (55%), Positives = 467/653 (71%), Gaps = 42/653 (6%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ + KYDEFLEVMK+FK+QR+DTAGVI RVKEL
Sbjct: 24 MVGGGSAQKLTTNDALAYLKAVKDKFQDQRGKYDEFLEVMKNFKSQRVDTAGVITRVKEL 83
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYK 117
FKGH++LILGFNTFLPKG+ ITL +D QP KK VEFEEAI+FV KIK RFQG+DRVYK
Sbjct: 84 FKGHQELILGFNTFLPKGFEITLQPEDGQPPLKKRVEFEEAISFVNKIKTRFQGDDRVYK 143
Query: 118 TFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLL 177
+FLDILNMYR++ K IT VYQEV+ LF+DH DLL EFTHFLPDTS A A S + +
Sbjct: 144 SFLDILNMYRRDSKSITEVYQEVAILFRDHSDLLVEFTHFLPDTS--ATASIPSVKTSV- 200
Query: 178 RDRSSAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRR 237
R+R ++ DK+++ T H D D +H D + +R + + +KE R +++
Sbjct: 201 RERGVSLA-------DKKDRIITPHPDHDYGTEHIDQDRERPIKKENKEHMRGTNKENEH 253
Query: 238 EERDRRERERDDRDYDNDGNLERLPHKKKSVH-RATDPGTAEPLHDADEKLDLLPNSSTC 296
RD RD++ E+ +KK+ +H R DP + + + +P+SST
Sbjct: 254 ---------RDARDFEPHSKKEQFLNKKQKLHIRGDDPAE---ISNQSKLSGAVPSSSTY 301
Query: 297 EDKSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGK 356
++K ++KS S LA +++VKEKL N +YQEFL+CL+++S+EII+R EL +LVG+L+G
Sbjct: 302 DEKGAMKSY-SQDLAIVDRVKEKL-NASEYQEFLRCLNLFSKEIISRPELQSLVGNLIGV 359
Query: 357 YADIMEGFDDFVTQCEKNEGFLAGVMNKKS----LWNEGHGQKPLKVEEKDRDRGRDDGV 412
Y D+M+ F +F+ QCEKNEG L+G++ K L EG +P ++D++ RDDG+
Sbjct: 360 YPDLMDSFIEFLVQCEKNEGLLSGILTKSKSTYLLQGEGKYPQPSLDNDRDQEHKRDDGL 419
Query: 413 KERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPK 472
++RD E +K+ I ++ LS E ++ L N P +++
Sbjct: 420 RDRDHEKERLEKAAANLKWAKPISELDLSNC-----EQCTPSYRLLPKN--YPISIAS-- 470
Query: 473 IQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATS 532
QKTE+G VLNDHWVSVTSGSEDYSF HMRKNQYEESLF+CEDDRFELDMLLES+N+T+
Sbjct: 471 -QKTEIGKLVLNDHWVSVTSGSEDYSFSHMRKNQYEESLFKCEDDRFELDMLLESVNSTT 529
Query: 533 KKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPV 592
K VEE++ K+N + + + PIR+E+HL+ALNLRCIERLYGDHGLDVM+VLKKN SLALPV
Sbjct: 530 KHVEELLTKINSNELKTNSPIRVEDHLTALNLRCIERLYGDHGLDVMDVLKKNVSLALPV 589
Query: 593 ILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
ILTRLKQKQEEWARCR DF+KVWAEIYAKNY+KSLDHRSFYFKQQD+K LS K
Sbjct: 590 ILTRLKQKQEEWARCRSDFDKVWAEIYAKNYYKSLDHRSFYFKQQDSKKLSMK 642
>ref|NP_177163.2| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 1362
Score = 667 bits (1721), Expect = 0.0
Identities = 364/674 (54%), Positives = 478/674 (70%), Gaps = 46/674 (6%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ ++KYDEFLEVMKDFKAQR+DT GVI RVKEL
Sbjct: 1 MVGGSSAQKLTTNDALAYLKAVKDKFQDKRDKYDEFLEVMKDFKAQRVDTTGVILRVKEL 60
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQ-KKPVEFEEAINFVGKIKNRFQGNDRVY 116
FKG+++LILGFNTFLPKG+ ITL +D+QP KKPVEFEEAI+FV KIK RFQG+DRVY
Sbjct: 61 FKGNRELILGFNTFLPKGFEITLRPEDDQPAAPKKPVEFEEAISFVNKIKTRFQGDDRVY 120
Query: 117 KTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPL 176
K+FLDILNMYRKE K IT VY EV+ LF+DH DLL EFTHFLPDTS A+ + S + P+
Sbjct: 121 KSFLDILNMYRKENKSITEVYHEVAILFRDHHDLLGEFTHFLPDTSATASTN-DSVKVPV 179
Query: 177 LRDRS-SAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRR-EREK 234
RDR ++ T RQ+ +DK+++ T H +R L ++ D + +R +++ KE+ RR +++
Sbjct: 180 -RDRGIKSLPTMRQIDLDKKDRIITSHPNRALKTENMDVDHERSLLKDSKEEVRRIDKKN 238
Query: 235 DRREERDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGT--AEPLHDADEKLDLLPN 292
D ++RDR+ D R D+D + E + KK + R D ++ + D+ +P+
Sbjct: 239 DFMDDRDRK----DYRGLDHDSHKEHFFNSKKKLIRKDDDSAEMSDQAREGDKFSGAIPS 294
Query: 293 SSTCEDKSSLKSLC---SPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLAL 349
SST ++K + S LAF+++VK KL D+ QEFL+CL++YS+EII++ EL +L
Sbjct: 295 SSTYDEKGFIIDFLESHSQELAFVDRVKAKLDTADN-QEFLRCLNLYSKEIISQPELQSL 353
Query: 350 VGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNK----------------KSLWNEGHG 393
V DL+G Y D+M+ F F+ QC+KN+G L+G+++K +SLW+EG
Sbjct: 354 VSDLIGVYPDLMDAFKVFLAQCDKNDGLLSGIVSKSKSSTFYNILLTYLFGQSLWSEGKC 413
Query: 394 QKPLKVEEKDRDRGRD--DGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL 451
+P K +KD DR R+ + +ERDRE +K I ++ LS E
Sbjct: 414 PQPTKSLDKDTDREREKIERYRERDREKERLEKVAASQKWAKPISELDLSNC-----EQC 468
Query: 452 *MNWTFLTVNNVLPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESL 511
++ L N +P QK E+G++VLNDHWVSVTSGSEDYSFKHMRKNQYEESL
Sbjct: 469 TPSYRRLPKNYPIPIAS-----QKMEIGSQVLNDHWVSVTSGSEDYSFKHMRKNQYEESL 523
Query: 512 FRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLY 571
F+CEDDRFELDMLLES+ + + +VEE++ K+N + + D PI IE+HL+ALNLRCIERLY
Sbjct: 524 FKCEDDRFELDMLLESVISATNRVEELLAKINSNELKTDTPICIEDHLTALNLRCIERLY 583
Query: 572 GDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRS 631
DHGLDV+++LKKNA LALPVILTRLKQKQEEWARCR +FNKVWA+IY KNYH+SLDHRS
Sbjct: 584 SDHGLDVLDLLKKNAYLALPVILTRLKQKQEEWARCRTEFNKVWADIYTKNYHRSLDHRS 643
Query: 632 FYFKQQDTKSLSTK 645
FYFKQQD+K+LSTK
Sbjct: 644 FYFKQQDSKNLSTK 657
>pir||C96723 hypothetical protein F20P5.21 [imported] - Arabidopsis thaliana
gi|2194132|gb|AAB61107.1| F20P5.21 gene product
[Arabidopsis thaliana]
Length = 1383
Score = 667 bits (1720), Expect = 0.0
Identities = 363/680 (53%), Positives = 483/680 (70%), Gaps = 47/680 (6%)
Query: 1 MTGG---QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKEL 57
M GG QKLTTNDAL+YLKAV++ FQ+ ++KYDEFLEVMKDFKAQR+DT GVI RVKEL
Sbjct: 1 MVGGSSAQKLTTNDALAYLKAVKDKFQDKRDKYDEFLEVMKDFKAQRVDTTGVILRVKEL 60
Query: 58 FKGHKDLILGFNTFLPKGYAITLPSDDEQPLQ-KKPVEFEEAINFVGKIKNRFQGNDRVY 116
FKG+++LILGFNTFLPKG+ ITL +D+QP KKPVEFEEAI+FV KIK RFQG+DRVY
Sbjct: 61 FKGNRELILGFNTFLPKGFEITLRPEDDQPAAPKKPVEFEEAISFVNKIKTRFQGDDRVY 120
Query: 117 KTFLDILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPL 176
K+FLDILNMYRKE K IT VY EV+ LF+DH DLL EFTHFLPDTS A+ + S + P+
Sbjct: 121 KSFLDILNMYRKENKSITEVYHEVAILFRDHHDLLGEFTHFLPDTSATASTN-DSVKVPV 179
Query: 177 LRDRS-SAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRR-EREK 234
RDR ++ T RQ+ +DK+++ T H +R L ++ D + +R +++ KE+ RR +++
Sbjct: 180 -RDRGIKSLPTMRQIDLDKKDRIITSHPNRALKTENMDVDHERSLLKDSKEEVRRIDKKN 238
Query: 235 DRREERDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGT--AEPLHDADEKLDLLPN 292
D ++RDR+ D R D+D + E + KK + R D ++ + D+ +P+
Sbjct: 239 DFMDDRDRK----DYRGLDHDSHKEHFFNSKKKLIRKDDDSAEMSDQAREGDKFSGAIPS 294
Query: 293 SSTCEDKSSLKSLC---SPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLAL 349
SST ++K + S LAF+++VK KL D+ QEFL+CL++YS+EII++ EL +L
Sbjct: 295 SSTYDEKGFIIDFLESHSQELAFVDRVKAKLDTADN-QEFLRCLNLYSKEIISQPELQSL 353
Query: 350 VGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVMNK----------------KSLWNEGHG 393
V DL+G Y D+M+ F F+ QC+KN+G L+G+++K +SLW+EG
Sbjct: 354 VSDLIGVYPDLMDAFKVFLAQCDKNDGLLSGIVSKSKSSTFYNILLTYLFGQSLWSEGKC 413
Query: 394 QKPLKVEEKDRDRGRD--DGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTS----- 446
+P K +KD DR R+ + +ERDRE +K I ++ LS +
Sbjct: 414 PQPTKSLDKDTDREREKIERYRERDREKERLEKVAASQKWAKPISELDLSNCEQCTPSYR 473
Query: 447 -M*ESL*MNWTFLTVNNVLPATVSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKN 505
+ ++L ++ F+ + +P QK E+G++VLNDHWVSVTSGSEDYSFKHMRKN
Sbjct: 474 RLPKNLNVHTYFVLLQYPIPIAS-----QKMEIGSQVLNDHWVSVTSGSEDYSFKHMRKN 528
Query: 506 QYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLR 565
QYEESLF+CEDDRFELDMLLES+ + + +VEE++ K+N + + D PI IE+HL+ALNLR
Sbjct: 529 QYEESLFKCEDDRFELDMLLESVISATNRVEELLAKINSNELKTDTPICIEDHLTALNLR 588
Query: 566 CIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHK 625
CIERLY DHGLDV+++LKKNA LALPVILTRLKQKQEEWARCR +FNKVWA+IY KNYH+
Sbjct: 589 CIERLYSDHGLDVLDLLKKNAYLALPVILTRLKQKQEEWARCRTEFNKVWADIYTKNYHR 648
Query: 626 SLDHRSFYFKQQDTKSLSTK 645
SLDHRSFYFKQQD+K+LSTK
Sbjct: 649 SLDHRSFYFKQQDSKNLSTK 668
>ref|NP_908404.1| putative co-repressor protein [Oryza sativa (japonica
cultivar-group)]
Length = 1589
Score = 642 bits (1656), Expect = 0.0
Identities = 375/660 (56%), Positives = 451/660 (67%), Gaps = 53/660 (8%)
Query: 2 TGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGH 61
T GQKLTTNDAL YLKAV++ FQ+ +EKY+EFLEVM+DFK++RIDT GVI RVK LF G+
Sbjct: 236 TAGQKLTTNDALVYLKAVKDKFQDKREKYEEFLEVMRDFKSERIDTNGVIIRVKTLFNGY 295
Query: 62 KDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLD 121
+LILGFNTFLPKGYAI L + KKPV+F EAINFV KIKNRFQ ++ VYK FLD
Sbjct: 296 PELILGFNTFLPKGYAIKLQEE------KKPVDFVEAINFVNKIKNRFQHDEHVYKAFLD 349
Query: 122 ILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRS 181
ILNMYRK+ K I VY EV+ LF DH DLLEEF HFLPDTS A A +R + RD
Sbjct: 350 ILNMYRKDNKSIQDVYHEVAVLFADHKDLLEEFQHFLPDTSVPPQA-VAPSRPGIRRDDR 408
Query: 182 SAMTTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRRE-ER 240
+++ + +KR+K HADR+ SVD PD LD + +R R KDR + +R
Sbjct: 409 TSLVPPASRN-EKRDKAHP-HADRE-SVDRPD--LDHVI--------QRRRPKDRHDYDR 455
Query: 241 DRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDAD-EKLDLLPNSSTCED- 298
++ E D +D D L+R P +K DP +A+ H E +L S++ D
Sbjct: 456 GDKDGELDSKDLDI--GLKRKPFPRKM----EDPTSADAHHGGPLENHGILGASASLYDN 509
Query: 299 KSSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYA 358
K +LKS+ + F EKVKEKL++ D YQEFLKCLHIYS+EIITR EL LV D+L ++
Sbjct: 510 KDALKSVYTQEFHFCEKVKEKLEH-DAYQEFLKCLHIYSQEIITRSELKNLVNDILQQHP 568
Query: 359 DIMEGFDDFVTQCEKNEGFLAGVMNKKSLWNEGH---GQKPLKVEEKDRDRGRDDGVKER 415
D+M+GF++F+ CE +GFLAGV +KKS N + GQ V+ ++R G KE
Sbjct: 569 DLMDGFNEFLEHCENIDGFLAGVFSKKSHVNTNYINEGQTGRIVKTEERKEGGKGTEKEP 628
Query: 416 DRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSY----- 470
DR + S K V K S L ++N T SY
Sbjct: 629 DRIEKVPAYKEAPSQKPVFSSKEKYIYKPVSE----------LDLSNCQRCTPSYRLLPK 678
Query: 471 -----PKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLL 525
P KTELGA VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLL
Sbjct: 679 HYPMPPAGNKTELGASVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLL 738
Query: 526 ESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKN 585
ES+NA +K+VEE+IEK+ D+ + D PIRI+EHL+ LNLRCIERLYGDHGLDVM+VL+KN
Sbjct: 739 ESVNAATKRVEELIEKMQDNSLKPDSPIRIDEHLTPLNLRCIERLYGDHGLDVMDVLRKN 798
Query: 586 ASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
AS+ALPVILTRLKQKQEEW+RCR DFNKVWAEIYAKNYHKSLDHRSFYFKQQDTK+LSTK
Sbjct: 799 ASVALPVILTRLKQKQEEWSRCRSDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKNLSTK 858
Score = 41.2 bits (95), Expect = 0.11
Identities = 27/68 (39%), Positives = 39/68 (56%), Gaps = 6/68 (8%)
Query: 92 PVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQDHGDLL 151
P+ FE+++ FV K+K N +Y + DIL R EL + A Y+E+ LFQ++ DL
Sbjct: 13 PLTFEDSLRFVKKVK---ACNYMLYLSLFDILG--RMELSRLEA-YRELQLLFQNYPDLH 66
Query: 152 EEFTHFLP 159
EE F P
Sbjct: 67 EELEKFRP 74
>gb|AAF03494.1| unknown protein [Arabidopsis thaliana]
Length = 1324
Score = 598 bits (1542), Expect = e-169
Identities = 344/655 (52%), Positives = 433/655 (65%), Gaps = 55/655 (8%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTTNDALSYL+ V+EMFQ+ +EKYD FLEVMKDFKAQR DT GVI RVKELFKGH +L
Sbjct: 51 QKLTTNDALSYLREVKEMFQDQREKYDRFLEVMKDFKAQRTDTGGVIARVKELFKGHNNL 110
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKGY ITL +D+ L KK VEFE+AINFV KIK RF+ ++ VYK+FL+ILN
Sbjct: 111 IYGFNTFLPKGYEITLIEEDDA-LPKKTVEFEQAINFVNKIKMRFKHDEHVYKSFLEILN 169
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT--SGAAAAHFASARNPLLRDRSS 182
MYRKE K I VY EVS LFQ H DLLE+FT FLP + S +AA H S DR S
Sbjct: 170 MYRKENKEIKEVYNEVSILFQGHLDLLEQFTRFLPASLPSHSAAQHSRSQAQQY-SDRGS 228
Query: 183 AMTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREE 239
QM V+K RE+ L D SV+ D D+ +++ +EQR+R +E
Sbjct: 229 DPPLLHQMQVEKERRRERAVALRGD--YSVERYDLNDDKTMVKIQREQRKRLD----KEN 282
Query: 240 RDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDK 299
R RR R+ DDR+ D NL P K+KS RA E L+ S++ +K
Sbjct: 283 RARRGRDLDDREAGQD-NLHHFPEKRKSSRRA-------------EALEAYSGSASHSEK 328
Query: 300 SSLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYAD 359
+LKS+ F EKVK++L + DDYQ FLKCL+I+S II R++L LV DLLGK+ D
Sbjct: 329 DNLKSMYKQAFVFCEKVKDRLCSQDDYQTFLKCLNIFSNGIIQRKDLQNLVSDLLGKFPD 388
Query: 360 IMEGFDDFVTQCEKNEGF-------LAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRD-DG 411
+M+ F+ F +CE G LAGVM+KK +E +P+KVEEK+ + + +
Sbjct: 389 LMDEFNQFFERCESITGTEIHGFQRLAGVMSKKLFSSEEQLSRPMKVEEKESEHKPELEA 448
Query: 412 VKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYP 471
VKE ++ +E SI ++ LS + +LPA P
Sbjct: 449 VKETEQCKKEY--------MGKSIQELDLSDCECCT-----------PSYRLLPADYPIP 489
Query: 472 -KIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINA 530
Q++ELGA+VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCEDDRFELDMLLES+++
Sbjct: 490 IASQRSELGAEVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCEDDRFELDMLLESVSS 549
Query: 531 TSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLAL 590
++ E ++ + + I RIE+H +ALNLRCIERLYGDHGLDV+++L KN + AL
Sbjct: 550 AARSAESLLNIITEKKISFSGSFRIEDHFTALNLRCIERLYGDHGLDVIDILNKNPATAL 609
Query: 591 PVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
PVILTRLKQKQ EW +CR+DF+KVWA +YAKN++KSLDHRSFYFKQQD+K+LS K
Sbjct: 610 PVILTRLKQKQGEWKKCRDDFDKVWANVYAKNHYKSLDHRSFYFKQQDSKNLSAK 664
>emb|CAC01821.1| transcriptional regulatory-like protein [Arabidopsis thaliana]
gi|15242192|ref|NP_197006.1| paired amphipathic helix
repeat-containing protein [Arabidopsis thaliana]
gi|11358896|pir||T51447 transcription regulator-like
protein - Arabidopsis thaliana
Length = 1377
Score = 595 bits (1535), Expect = e-168
Identities = 337/686 (49%), Positives = 441/686 (64%), Gaps = 81/686 (11%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTT+DAL+YLK V+EMFQ+ ++KYD FLEVMKDFKAQ+ DT+GVI RVKELFKGH +L
Sbjct: 46 QKLTTDDALTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNL 105
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKG+ ITL DD + KK VEFEEAI+FV KIK RFQ N+ VYK+FL+ILN
Sbjct: 106 IFGFNTFLPKGFEITL--DDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILN 163
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGA-AAAHFASARNPLLRDRSSA 183
MYRK+ K IT VY EVS LF+DH DLLEEFT FLPD+ A ++ DR S
Sbjct: 164 MYRKDNKDITEVYNEVSTLFEDHSDLLEEFTRFLPDSLAPHTEAQLLRSQAQRYDDRGSG 223
Query: 184 MTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREER 240
R+M ++K RE+T DRD SVD D D+ +++ ++QR+R +KD RE
Sbjct: 224 PPLVRRMFMEKDRRRERTVASRGDRDHSVDRSDLNDDKSMVKMHRDQRKRV-DKDNRE-- 280
Query: 241 DRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKS 300
RR R+ +D + + D NL+ K+KS R E + ++ +K+
Sbjct: 281 -RRSRDLEDGEAEQD-NLQHFSEKRKSSRRM-------------EGFEAYSGPASHSEKN 325
Query: 301 SLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADI 360
+LKS+ + F EKVKE+L + DDYQ FLKCL+++S II R++L LV D+LGK+ D+
Sbjct: 326 NLKSMYNQAFLFCEKVKERLCSQDDYQAFLKCLNMFSNGIIQRKDLQNLVSDVLGKFPDL 385
Query: 361 MEGFDDFVTQCEKNEGF--LAGVMNK---------------------------------- 384
M+ F+ F +CE +GF LAGVM+K
Sbjct: 386 MDEFNQFFERCESIDGFQHLAGVMSKSRQQSPSFLSMSILFSFFSYVIGIEITLPGTLAA 445
Query: 385 KSLWNEGHGQKPLKVEEKDRDRGRD-DGVKERDRELRERDKSTGISNKDVSIPKVSLSKD 443
+SL +E + + +K EEKDR+ RD + KE++R +DK G S +++ +
Sbjct: 446 ESLGSEENLSRSVKGEEKDREHKRDVEAAKEKERS---KDKYMGKSIQELDLSDCERCTP 502
Query: 444 KTSM*ESL*MNWTFLTVNNVLPATVSYPKIQ-KTELGAKVLNDHWVSVTSGSEDYSFKHM 502
+ LP P ++ + + GA VLNDHWVSVTSGSEDYSFKHM
Sbjct: 503 SYRL----------------LPPDYPIPSVRHRQKSGAAVLNDHWVSVTSGSEDYSFKHM 546
Query: 503 RKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSAL 562
R+NQYEESLFRCEDDRFELDMLLES+ + +K EE++ + D I + RIE+H +AL
Sbjct: 547 RRNQYEESLFRCEDDRFELDMLLESVGSAAKSAEELLNIIIDKKISFEGSFRIEDHFTAL 606
Query: 563 NLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKN 622
NLRCIERLYGDHGLDV ++++KN + ALPVILTRLKQKQ+EW +CRE FN VWA++YAKN
Sbjct: 607 NLRCIERLYGDHGLDVTDLIRKNPAAALPVILTRLKQKQDEWTKCREGFNVVWADVYAKN 666
Query: 623 YHKSLDHRSFYFKQQDTKSLSTKGKI 648
++KSLDHRSFYFKQQD+K+LS K +
Sbjct: 667 HYKSLDHRSFYFKQQDSKNLSAKALV 692
>ref|NP_186781.3| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 1378
Score = 593 bits (1529), Expect = e-168
Identities = 344/657 (52%), Positives = 435/657 (65%), Gaps = 57/657 (8%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTTNDALSYL+ V+EMFQ+ +EKYD FLEVMKDFKAQR DT GVI RVKELFKGH +L
Sbjct: 51 QKLTTNDALSYLREVKEMFQDQREKYDRFLEVMKDFKAQRTDTGGVIARVKELFKGHNNL 110
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKGY ITL +D+ L KK VEFE+AINFV KIK RF+ ++ VYK+FL+ILN
Sbjct: 111 IYGFNTFLPKGYEITLIEEDDA-LPKKTVEFEQAINFVNKIKMRFKHDEHVYKSFLEILN 169
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT--SGAAAAHFASARNPLLRDRSS 182
MYRKE K I VY EVS LFQ H DLLE+FT FLP + S +AA H S DR S
Sbjct: 170 MYRKENKEIKEVYNEVSILFQGHLDLLEQFTRFLPASLPSHSAAQHSRSQAQQY-SDRGS 228
Query: 183 AMTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREE 239
QM V+K RE+ L D SV+ D D+ +++ +EQR+R +E
Sbjct: 229 DPPLLHQMQVEKERRRERAVALRGD--YSVERYDLNDDKTMVKIQREQRKRLD----KEN 282
Query: 240 RDRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDK 299
R RR R+ DDR+ D NL P K+KS RA E L+ S++ +K
Sbjct: 283 RARRGRDLDDREAGQD-NLHHFPEKRKSSRRA-------------EALEAYSGSASHSEK 328
Query: 300 SSLKS------LCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDL 353
+LK+ + F EKVK++L + DDYQ FLKCL+I+S II R++L LV DL
Sbjct: 329 DNLKNVYFVLGMYKQAFVFCEKVKDRLCSQDDYQTFLKCLNIFSNGIIQRKDLQNLVSDL 388
Query: 354 LGKYADIMEGFDDFVTQCEK-NEGF--LAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRD- 409
LGK+ D+M+ F+ F +CE +GF LAGVM+KK +E +P+KVEEK+ + +
Sbjct: 389 LGKFPDLMDEFNQFFERCESITDGFQRLAGVMSKKLFSSEEQLSRPMKVEEKESEHKPEL 448
Query: 410 DGVKERDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVS 469
+ VKE ++ +E SI ++ LS + +LPA
Sbjct: 449 EAVKETEQCKKEY--------MGKSIQELDLSDCECCT-----------PSYRLLPADYP 489
Query: 470 YP-KIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESI 528
P Q++ELGA+VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCEDDRFELDMLLES+
Sbjct: 490 IPIASQRSELGAEVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCEDDRFELDMLLESV 549
Query: 529 NATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASL 588
++ ++ E ++ + + I RIE+H +ALNLRCIERLYGDHGLDV+++L KN +
Sbjct: 550 SSAARSAESLLNIITEKKISFSGSFRIEDHFTALNLRCIERLYGDHGLDVIDILNKNPAT 609
Query: 589 ALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
ALPVILTRLKQKQ EW +CR+DF+KVWA +YAKN++KSLDHRSFYFKQQD+K+LS K
Sbjct: 610 ALPVILTRLKQKQGEWKKCRDDFDKVWANVYAKNHYKSLDHRSFYFKQQDSKNLSAK 666
>ref|XP_549869.1| transcriptional co-repressor -like [Oryza sativa (japonica
cultivar-group)] gi|52076211|dbj|BAD44865.1|
transcriptional co-repressor -like [Oryza sativa
(japonica cultivar-group)]
Length = 1243
Score = 470 bits (1209), Expect = e-131
Identities = 285/534 (53%), Positives = 347/534 (64%), Gaps = 52/534 (9%)
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAM 184
MYRK+ K I VY EV+ LF DH DLLEEF HFLPDTS A A +R + RD +++
Sbjct: 1 MYRKDNKSIQDVYHEVAVLFADHKDLLEEFQHFLPDTSVPPQA-VAPSRPGIRRDDRTSL 59
Query: 185 TTGRQMHVDKREKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRRE-ERDRR 243
+ +KR+K HADR+ SVD PD LD + +R R KDR + +R +
Sbjct: 60 VPPASRN-EKRDKAHP-HADRE-SVDRPD--LDHVI--------QRRRPKDRHDYDRGDK 106
Query: 244 ERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDAD-EKLDLLPNSSTCED-KSS 301
+ E D +D D L+R P +K DP +A+ H E +L S++ D K +
Sbjct: 107 DGELDSKDLDI--GLKRKPFPRKM----EDPTSADAHHGGPLENHGILGASASLYDNKDA 160
Query: 302 LKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADIM 361
LKS+ + F EKVKEKL++ D YQEFLKCLHIYS+EIITR EL LV D+L ++ D+M
Sbjct: 161 LKSVYTQEFHFCEKVKEKLEH-DAYQEFLKCLHIYSQEIITRSELKNLVNDILQQHPDLM 219
Query: 362 EGFDDFVTQCEKNEGFLAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRDDGVKERDRELRE 421
+GF++F+ CE +GFLAGV +K+ Q V+ ++R G KE DR +
Sbjct: 220 DGFNEFLEHCENIDGFLAGVFSKR--------QTGRIVKTEERKEGGKGTEKEPDRIEKV 271
Query: 422 RDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSY----------P 471
S K V K S L ++N T SY P
Sbjct: 272 PAYKEAPSQKPVFSSKEKYIYKPVSE----------LDLSNCQRCTPSYRLLPKHYPMPP 321
Query: 472 KIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINAT 531
KTELGA VLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLES+NA
Sbjct: 322 AGNKTELGASVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVNAA 381
Query: 532 SKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALP 591
+K+VEE+IEK+ D+ + D PIRI+EHL+ LNLRCIERLYGDHGLDVM+VL+KNAS+ALP
Sbjct: 382 TKRVEELIEKMQDNSLKPDSPIRIDEHLTPLNLRCIERLYGDHGLDVMDVLRKNASVALP 441
Query: 592 VILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
VILTRLKQKQEEW+RCR DFNKVWAEIYAKNYHKSLDHRSFYFKQQDTK+LSTK
Sbjct: 442 VILTRLKQKQEEWSRCRSDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKNLSTK 495
>dbj|BAC43533.1| putative transcriptional regulatory protein [Arabidopsis thaliana]
Length = 543
Score = 441 bits (1133), Expect = e-122
Identities = 256/523 (48%), Positives = 337/523 (63%), Gaps = 47/523 (8%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTT+DAL+YLK V+EMFQ+ ++KYD FLEVMKDFKAQ+ DT+GVI RVKELFKGH +L
Sbjct: 46 QKLTTDDALTYLKEVKEMFQDQRDKYDMFLEVMKDFKAQKTDTSGVISRVKELFKGHNNL 105
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
I GFNTFLPKG+ ITL DD + KK VEFEEAI+FV KIK RFQ N+ VYK+FL+ILN
Sbjct: 106 IFGFNTFLPKGFEITL--DDVEAPSKKTVEFEEAISFVNKIKTRFQHNELVYKSFLEILN 163
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT-SGAAAAHFASARNPLLRDRSSA 183
MYRK+ K IT VY EVS LF+DH DLLEEFT FLPD+ + A ++ DR S
Sbjct: 164 MYRKDNKDITEVYNEVSTLFEDHSDLLEEFTRFLPDSLAPHTEAQLLRSQAQRYDDRGSG 223
Query: 184 MTTGRQMHVDK---REKTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREER 240
R+M ++K RE+T DRD SVD D D+ +++ ++QR+R +KD RE
Sbjct: 224 PPLVRRMFMEKDRRRERTVASRGDRDHSVDRSDLNDDKSMVKMHRDQRKRV-DKDNRE-- 280
Query: 241 DRRERERDDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKS 300
RR R+ +D + + D NL+ K+KS R E + ++ +K+
Sbjct: 281 -RRSRDLEDGEAEQD-NLQHFSEKRKSSRRM-------------EGFEAYSGPASHSEKN 325
Query: 301 SLKSLCSPVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQELLALVGDLLGKYADI 360
+LKS+ + F EKVKE+L + DDYQ FLKCL+++S II R++L LV D+LGK+ D+
Sbjct: 326 NLKSMYNQAFLFCEKVKERLCSQDDYQAFLKCLNMFSNGIIQRKDLQNLVSDVLGKFPDL 385
Query: 361 MEGFDDFVTQCEKNEGF--LAGVMNKKSLWNEGHGQKPLKVEEKDRDRGRD-DGVKERDR 417
M+ F+ F +CE +GF LAGVM+KKSL +E + + +K EEKDR+ RD + KE++
Sbjct: 386 MDEFNQFFERCESIDGFQHLAGVMSKKSLGSEENLSRSVKGEEKDREHKRDVEAAKEKE- 444
Query: 418 ELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPKIQ-KT 476
R +DK G S +++ + +LP P ++ +
Sbjct: 445 --RSKDKYMGKSIQELDLSDCERCTPS----------------YRLLPPDYPIPSVRHRQ 486
Query: 477 ELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRF 519
+ GA VLNDHWVSVTSGSEDYSFKHMR+NQYEESLFRCEDDRF
Sbjct: 487 KSGAAVLNDHWVSVTSGSEDYSFKHMRRNQYEESLFRCEDDRF 529
>ref|XP_493857.1| Similar to Arabidopsis chromosome I BAC genomic sequence
(AC002396); unknown protein [Oryza sativa]
Length = 1056
Score = 303 bits (775), Expect = 2e-80
Identities = 178/353 (50%), Positives = 224/353 (63%), Gaps = 31/353 (8%)
Query: 312 FLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQELLALVGDLLGKYADIMEGFDDFVTQ 370
F+ K+K + + D Y+ FL L++Y + Q++ V L Y D++E F F+
Sbjct: 112 FVNKIKARFQQEDHVYKSFLGILNMYRLHNKSIQDVYGEV-PLFRDYPDLLEEFKHFLPD 170
Query: 371 CEKNEGFLAGVMNKKSLWNEGHGQ-----------KPLKVEEKDRDRGRDDGVKE----- 414
+ V S ++ G K + EK +++ + +E
Sbjct: 171 TSTAPEPVT-VPRGVSSRHDDRGPLMPSARNAQIIKEFRFCEKVKEKLEPEAYQEFLKCL 229
Query: 415 --RDRELRERDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVNNVLPATVSYPK 472
+E+ R + + N I ++ LS + ++ L N +P P
Sbjct: 230 HIYSQEIITRSELKNLYNLCKPISELDLSNCQRCT-----PSYRLLPKNYPMP-----PA 279
Query: 473 IQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATS 532
+T+LGA VLND WVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLES+
Sbjct: 280 SCRTDLGASVLNDLWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVIVAI 339
Query: 533 KKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPV 592
K+VEE+IEK+ D+ I D PIRI+EHL+ LNLRCIERLYGDHGLDVM+VL+KNAS+ALPV
Sbjct: 340 KRVEELIEKMQDNSIKPDSPIRIDEHLTPLNLRCIERLYGDHGLDVMDVLRKNASVALPV 399
Query: 593 ILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
ILTRLKQKQEEW+RCR DFNKVWAEIYAKNYHKSLDHRSFYFKQQDTK+LSTK
Sbjct: 400 ILTRLKQKQEEWSRCRSDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKNLSTK 452
Score = 207 bits (526), Expect = 1e-51
Identities = 111/160 (69%), Positives = 125/160 (77%), Gaps = 6/160 (3%)
Query: 5 QKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDL 64
QKLTTNDAL YLKAV++ FQ+ ++KYDEFLEVM+DFK+ RIDTAGVI RVK LF GH +L
Sbjct: 21 QKLTTNDALLYLKAVKDKFQDKRDKYDEFLEVMRDFKSGRIDTAGVIIRVKTLFNGHHEL 80
Query: 65 ILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILN 124
ILGFN FLPKG+AI L Q L+KKPV+F EAINFV KIK RFQ D VYK+FL ILN
Sbjct: 81 ILGFNAFLPKGFAIKL-----QDLEKKPVDFMEAINFVNKIKARFQQEDHVYKSFLGILN 135
Query: 125 MYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGA 164
MYR K I VY EV LF+D+ DLLEEF HFLPDTS A
Sbjct: 136 MYRLHNKSIQDVYGEV-PLFRDYPDLLEEFKHFLPDTSTA 174
Score = 38.9 bits (89), Expect = 0.55
Identities = 22/80 (27%), Positives = 38/80 (47%), Gaps = 1/80 (1%)
Query: 81 PSDDEQ-PLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQE 139
PS Q P Q + + +A+ ++ +K++FQ Y FL+++ ++ V
Sbjct: 10 PSQQGQGPSQNQKLTTNDALLYLKAVKDKFQDKRDKYDEFLEVMRDFKSGRIDTAGVIIR 69
Query: 140 VSALFQDHGDLLEEFTHFLP 159
V LF H +L+ F FLP
Sbjct: 70 VKTLFNGHHELILGFNAFLP 89
>ref|NP_172515.1| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 1173
Score = 282 bits (722), Expect = 2e-74
Identities = 177/398 (44%), Positives = 234/398 (58%), Gaps = 43/398 (10%)
Query: 286 KLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQ 344
K+ LLP E+K ++ + F+ K+K + + + Y+ FL L++Y +E +
Sbjct: 150 KITLLPE----EEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSIS 205
Query: 345 ELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVM--NKKSLWNEGHGQKPLKVEEK 402
E+ V L + D++ F +F+ C ++ + +K + H K K K
Sbjct: 206 EVYEEVTMLFKGHEDLLMEFVNFLPNCPESAPSTKNAVPRHKGTATTAMHSDKKRKQRCK 265
Query: 403 DRDRG-----RDDGVKE------------------RDRELRE---------RDKSTGISN 430
D R+DG + RD E RE +KS +
Sbjct: 266 LEDYSGHSDQREDGDENLVTCSADSPVGEGQPGYFRDYENREDTETDTADRTEKSAASGS 325
Query: 431 KDVSIPKVSLSKDKTSM*ESL*MNWTFLTVN-NVLPA--TVSYPKIQKTELGAKVLNDHW 487
+D+ K + T + E T T + +LP V P + T LG K LNDH
Sbjct: 326 QDIGNHKSTTKYVGTPINELDLSECTQCTPSYRLLPKDYAVEIPSYRNT-LGKKTLNDHL 384
Query: 488 VSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKVNDDII 547
VSVTSGSEDYSF HMRKNQYEESLFRCEDDR+E+DMLL S+++ K+VE ++EK+N++ I
Sbjct: 385 VSVTSGSEDYSFSHMRKNQYEESLFRCEDDRYEMDMLLGSVSSAIKQVEILLEKMNNNTI 444
Query: 548 PGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQKQEEWARC 607
D I IE+HLSA+NLRCIERLYGD+GLDVM++LKKN ALPVILTRLKQKQEEWARC
Sbjct: 445 SVDSTICIEKHLSAMNLRCIERLYGDNGLDVMDLLKKNMHSALPVILTRLKQKQEEWARC 504
Query: 608 REDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTK
Sbjct: 505 HSDFQKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTK 542
Score = 201 bits (512), Expect = 5e-50
Identities = 118/255 (46%), Positives = 160/255 (62%), Gaps = 24/255 (9%)
Query: 9 TNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGF 68
T DAL+YLKAV+++F ++KEKY+ FLE+MK+FKAQ IDT GVIER+K LFKG++DL+LGF
Sbjct: 82 TIDALTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGF 141
Query: 69 NTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRK 128
NTFLPKGY ITL ++E+P K V+F++AI FV KIK RF ++ YK FLDILN+YRK
Sbjct: 142 NTFLPKGYKITLLPEEEKP--KIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRK 199
Query: 129 ELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGR 188
E K I+ VY+EV+ LF+ H DLL EF +FLP+ +A S +N + R + +A T
Sbjct: 200 EKKSISEVYEEVTMLFKGHEDLLMEFVNFLPNCPESA----PSTKNAVPRHKGTATTA-- 253
Query: 189 QMHVDKREKTT------TLHADR---------DLSVDHPDPELDRGVMRTDKEQRRRERE 233
MH DK+ K + H+D+ S D P E G R + + E +
Sbjct: 254 -MHSDKKRKQRCKLEDYSGHSDQREDGDENLVTCSADSPVGEGQPGYFRDYENREDTETD 312
Query: 234 KDRREERDRRERERD 248
R E+ +D
Sbjct: 313 TADRTEKSAASGSQD 327
>ref|NP_176197.2| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 1159
Score = 279 bits (714), Expect = 2e-73
Identities = 133/174 (76%), Positives = 153/174 (87%), Gaps = 3/174 (1%)
Query: 475 KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKK 534
+ LG KVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLES++A K+
Sbjct: 338 RNSLGEKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESVSAAIKR 397
Query: 535 VEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVIL 594
VE ++EK+N++ I + PI I EHLS +CIERLYGD+GLDVM+ LKKN+ +ALPVIL
Sbjct: 398 VESLLEKINNNTISIETPICIREHLSG---KCIERLYGDYGLDVMDFLKKNSHIALPVIL 454
Query: 595 TRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKGKI 648
TRLKQKQEEWARCR DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTKG +
Sbjct: 455 TRLKQKQEEWARCRADFRKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTKGLV 508
Score = 235 bits (599), Expect = 4e-60
Identities = 127/213 (59%), Positives = 153/213 (71%), Gaps = 5/213 (2%)
Query: 2 TGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGH 61
TGG LTT DAL+YLKAV++MFQ++KEKY+ FL VMKDFKAQR+DT GVI RVK+LFKG+
Sbjct: 38 TGG--LTTVDALTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGY 95
Query: 62 KDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLD 121
DL+LGFNTFLPKGY ITL +DE+P KKPV+F+ AI FV +IK RF G+DR YK FLD
Sbjct: 96 DDLLLGFNTFLPKGYKITLQPEDEKP--KKPVDFQVAIEFVNRIKARFGGDDRAYKKFLD 153
Query: 122 ILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHF-ASARNPLLRDR 180
ILNMYRKE K I VYQEV+ LFQDH DLL EF HFLPD G+ + + RN + RDR
Sbjct: 154 ILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHFLPDFRGSVSVNDPLFQRNTIPRDR 213
Query: 181 SSAMTTGRQMHVDKREKTTTLHADRDLSVDHPD 213
+S H +K+ K + +LS D
Sbjct: 214 NSTFPGMHPKHFEKKIKRSRHDEYTELSDQRED 246
>gb|AAD39565.1| T10O24.5 [Arabidopsis thaliana] gi|25402600|pir||C86238 protein
T10O24.5 [imported] - Arabidopsis thaliana
Length = 1164
Score = 275 bits (702), Expect = 5e-72
Identities = 177/418 (42%), Positives = 234/418 (55%), Gaps = 63/418 (15%)
Query: 286 KLDLLPNSSTCEDKSSLKSLCSPVLAFLEKVKEKLKNPDD-YQEFLKCLHIYSREIITRQ 344
K+ LLP E+K ++ + F+ K+K + + + Y+ FL L++Y +E +
Sbjct: 150 KITLLPE----EEKPKIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRKEKKSIS 205
Query: 345 ELLALVGDLLGKYADIMEGFDDFVTQCEKNEGFLAGVM--NKKSLWNEGHGQKPLKVEEK 402
E+ V L + D++ F +F+ C ++ + +K + H K K K
Sbjct: 206 EVYEEVTMLFKGHEDLLMEFVNFLPNCPESAPSTKNAVPRHKGTATTAMHSDKKRKQRCK 265
Query: 403 DRDRG-----RDDGVKE--------------------------------------RDREL 419
D R+DG + RD E
Sbjct: 266 LEDYSGHSDQREDGDENLVTCSAGNSLEFSGLYPVLRSMLIFADSPVGEGQPGYFRDYEN 325
Query: 420 RE---------RDKSTGISNKDVSIPKVSLSKDKTSM*ESL*MNWTFLTVN-NVLPA--T 467
RE +KS ++D+ K + T + E T T + +LP
Sbjct: 326 REDTETDTADRTEKSAASGSQDIGNHKSTTKYVGTPINELDLSECTQCTPSYRLLPKDYA 385
Query: 468 VSYPKIQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLES 527
V P + T LG K LNDH VSVTSGSEDYSF HMRKNQYEESLFRCEDDR+E+DMLL S
Sbjct: 386 VEIPSYRNT-LGKKTLNDHLVSVTSGSEDYSFSHMRKNQYEESLFRCEDDRYEMDMLLGS 444
Query: 528 INATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNAS 587
+++ K+VE ++EK+N++ I D I IE+HLSA+NLRCIERLYGD+GLDVM++LKKN
Sbjct: 445 VSSAIKQVEILLEKMNNNTISVDSTICIEKHLSAMNLRCIERLYGDNGLDVMDLLKKNMH 504
Query: 588 LALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTK 645
ALPVILTRLKQKQEEWARC DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTK
Sbjct: 505 SALPVILTRLKQKQEEWARCHSDFQKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTK 562
Score = 201 bits (510), Expect = 8e-50
Identities = 106/189 (56%), Positives = 140/189 (73%), Gaps = 9/189 (4%)
Query: 9 TNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGF 68
T DAL+YLKAV+++F ++KEKY+ FLE+MK+FKAQ IDT GVIER+K LFKG++DL+LGF
Sbjct: 82 TIDALTYLKAVKDIFHDNKEKYESFLELMKEFKAQTIDTNGVIERIKVLFKGYRDLLLGF 141
Query: 69 NTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRK 128
NTFLPKGY ITL ++E+P K V+F++AI FV KIK RF ++ YK FLDILN+YRK
Sbjct: 142 NTFLPKGYKITLLPEEEKP--KIRVDFKDAIGFVTKIKTRFGDDEHAYKRFLDILNLYRK 199
Query: 129 ELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGR 188
E K I+ VY+EV+ LF+ H DLL EF +FLP+ +A S +N + R + +A T
Sbjct: 200 EKKSISEVYEEVTMLFKGHEDLLMEFVNFLPNCPESA----PSTKNAVPRHKGTATTA-- 253
Query: 189 QMHVDKREK 197
MH DK+ K
Sbjct: 254 -MHSDKKRK 261
>gb|AAD39330.1| Hypothetical protein [Arabidopsis thaliana] gi|25404243|pir||A96623
hypothetical protein F23H11.20 [imported] - Arabidopsis
thaliana
Length = 1108
Score = 235 bits (599), Expect = 4e-60
Identities = 127/213 (59%), Positives = 153/213 (71%), Gaps = 5/213 (2%)
Query: 2 TGGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGH 61
TGG LTT DAL+YLKAV++MFQ++KEKY+ FL VMKDFKAQR+DT GVI RVK+LFKG+
Sbjct: 38 TGG--LTTVDALTYLKAVKDMFQDNKEKYETFLGVMKDFKAQRVDTNGVIARVKDLFKGY 95
Query: 62 KDLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLD 121
DL+LGFNTFLPKGY ITL +DE+P KKPV+F+ AI FV +IK RF G+DR YK FLD
Sbjct: 96 DDLLLGFNTFLPKGYKITLQPEDEKP--KKPVDFQVAIEFVNRIKARFGGDDRAYKKFLD 153
Query: 122 ILNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHF-ASARNPLLRDR 180
ILNMYRKE K I VYQEV+ LFQDH DLL EF HFLPD G+ + + RN + RDR
Sbjct: 154 ILNMYRKETKSINEVYQEVTLLFQDHEDLLGEFVHFLPDFRGSVSVNDPLFQRNTIPRDR 213
Query: 181 SSAMTTGRQMHVDKREKTTTLHADRDLSVDHPD 213
+S H +K+ K + +LS D
Sbjct: 214 NSTFPGMHPKHFEKKIKRSRHDEYTELSDQRED 246
Score = 214 bits (546), Expect = 6e-54
Identities = 109/174 (62%), Positives = 119/174 (67%), Gaps = 47/174 (27%)
Query: 475 KTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKK 534
+ LG KVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDR
Sbjct: 343 RNSLGEKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDR---------------- 386
Query: 535 VEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVIL 594
CIERLYGD+GLDVM+ LKKN+ +ALPVIL
Sbjct: 387 -------------------------------CIERLYGDYGLDVMDFLKKNSHIALPVIL 415
Query: 595 TRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKGKI 648
TRLKQKQEEWARCR DF KVWAE+YAKN+HKSLDHRSFYFKQQD+K+LSTKG +
Sbjct: 416 TRLKQKQEEWARCRADFRKVWAEVYAKNHHKSLDHRSFYFKQQDSKNLSTKGLV 469
>ref|NP_173830.1| paired amphipathic helix repeat-containing protein [Arabidopsis
thaliana]
Length = 196
Score = 209 bits (531), Expect = 3e-52
Identities = 105/159 (66%), Positives = 123/159 (77%), Gaps = 3/159 (1%)
Query: 3 GGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHK 62
G K TTNDAL YL+AV+ FQ +EKYDEFL++M D+K QRID +GVI R+KEL K +
Sbjct: 6 GAHKPTTNDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMKELLKEQQ 65
Query: 63 DLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDI 122
L+LGFN FLP GY IT EQP QKKPVE EAI+F+ KIK RFQG+DRVYK+ LDI
Sbjct: 66 GLLLGFNAFLPNGYMIT---HHEQPSQKKPVELGEAISFINKIKTRFQGDDRVYKSVLDI 122
Query: 123 LNMYRKELKPITAVYQEVSALFQDHGDLLEEFTHFLPDT 161
LNMYRK+ KPITAVY+EV+ LF DH +LL EFTHFLP T
Sbjct: 123 LNMYRKDRKPITAVYREVAILFLDHNNLLVEFTHFLPAT 161
>pir||T00650 hypothetical protein F3I6.13 - Arabidopsis thaliana
gi|2829872|gb|AAC00580.1| Hypothetical protein
[Arabidopsis thaliana]
Length = 173
Score = 179 bits (454), Expect = 3e-43
Identities = 90/137 (65%), Positives = 106/137 (76%), Gaps = 3/137 (2%)
Query: 3 GGQKLTTNDALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHK 62
G K TTNDAL YL+AV+ FQ +EKYDEFL++M D+K QRID +GVI R+KEL K +
Sbjct: 6 GAHKPTTNDALKYLRAVKAKFQGQREKYDEFLQIMIDYKTQRIDISGVIIRMKELLKEQQ 65
Query: 63 DLILGFNTFLPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDI 122
L+LGFN FLP GY IT EQP QKKPVE EAI+F+ KIK RFQG+DRVYK+ LDI
Sbjct: 66 GLLLGFNAFLPNGYMIT---HHEQPSQKKPVELGEAISFINKIKTRFQGDDRVYKSVLDI 122
Query: 123 LNMYRKELKPITAVYQE 139
LNMYRK+ KPITAVY+E
Sbjct: 123 LNMYRKDRKPITAVYRE 139
>gb|AAN74748.1| hypothetical protein [Marchantia polymorpha]
Length = 366
Score = 172 bits (435), Expect = 4e-41
Identities = 84/165 (50%), Positives = 118/165 (70%), Gaps = 9/165 (5%)
Query: 467 TVSYPK---IQKTELGAKVLNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDM 523
T SYPK +T+L +VLND WVS + E FKH+ KN+YEE+LFRCEDD+FE D+
Sbjct: 206 TKSYPKPICTHRTDLAREVLNDSWVSTSQSKE---FKHIEKNKYEENLFRCEDDQFETDV 262
Query: 524 LLESINATSKKVEEIIEKVNDDIIPGDIPIRIEEHLSALNLRCIERLYGDHGLDVMEVLK 583
LESI + ++V E++E + D P + E+HL+ +N RCIER+YG HGL+V++ ++
Sbjct: 263 FLESIKDSIRRVTELLETLED---PSLSKLNFEDHLTPINFRCIERIYGKHGLEVVDQVR 319
Query: 584 KNASLALPVILTRLKQKQEEWARCREDFNKVWAEIYAKNYHKSLD 628
+N S+ALP+IL RLKQKQ+E + R N+VWA++YAKNYH SL+
Sbjct: 320 RNDSVALPIILNRLKQKQDEVSSFRTKMNEVWAKVYAKNYHTSLN 364
Score = 95.9 bits (237), Expect = 4e-18
Identities = 68/194 (35%), Positives = 102/194 (52%), Gaps = 28/194 (14%)
Query: 96 EEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELKPITAVYQEVSALFQ-----DHGDL 150
++ INF+ K+K RF ++ VYK FL+ILNMYRK KPI+ +YQEV+ LF +H DL
Sbjct: 95 DQTINFINKVKTRFSADEHVYKAFLEILNMYRKGNKPISEMYQEVATLFSEHADGEHADL 154
Query: 151 LEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMHVDKREKTTTLHADRDLSVD 210
LEEFT F PD+SGA P R + + ++ + E+ TT + R L+
Sbjct: 155 LEEFTSFRPDSSGAPVGPLL----PTSRKKKYSNKPISELDMSNCERCTTSY--RLLTKS 208
Query: 211 HPDP------ELDRGVMR-----TDKEQRRREREKDRREERDRRERERDDRDYDNDGNLE 259
+P P +L R V+ T + + + EK++ EE R +D ++ D LE
Sbjct: 209 YPKPICTHRTDLAREVLNDSWVSTSQSKEFKHIEKNKYEENLFR---CEDDQFETDVFLE 265
Query: 260 RLPHKKKSVHRATD 273
+ K S+ R T+
Sbjct: 266 SI---KDSIRRVTE 276
>gb|EAK88210.1| Sin3 like paired amphipathic helix containing protein, transcripts
identified by EST [Cryptosporidium parvum]
gi|66357980|ref|XP_626168.1| Sin3 like paired
amphipathic helix containing protein, transcripts
identified by EST [Cryptosporidium parvum]
Length = 1434
Score = 169 bits (427), Expect = 3e-40
Identities = 83/167 (49%), Positives = 116/167 (68%), Gaps = 4/167 (2%)
Query: 483 LNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKV 542
LND WVS+ GSED+SFKHMRKN YEE+LF+CED+RFELDM++E+ +T +E I E++
Sbjct: 401 LNDKWVSIPQGSEDFSFKHMRKNVYEENLFKCEDERFELDMVIENNRSTINALEPIAEEI 460
Query: 543 NDDIIPGDIP--IRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQK 600
+ + P D ++ S ++L+ I R+YGD+G +++E+LK+ +PVIL RL+QK
Sbjct: 461 S-KLSPEDKKNFKLVKPPFSIIHLKAISRIYGDNGPEILELLKRTPYSCIPVILNRLRQK 519
Query: 601 QEEWARCREDFNK-VWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKG 646
EEW R N+ VW +I KNY KS DHRSFYF+Q D K+ + KG
Sbjct: 520 DEEWTHARHLMNQGVWRDIQTKNYFKSFDHRSFYFRQVDKKNTNVKG 566
Score = 96.3 bits (238), Expect = 3e-18
Identities = 95/343 (27%), Positives = 149/343 (42%), Gaps = 34/343 (9%)
Query: 12 ALSYLKAVREMFQNDKEKYDEFLEVMKDFKAQRIDTAGVIERVKELFKGHKDLILGFNTF 71
A YL +R D E Y EFL +M+DFK I+ VI++V ELFK LI FN F
Sbjct: 14 ARDYLSRLRSKCGEDTELYQEFLRIMRDFKHGSINARMVIDQVAELFKKDTSLIAEFNNF 73
Query: 72 LPKGYAITLPSDDEQPLQKKPVEFEEAINFVGKIKNRFQGNDRVYKTFLDILNMYRKELK 131
LP+ + +P +D E A FV K+K+ +Y FL +L+ Y+ K
Sbjct: 74 LPEELRLQIPQND----------CEYAAAFVKKVKDIAPA---IYNDFLMLLSKYKDGEK 120
Query: 132 PITAVYQEVSALFQDHGDLLEEFTHFLPDTSGAAAAHFASARNPLLRDRSSAMTTGRQMH 191
+ V + S+LF + DLLEEF F+P+ SG + +S + ++ S + +
Sbjct: 121 SVNEVCELSSSLFASYPDLLEEFVLFIPELSGKTESLISSNKQSTEKNLSDSQND----Y 176
Query: 192 VDKRE----KTTTLHADRDLSVDHPDPELDRGVMRTDKEQRRREREKDRREERDRRERER 247
+DK E +T+H+ S P + R + E DR E ++
Sbjct: 177 LDKSEFLSYLASTVHSISTKS--EPPKVVTRKASSNLPYEMTVVPELDRILEDQKKRLSN 234
Query: 248 DDRDYDNDGNLERLPHKKKSVHRATDPGTAEPLHDADEKLDLLPNSSTCEDKSSLKSLCS 307
+ NL+ + V T + A + P + + + +
Sbjct: 235 AEEGSGITDNLKSEGVNQGIVSNEKGYSTQKA---APQSYYYNPEDMSIWE-ADYRIFEE 290
Query: 308 PVLAFLEKVKEKLKNPDDYQEFLKCLHIYSREIITRQE-LLAL 349
+AF E + D YQ+FLK +H+Y+R ++T E LLAL
Sbjct: 291 VYIAFGENCQ------DYYQDFLKLIHLYTRGVLTVVETLLAL 327
>ref|XP_666929.1| transcriptional regulatory-like protein [Cryptosporidium hominis]
gi|54658007|gb|EAL36701.1| transcriptional
regulatory-like protein [Cryptosporidium hominis]
Length = 1157
Score = 169 bits (427), Expect = 3e-40
Identities = 83/167 (49%), Positives = 116/167 (68%), Gaps = 4/167 (2%)
Query: 483 LNDHWVSVTSGSEDYSFKHMRKNQYEESLFRCEDDRFELDMLLESINATSKKVEEIIEKV 542
LND WVS+ GSED+SFKHMRKN YEE+LF+CED+RFELDM++E+ +T +E I E++
Sbjct: 124 LNDKWVSIPQGSEDFSFKHMRKNVYEENLFKCEDERFELDMVIENNRSTINALEPIAEEI 183
Query: 543 NDDIIPGDIP--IRIEEHLSALNLRCIERLYGDHGLDVMEVLKKNASLALPVILTRLKQK 600
+ + P D ++ S ++L+ I R+YGD+G +++E+LK+ +PVIL RL+QK
Sbjct: 184 S-KLSPEDKKNFKLVKPPFSIIHLKAISRIYGDNGPEILELLKRTPYSCIPVILNRLRQK 242
Query: 601 QEEWARCREDFNK-VWAEIYAKNYHKSLDHRSFYFKQQDTKSLSTKG 646
EEW R N+ VW +I KNY KS DHRSFYF+Q D K+ + KG
Sbjct: 243 DEEWTHARHLMNQGVWRDIQTKNYFKSFDHRSFYFRQVDKKNTNVKG 289
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,150,150,047
Number of Sequences: 2540612
Number of extensions: 51453323
Number of successful extensions: 305538
Number of sequences better than 10.0: 1818
Number of HSP's better than 10.0 without gapping: 653
Number of HSP's successfully gapped in prelim test: 1297
Number of HSP's that attempted gapping in prelim test: 263008
Number of HSP's gapped (non-prelim): 15480
length of query: 670
length of database: 863,360,394
effective HSP length: 135
effective length of query: 535
effective length of database: 520,377,774
effective search space: 278402109090
effective search space used: 278402109090
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146588.6


