

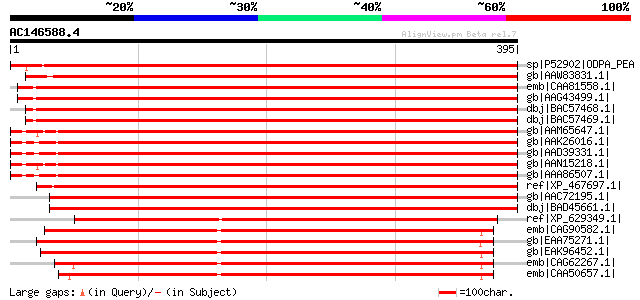
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146588.4 - phase: 0
(395 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component alpha sub... 662 0.0
gb|AAW83831.1| E1 alpha subunit of pyruvate dehydrogenase [Petun... 659 0.0
emb|CAA81558.1| E1 alpha subunit of pyruvate dehydrogenase precu... 649 0.0
gb|AAG43499.1| pyruvate dehydrogenase [Lycopersicon esculentum] 645 0.0
dbj|BAC57468.1| pyruvate dehydrogenase E1alpha subunit [Beta vul... 634 e-180
dbj|BAC57469.1| pyruvate dehydrogenase E1 alpha subunit [Beta vu... 630 e-179
gb|AAM65647.1| pyruvate dehydrogenase E1 alpha subunit [Arabidop... 627 e-178
gb|AAK26016.1| putative pyruvate dehydrogenase e1 alpha subunit ... 626 e-178
gb|AAD39331.1| pyruvate dehydrogenase E1 alpha subunit [Arabidop... 624 e-177
gb|AAN15218.1| pyruvate dehydrogenase E1a-like subunit IAR4 [Ara... 623 e-177
gb|AAA86507.1| pyruvate dehydrogenase E1 alpha subunit gi|211753... 621 e-176
ref|XP_467697.1| putative pyruvate dehydrogenase E1 alpha subuni... 620 e-176
gb|AAC72195.1| pyruvate dehydrogenase E1 alpha subunit [Zea mays] 610 e-173
dbj|BAD45661.1| putative pyruvate dehydrogenase E1 alpha subunit... 591 e-167
ref|XP_629349.1| pyruvate dehydrogenase E1 subunit alpha [Dictyo... 392 e-107
emb|CAG90582.1| unnamed protein product [Debaryomyces hansenii C... 386 e-106
gb|EAA75271.1| conserved hypothetical protein [Gibberella zeae P... 379 e-104
gb|EAK96452.1| hypothetical protein CaO19.10609 [Candida albican... 377 e-103
emb|CAG62267.1| unnamed protein product [Candida glabrata CBS138... 375 e-102
emb|CAA50657.1| PDA1 [Saccharomyces cerevisiae] gi|37362644|ref|... 371 e-101
>sp|P52902|ODPA_PEA Pyruvate dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A) gi|1263302|gb|AAA97411.1| pyruvate
dehydrogenase E1 alpha subunit gi|7431564|pir||T06531
pyruvate dehydrogenase (lipoamide) (EC 1.2.4.1) complex
E1 alpha chain - garden pea
Length = 397
Score = 662 bits (1708), Expect = 0.0
Identities = 322/398 (80%), Positives = 354/398 (88%), Gaps = 4/398 (1%)
Query: 1 MALSRFSSSQF---GSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTT 57
MALSR SSS GS L P+ + + +R ISS +T TLT+ETS+PFT+HNC+PPS +
Sbjct: 1 MALSRLSSSSSSSNGSNLFNPFSAAFTL-NRPISSDTTATLTIETSLPFTAHNCDPPSRS 59
Query: 58 VQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDC 117
V T+ SEL+SFF M LMRRMEIAADSLYKA LIRGFCHLYDGQEAVAVGMEA +KDC
Sbjct: 60 VTTSPSELLSFFRTMALMRRMEIAADSLYKANLIRGFCHLYDGQEAVAVGMEAGTTKKDC 119
Query: 118 VITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQI 177
+ITAYRDHCTFL RGGTL+ V++ELMGR+DGCSKGKGGSMHFY+K+ GFYGGHGIVGAQ+
Sbjct: 120 IITAYRDHCTFLGRGGTLLRVYAELMGRRDGCSKGKGGSMHFYKKDSGFYGGHGIVGAQV 179
Query: 178 PLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGT 237
PLG GLAFGQKY KD +VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGT
Sbjct: 180 PLGCGLAFGQKYLKDESVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGT 239
Query: 238 AEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGH 297
A WRSAKSPAY+KRGDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGH
Sbjct: 240 ATWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGH 299
Query: 298 SMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAK 357
SMSDPGSTYRTRDEISGVRQERDPIERVRKL+L+HDI+TEKELKD EKE RK+VDEAIAK
Sbjct: 300 SMSDPGSTYRTRDEISGVRQERDPIERVRKLLLSHDIATEKELKDTEKEVRKEVDEAIAK 359
Query: 358 AKESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
AK+S MPDPSDLF+NVYVKG GVEA G DRKEV+ TLP
Sbjct: 360 AKDSPMPDPSDLFSNVYVKGYGVEAFGVDRKEVRVTLP 397
>gb|AAW83831.1| E1 alpha subunit of pyruvate dehydrogenase [Petunia x hybrida]
Length = 390
Score = 659 bits (1699), Expect = 0.0
Identities = 317/384 (82%), Positives = 347/384 (89%), Gaps = 5/384 (1%)
Query: 13 STLLKPYFLS-SAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFND 71
S LKP + S RH S +T TLT+ETS+PFT HN +PPS TV+T +EL++FF D
Sbjct: 11 SKFLKPLTTAVSTTRHLS----TTNTLTIETSLPFTGHNIDPPSRTVETNPNELLTFFKD 66
Query: 72 MVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCR 131
M MRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GME+AI +KDC+ITAYRDHC FL R
Sbjct: 67 MAEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMESAITKKDCIITAYRDHCIFLSR 126
Query: 132 GGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNK 191
GGTL E FSELMGRKDGCSKGKGGSMHFY+K+ GFYGGHGIVGAQ+PLG+GLAF QKY+K
Sbjct: 127 GGTLFECFSELMGRKDGCSKGKGGSMHFYKKDSGFYGGHGIVGAQVPLGIGLAFAQKYSK 186
Query: 192 DPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKR 251
+ +VTF +YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKSP+YYKR
Sbjct: 187 EDHVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPSYYKR 246
Query: 252 GDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDE 311
GDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGHSMSDPGSTYRTRDE
Sbjct: 247 GDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGHSMSDPGSTYRTRDE 306
Query: 312 ISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFT 371
ISGVRQERDPIER+RKL+LAHDI+TEKELKDIEKE RK VDEAIAKAKES MPDPS+LFT
Sbjct: 307 ISGVRQERDPIERIRKLILAHDIATEKELKDIEKEKRKIVDEAIAKAKESAMPDPSELFT 366
Query: 372 NVYVKGLGVEACGADRKEVKATLP 395
NVYVKG GVEACGADRKEV+ATLP
Sbjct: 367 NVYVKGFGVEACGADRKEVRATLP 390
>emb|CAA81558.1| E1 alpha subunit of pyruvate dehydrogenase precursor [Solanum
tuberosum] gi|1709453|sp|P52903|ODPA_SOLTU Pyruvate
dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A) gi|7431569|pir||T07372 pyruvate
dehydrogenase (lipoamide) (EC 1.2.4.1) E1 alpha chain -
potato
Length = 391
Score = 649 bits (1675), Expect = 0.0
Identities = 314/390 (80%), Positives = 352/390 (89%), Gaps = 3/390 (0%)
Query: 7 SSSQFGSTLLKPYFLSSAI-RHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASEL 65
S+S+ + ++KP LS+A+ R +SS ST T+TVETS+PFTSHN +PPS +V+T+ EL
Sbjct: 4 STSRAINHIMKP--LSAAVCATRRLSSDSTATITVETSLPFTSHNIDPPSRSVETSPKEL 61
Query: 66 MSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDH 125
M+FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI +KDC+ITAYRDH
Sbjct: 62 MTFFKDMTEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDCIITAYRDH 121
Query: 126 CTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAF 185
C FL RGGTLVE F+ELMGR+DGCS+GKGGSMHFY+KE GFYGGHGIVGAQ+PLG+GLAF
Sbjct: 122 CIFLGRGGTLVEAFAELMGRRDGCSRGKGGSMHFYKKESGFYGGHGIVGAQVPLGIGLAF 181
Query: 186 GQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKS 245
QKY K+ VTF +YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKS
Sbjct: 182 AQKYKKEDYVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKS 241
Query: 246 PAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGST 305
PAYYKRGDY PGL+VDGMDV AVKQAC FAK+HALKNGP+ILEMDTYRYHGHSMSDPGST
Sbjct: 242 PAYYKRGDYVPGLRVDGMDVFAVKQACTFAKQHALKNGPIILEMDTYRYHGHSMSDPGST 301
Query: 306 YRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPD 365
YRTRDEISGVRQERDP+ER+R L+LAH+I+TE ELKDIEKE RK VDEAIAKAKES MPD
Sbjct: 302 YRTRDEISGVRQERDPVERIRSLILAHNIATEAELKDIEKENRKVVDEAIAKAKESPMPD 361
Query: 366 PSDLFTNVYVKGLGVEACGADRKEVKATLP 395
PS+LFTNVYVKG GVEA GADRKE++ATLP
Sbjct: 362 PSELFTNVYVKGFGVEAYGADRKELRATLP 391
>gb|AAG43499.1| pyruvate dehydrogenase [Lycopersicon esculentum]
Length = 391
Score = 645 bits (1665), Expect = 0.0
Identities = 313/390 (80%), Positives = 350/390 (89%), Gaps = 3/390 (0%)
Query: 7 SSSQFGSTLLKPYFLSSAI-RHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASEL 65
S+S+ + ++KP LS A+ R +SS ST T+TVETS+PFTSHN +PPS +V+T+ EL
Sbjct: 4 STSRAINHIMKP--LSRAVCATRRLSSDSTATITVETSLPFTSHNVDPPSRSVETSPMEL 61
Query: 66 MSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDH 125
M+FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI +KDC+ITAYRDH
Sbjct: 62 MTFFKDMTEMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDCIITAYRDH 121
Query: 126 CTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAF 185
C FL RGGTLVE F+ELMGR+DGCS+GKGGSMHFY+KE GFYGGHGIVGAQ+PLG+GLAF
Sbjct: 122 CIFLGRGGTLVESFAELMGRRDGCSRGKGGSMHFYKKESGFYGGHGIVGAQVPLGIGLAF 181
Query: 186 GQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKS 245
QKY K+ VTF +YGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKS
Sbjct: 182 AQKYKKEDYVTFAMYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKS 241
Query: 246 PAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGST 305
PAYYKRGDY PGL+VDGMDV AVKQAC FAK+HALKNGP+ILEMDTYRYHGHSMSDPGST
Sbjct: 242 PAYYKRGDYVPGLRVDGMDVFAVKQACAFAKQHALKNGPIILEMDTYRYHGHSMSDPGST 301
Query: 306 YRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPD 365
YRTRDEISGVRQERDP+ER+R L+LAH+I+TE ELKDIEKE RK VDEAI KAKES MPD
Sbjct: 302 YRTRDEISGVRQERDPVERIRSLILAHNIATEAELKDIEKENRKVVDEAIRKAKESPMPD 361
Query: 366 PSDLFTNVYVKGLGVEACGADRKEVKATLP 395
PS+LFTNVYVKG GVEA GADRKE++ATLP
Sbjct: 362 PSELFTNVYVKGFGVEAYGADRKELRATLP 391
>dbj|BAC57468.1| pyruvate dehydrogenase E1alpha subunit [Beta vulgaris]
Length = 395
Score = 634 bits (1635), Expect = e-180
Identities = 307/384 (79%), Positives = 346/384 (89%), Gaps = 3/384 (0%)
Query: 13 STLLKPYFLSSAIRH-RSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFND 71
S++L P L+S H R +SS ST+TLT+ETS+PF SH EPPS +V TT +ELM++F D
Sbjct: 14 SSVLLP--LTSTHTHSRRLSSDSTKTLTIETSVPFKSHIVEPPSRSVDTTPAELMTYFRD 71
Query: 72 MVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCR 131
M LMRRMEIA+DSLYKAKLIRGFCHLYDGQEAVAVGMEAAI +KD +ITAYRDHC +L R
Sbjct: 72 MALMRRMEIASDSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDNIITAYRDHCIYLAR 131
Query: 132 GGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNK 191
GG+L+ F+ELMGR+DGCS+GKGGSMHFY+K+ GFYGGHGIVGAQ+PLGVGLAF QKYNK
Sbjct: 132 GGSLLSAFAELMGRQDGCSRGKGGSMHFYKKDSGFYGGHGIVGAQVPLGVGLAFAQKYNK 191
Query: 192 DPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKR 251
+ V+F LYGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKSP+YYKR
Sbjct: 192 EDCVSFALYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPSYYKR 251
Query: 252 GDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDE 311
GDY PGLKVDGMDVLAVKQACKFAKE+ LKNGP+ILEMDTYRYHGHSMSDPGSTYRTRDE
Sbjct: 252 GDYVPGLKVDGMDVLAVKQACKFAKEYVLKNGPIILEMDTYRYHGHSMSDPGSTYRTRDE 311
Query: 312 ISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFT 371
ISG+RQERDPIERVRKL+LAHDI+ EKELKDIEKE RK+VDEAIAKAKES MPD S+LFT
Sbjct: 312 ISGIRQERDPIERVRKLLLAHDIAGEKELKDIEKEIRKEVDEAIAKAKESPMPDTSELFT 371
Query: 372 NVYVKGLGVEACGADRKEVKATLP 395
N+YVKG GVE+ GADRK ++ TLP
Sbjct: 372 NIYVKGYGVESFGADRKVLRTTLP 395
>dbj|BAC57469.1| pyruvate dehydrogenase E1 alpha subunit [Beta vulgaris]
Length = 395
Score = 630 bits (1626), Expect = e-179
Identities = 306/384 (79%), Positives = 345/384 (89%), Gaps = 3/384 (0%)
Query: 13 STLLKPYFLSSAIRH-RSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFND 71
S+LL P L+S H R +SS ST+TLT+ETS+PF SH EPPS +V TT +ELM++F D
Sbjct: 14 SSLLLP--LTSTHTHSRRLSSDSTKTLTIETSVPFKSHIVEPPSRSVDTTPAELMTYFRD 71
Query: 72 MVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCR 131
M LMRRMEIA+DSLYKAKLIRGFCHLYDGQEAVAVGMEAAI +KD +ITAYRDHC +L R
Sbjct: 72 MALMRRMEIASDSLYKAKLIRGFCHLYDGQEAVAVGMEAAITKKDNIITAYRDHCIYLAR 131
Query: 132 GGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNK 191
GG+L+ F+EL+GR+DGCS+GKGGSMHFY+K+ GFYGGHGIVGAQ+PLGVGLAF QKYNK
Sbjct: 132 GGSLLSAFAELVGRQDGCSRGKGGSMHFYKKDSGFYGGHGIVGAQVPLGVGLAFAQKYNK 191
Query: 192 DPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKR 251
+ V+F LYGDGAANQGQLFEALN+AALWDLPAILVCENNHYGMGTAEWR+AKSP+YYKR
Sbjct: 192 EDCVSFALYGDGAANQGQLFEALNMAALWDLPAILVCENNHYGMGTAEWRAAKSPSYYKR 251
Query: 252 GDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDE 311
GDY PGLKVDGMDVLAVKQACK AKE+ LKNGP+ILEMDTYRYHGHSMSDPGSTYRTRDE
Sbjct: 252 GDYVPGLKVDGMDVLAVKQACKSAKEYVLKNGPIILEMDTYRYHGHSMSDPGSTYRTRDE 311
Query: 312 ISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFT 371
ISG+RQERDPIERVRKL+LAHDI+ EKELKDIEKE RK+VDEAIAKAKES MPD S+LFT
Sbjct: 312 ISGIRQERDPIERVRKLLLAHDIAGEKELKDIEKEIRKEVDEAIAKAKESPMPDTSELFT 371
Query: 372 NVYVKGLGVEACGADRKEVKATLP 395
N+YVKG GVE+ GADRK ++ TLP
Sbjct: 372 NIYVKGYGVESFGADRKVLRTTLP 395
>gb|AAM65647.1| pyruvate dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|15293169|gb|AAK93695.1| putative pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|13430606|gb|AAK25925.1| putative pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|15221692|ref|NP_173828.1| pyruvate dehydrogenase E1
component alpha subunit, mitochondrial, putative
[Arabidopsis thaliana] gi|7431563|pir||T00648 pyruvate
dehydrogenase (lipoamide) (EC 1.2.4.1) E1 alpha chain -
Arabidopsis thaliana gi|2829869|gb|AAC00577.1| pyruvate
dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
Length = 393
Score = 627 bits (1617), Expect = e-178
Identities = 309/397 (77%), Positives = 348/397 (86%), Gaps = 6/397 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYF--LSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTV 58
MALSR SS +T LKP L S+IR R +S+ S+ +T+ET++PFTSH CE PS +V
Sbjct: 1 MALSRLSSRS--NTFLKPAITALPSSIR-RHVSTDSSP-ITIETAVPFTSHLCESPSRSV 56
Query: 59 QTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCV 118
+T++ E+++FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEA+AVGMEAAI +KD +
Sbjct: 57 ETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITKKDAI 116
Query: 119 ITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIP 178
IT+YRDHCTF+ RGG LV+ FSELMGRK GCS GKGGSMHFY+K+ FYGGHGIVGAQIP
Sbjct: 117 ITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVGAQIP 176
Query: 179 LGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTA 238
LG GLAF QKYNKD VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTA
Sbjct: 177 LGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTA 236
Query: 239 EWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHS 298
WRSAKSPAY+KRGDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGHS
Sbjct: 237 TWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGHS 296
Query: 299 MSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKA 358
MSDPGSTYRTRDEISGVRQ RDPIERVRKL+L HDI+TEKELKD+EKE RK+VD+A+A+A
Sbjct: 297 MSDPGSTYRTRDEISGVRQVRDPIERVRKLLLTHDIATEKELKDMEKEIRKEVDDAVAQA 356
Query: 359 KESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
KES +PD S+LFTN+YVK GVE+ GADRKE+K TLP
Sbjct: 357 KESPIPDASELFTNMYVKDCGVESFGADRKELKVTLP 393
>gb|AAK26016.1| putative pyruvate dehydrogenase e1 alpha subunit [Arabidopsis
thaliana]
Length = 389
Score = 626 bits (1614), Expect = e-178
Identities = 303/395 (76%), Positives = 347/395 (87%), Gaps = 6/395 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR SS + + +P+ SA R IS+ +T +T+ETS+PFT+H C+PPS +V++
Sbjct: 1 MALSRLSSRS--NIITRPF---SAAFSRLISTDTTP-ITIETSLPFTAHLCDPPSRSVES 54
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
++ EL+ FF M LMRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEAAI +KD +IT
Sbjct: 55 SSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKDAIIT 114
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHC FL RGG+L EVFSELMGR+ GCSKGKGGSMHFY+KE FYGGHGIVGAQ+PLG
Sbjct: 115 AYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLG 174
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
G+AF QKYNK+ VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTAEW
Sbjct: 175 CGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEW 234
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
R+AKSP+YYKRGDY PGLKVDGMD AVKQACKFAK+HAL+ GP+ILEMDTYRYHGHSMS
Sbjct: 235 RAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGPIILEMDTYRYHGHSMS 294
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIER++KLVL+HD++TEKELKD+EKE RK+VD+AIAKAK+
Sbjct: 295 DPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRKEVDDAIAKAKD 354
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
MP+PSDLFTNVYVKG G E+ G DRKEVKA+LP
Sbjct: 355 CPMPEPSDLFTNVYVKGFGTESFGPDRKEVKASLP 389
>gb|AAD39331.1| pyruvate dehydrogenase E1 alpha subunit [Arabidopsis thaliana]
gi|24030439|gb|AAN41374.1| putative pyruvate
dehydrogenase e1 alpha subunit [Arabidopsis thaliana]
gi|21593256|gb|AAM65205.1| pyruvate dehydrogenase e1
alpha subunit, putative [Arabidopsis thaliana]
gi|15218940|ref|NP_176198.1| pyruvate dehydrogenase E1
component alpha subunit, mitochondrial (PDHE1-A)
[Arabidopsis thaliana] gi|25284401|pir||B96623 pyruvate
dehydrogenase E1 alpha subunit [imported] - Arabidopsis
thaliana gi|27735220|sp|P52901|ODPA_ARATH Pyruvate
dehydrogenase E1 component alpha subunit, mitochondrial
precursor (PDHE1-A)
Length = 389
Score = 624 bits (1610), Expect = e-177
Identities = 302/395 (76%), Positives = 347/395 (87%), Gaps = 6/395 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR SS + + +P+ SA R IS+ +T +T+ETS+PFT+H C+PPS +V++
Sbjct: 1 MALSRLSSRS--NIITRPF---SAAFSRLISTDTTP-ITIETSLPFTAHLCDPPSRSVES 54
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
++ EL+ FF M LMRRMEIAADSLYKAKLIRGFCHLYDGQEAVA+GMEAAI +KD +IT
Sbjct: 55 SSQELLDFFRTMALMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAIGMEAAITKKDAIIT 114
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHC FL RGG+L EVFSELMGR+ GCSKGKGGSMHFY+KE FYGGHGIVGAQ+PLG
Sbjct: 115 AYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLG 174
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
G+AF QKYNK+ VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTAEW
Sbjct: 175 CGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEW 234
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
R+AKSP+YYKRGDY PGLKVDGMD AVKQACKFAK+HAL+ GP+ILEMDTYRYHGHSMS
Sbjct: 235 RAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGPIILEMDTYRYHGHSMS 294
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIER++KLVL+HD++TEKELKD+EKE RK+VD+AIAKAK+
Sbjct: 295 DPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRKEVDDAIAKAKD 354
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
MP+PS+LFTNVYVKG G E+ G DRKEVKA+LP
Sbjct: 355 CPMPEPSELFTNVYVKGFGTESFGPDRKEVKASLP 389
>gb|AAN15218.1| pyruvate dehydrogenase E1a-like subunit IAR4 [Arabidopsis thaliana]
Length = 393
Score = 623 bits (1607), Expect = e-177
Identities = 308/397 (77%), Positives = 347/397 (86%), Gaps = 6/397 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYF--LSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTV 58
MALSR SS +T LKP L S+IR R +S+ S+ +T+ET++PFTSH CE PS +V
Sbjct: 1 MALSRLSSRS--NTFLKPAITALPSSIR-RHVSTDSSP-ITIETAVPFTSHLCESPSRSV 56
Query: 59 QTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCV 118
+T++ E+++FF DM MRRMEIAADSLYKAKLIRGFCHLYDGQEA+AVGMEAAI +KD +
Sbjct: 57 ETSSEEILAFFRDMARMRRMEIAADSLYKAKLIRGFCHLYDGQEALAVGMEAAITKKDAI 116
Query: 119 ITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIP 178
IT+YRDHCTF+ RGG LV+ FSELMGRK GCS GKGGSMHFY+K+ FYGGHGIVGAQIP
Sbjct: 117 ITSYRDHCTFIGRGGKLVDAFSELMGRKTGCSHGKGGSMHFYKKDASFYGGHGIVGAQIP 176
Query: 179 LGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTA 238
LG GLAF QKYNKD VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNH GMGTA
Sbjct: 177 LGCGLAFAQKYNKDEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHDGMGTA 236
Query: 239 EWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHS 298
WRSAKSPAY+KRGDY PGLKVDGMD LAVKQACKFAKEHALKNGP+ILEMDTYRYHGHS
Sbjct: 237 TWRSAKSPAYFKRGDYVPGLKVDGMDALAVKQACKFAKEHALKNGPIILEMDTYRYHGHS 296
Query: 299 MSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKA 358
MSDPGSTYRTRDEISGVRQ RDPIERVRKL+L HDI+TEKELKD+EKE RK+VD+A+A+A
Sbjct: 297 MSDPGSTYRTRDEISGVRQVRDPIERVRKLLLTHDIATEKELKDMEKEIRKEVDDAVAQA 356
Query: 359 KESQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
KES +PD S+LFTN+YVK GVE+ GADRKE+K TLP
Sbjct: 357 KESPIPDASELFTNMYVKDCGVESFGADRKELKVTLP 393
>gb|AAA86507.1| pyruvate dehydrogenase E1 alpha subunit gi|2117533|pir||JC4358
pyruvate dehydrogenase (lipoamide) (EC 1.2.4.1) alpha
chain precursor - Arabidopsis thaliana
Length = 389
Score = 621 bits (1602), Expect = e-176
Identities = 300/395 (75%), Positives = 346/395 (86%), Gaps = 6/395 (1%)
Query: 1 MALSRFSSSQFGSTLLKPYFLSSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQT 60
MALSR SS + + +P+ SA R IS+ +T +T+ETS+PFT+H C+PPS +V++
Sbjct: 1 MALSRLSSRS--NIITRPF---SAAFSRLISTDTTP-ITIETSLPFTAHLCDPPSRSVES 54
Query: 61 TASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVIT 120
++ EL+ FF M LMRRMEIAADSLYKA +IRGFCHLYDGQEAVA+GMEAAI +KD +IT
Sbjct: 55 SSQELLDFFRTMALMRRMEIAADSLYKANVIRGFCHLYDGQEAVAIGMEAAITKKDAIIT 114
Query: 121 AYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLG 180
AYRDHC FL RGG+L EVFSELMGR+ GCSKGKGGSMHFY+KE FYGGHGIVGAQ+PLG
Sbjct: 115 AYRDHCIFLGRGGSLHEVFSELMGRQAGCSKGKGGSMHFYKKESSFYGGHGIVGAQVPLG 174
Query: 181 VGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEW 240
G+AF QKYNK+ VTF LYGDGAANQGQLFEALNI+ALWDLPAILVCENNHYGMGTAEW
Sbjct: 175 CGIAFAQKYNKEEAVTFALYGDGAANQGQLFEALNISALWDLPAILVCENNHYGMGTAEW 234
Query: 241 RSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMS 300
R+AKSP+YYKRGDY PGLKVDGMD AVKQACKFAK+HAL+ GP+ILEMDTYRYHGHSMS
Sbjct: 235 RAAKSPSYYKRGDYVPGLKVDGMDAFAVKQACKFAKQHALEKGPIILEMDTYRYHGHSMS 294
Query: 301 DPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKE 360
DPGSTYRTRDEISGVRQERDPIER++KLVL+HD++TEKELKD+EKE RK+VD+AIAKAK+
Sbjct: 295 DPGSTYRTRDEISGVRQERDPIERIKKLVLSHDLATEKELKDMEKEIRKEVDDAIAKAKD 354
Query: 361 SQMPDPSDLFTNVYVKGLGVEACGADRKEVKATLP 395
MP+PS+LFTNVYVKG G E+ G DRKEVKA+LP
Sbjct: 355 CPMPEPSELFTNVYVKGFGTESFGPDRKEVKASLP 389
>ref|XP_467697.1| putative pyruvate dehydrogenase E1 alpha subunit [Oryza sativa
(japonica cultivar-group)] gi|51964350|ref|XP_506960.1|
PREDICTED P0684F11.25 gene product [Oryza sativa
(japonica cultivar-group)] gi|46390562|dbj|BAD16048.1|
putative pyruvate dehydrogenase E1 alpha subunit [Oryza
sativa (japonica cultivar-group)]
Length = 390
Score = 620 bits (1600), Expect = e-176
Identities = 299/374 (79%), Positives = 332/374 (87%), Gaps = 1/374 (0%)
Query: 22 SSAIRHRSISSSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIA 81
++ I RSIS S T LT+ETS+PFTSH +PPS V TT +EL++FF DM +MRRMEIA
Sbjct: 18 TALIAARSISDS-TAPLTIETSVPFTSHIVDPPSRDVTTTPAELLTFFRDMSVMRRMEIA 76
Query: 82 ADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSE 141
ADSLYKAKLIRGFCHLYDGQEAVAVGMEAAI R D +ITAYRDHCT+L RGG LV F+E
Sbjct: 77 ADSLYKAKLIRGFCHLYDGQEAVAVGMEAAITRSDSIITAYRDHCTYLARGGDLVSAFAE 136
Query: 142 LMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYG 201
LMGR+ GCS+GKGGSMHFY+K+ FYGGHGIVGAQ+PLG GLAF QKY K+ TF LYG
Sbjct: 137 LMGRQAGCSRGKGGSMHFYKKDANFYGGHGIVGAQVPLGCGLAFAQKYRKEETATFALYG 196
Query: 202 DGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVD 261
DGAANQGQLFEALNI+ALW LPAILVCENNHYGMGTAEWR+AKSPAYYKRGDY PGLKVD
Sbjct: 197 DGAANQGQLFEALNISALWKLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVD 256
Query: 262 GMDVLAVKQACKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDP 321
GMDVLAVKQACKFAKEHA+ NGP++LEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDP
Sbjct: 257 GMDVLAVKQACKFAKEHAIANGPIVLEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDP 316
Query: 322 IERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVE 381
IERVRKL+LAHD++T ELKD+EKE RK+VD+AIAKAKES MPD S+LFTNVYVKG GVE
Sbjct: 317 IERVRKLILAHDLATAAELKDMEKEIRKEVDDAIAKAKESPMPDTSELFTNVYVKGFGVE 376
Query: 382 ACGADRKEVKATLP 395
+ GADRKE++ATLP
Sbjct: 377 SFGADRKELRATLP 390
>gb|AAC72195.1| pyruvate dehydrogenase E1 alpha subunit [Zea mays]
Length = 392
Score = 610 bits (1572), Expect = e-173
Identities = 291/364 (79%), Positives = 322/364 (87%)
Query: 32 SSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLI 91
S ST LT+ETS+PFTSH +PPS V TT +EL++FF DM LMRRMEIAADSLYKAKLI
Sbjct: 29 SDSTAALTIETSVPFTSHLVDPPSRDVTTTPAELVTFFRDMSLMRRMEIAADSLYKAKLI 88
Query: 92 RGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSK 151
RGFCHLYDGQEAVAVGMEAAI R D +ITAYRDHCT+L RGG LV FSELMGR+ GCS+
Sbjct: 89 RGFCHLYDGQEAVAVGMEAAITRSDSIITAYRDHCTYLARGGDLVSAFSELMGREAGCSR 148
Query: 152 GKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLF 211
GKGGSMHFY+K+ FYGGHGIVGAQ+PLG GLAF QKY K+ TF LYGDGAANQGQLF
Sbjct: 149 GKGGSMHFYKKDANFYGGHGIVGAQVPLGCGLAFAQKYKKEDTATFALYGDGAANQGQLF 208
Query: 212 EALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQA 271
EALNI+ALW LPAILVCENNHYGMGTAEWR+AKSPAYYKRGDY PGLKVDGMDVLAVKQA
Sbjct: 209 EALNISALWKLPAILVCENNHYGMGTAEWRAAKSPAYYKRGDYVPGLKVDGMDVLAVKQA 268
Query: 272 CKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
CKFAK+HA+ NGP++LEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKL+L
Sbjct: 269 CKFAKDHAVANGPIVLEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLLLT 328
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADRKEVK 391
HD++ ELK++EKE RKQVD+AIAKAKES MPD S+LFTNVY KG GVE+ G DRKE++
Sbjct: 329 HDLANAAELKNMEKEIRKQVDDAIAKAKESSMPDTSELFTNVYKKGFGVESFGPDRKEMR 388
Query: 392 ATLP 395
A+LP
Sbjct: 389 ASLP 392
>dbj|BAD45661.1| putative pyruvate dehydrogenase E1 alpha subunit [Oryza sativa
(japonica cultivar-group)]
Length = 398
Score = 591 bits (1524), Expect = e-167
Identities = 281/364 (77%), Positives = 314/364 (86%)
Query: 32 SSSTETLTVETSIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLI 91
S STE LT+ETS+P+ SH +PP V TTA EL +FF DM MRR EIAADSLYKAKLI
Sbjct: 35 SDSTEPLTIETSVPYKSHIVDPPPREVATTARELATFFRDMSAMRRAEIAADSLYKAKLI 94
Query: 92 RGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSK 151
RGFCHLYDGQEAVAVGMEAA R D +ITAYRDHC +L RGG L +F+ELMGR+ GCS+
Sbjct: 95 RGFCHLYDGQEAVAVGMEAATTRADAIITAYRDHCAYLARGGDLAALFAELMGRRGGCSR 154
Query: 152 GKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLF 211
GKGGSMH Y+K+ FYGGHGIVGAQ+PLG GLAF Q+Y K+ VTF LYGDGAANQGQLF
Sbjct: 155 GKGGSMHLYKKDANFYGGHGIVGAQVPLGCGLAFAQRYRKEAAVTFDLYGDGAANQGQLF 214
Query: 212 EALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQA 271
EALN+AALW LP +LVCENNHYGMGTAEWR++KSPAYYKRGDY PGLKVDGMDVLAVKQA
Sbjct: 215 EALNMAALWKLPVVLVCENNHYGMGTAEWRASKSPAYYKRGDYVPGLKVDGMDVLAVKQA 274
Query: 272 CKFAKEHALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
CKFAK+HAL+NGP+ILEMDTYRYHGHSMSDPGSTYRTRDEI+G+RQERDPIERVRKL+LA
Sbjct: 275 CKFAKQHALENGPIILEMDTYRYHGHSMSDPGSTYRTRDEIAGIRQERDPIERVRKLLLA 334
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQMPDPSDLFTNVYVKGLGVEACGADRKEVK 391
HD +T +ELKD+EKE RKQVD AIAKAKES MPDPS+LFTNVYV G+E+ G DRK V+
Sbjct: 335 HDFATTQELKDMEKEIRKQVDTAIAKAKESPMPDPSELFTNVYVNDCGLESFGVDRKVVR 394
Query: 392 ATLP 395
LP
Sbjct: 395 TVLP 398
>ref|XP_629349.1| pyruvate dehydrogenase E1 subunit alpha [Dictyostelium discoideum]
gi|60462647|gb|EAL60849.1| pyruvate dehydrogenase E1
alpha subunit [Dictyostelium discoideum]
Length = 377
Score = 392 bits (1006), Expect = e-107
Identities = 193/331 (58%), Positives = 241/331 (72%), Gaps = 3/331 (0%)
Query: 51 CEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFCHLYDGQEAVAVGMEA 110
C+ PS + T EL+SFF +M RR+E D LYK KLIRGFCHLY GQEAV G+E+
Sbjct: 38 CDGPSDSTVTNKDELISFFTEMSRFRRLETVCDGLYKKKLIRGFCHLYTGQEAVCAGLES 97
Query: 111 AINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGGSMHFYRKEGGFYGGH 170
AI + D +ITAYRDH L RG T E+F+EL+ ++ GCSKGKGGSMH + K FYGG+
Sbjct: 98 AITKDDHIITAYRDHTYMLSRGATPEEIFAELLMKETGCSKGKGGSMHMFTKN--FYGGN 155
Query: 171 GIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALNIAALWDLPAILVCEN 230
GIVGAQ PLG G+AF QKYNK NV +YGDGAANQGQLFEA N+A+LW LP I +CEN
Sbjct: 156 GIVGAQCPLGAGIAFAQKYNKTGNVCLAMYGDGAANQGQLFEAFNMASLWKLPVIFICEN 215
Query: 231 NHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFAKEHA-LKNGPLILEM 289
N YGMGT++ RS +Y RG Y GLKVDGMDV AVK+A K+A E NGP+ILEM
Sbjct: 216 NKYGMGTSQKRSTAGHDFYTRGHYVAGLKVDGMDVFAVKEAGKYAAEWCRAGNGPIILEM 275
Query: 290 DTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDISTEKELKDIEKEARK 349
DTYRY GHSMSDPG TYRTR+E++ VRQ RDPIE +R+++L + I+TE +L IE+ R
Sbjct: 276 DTYRYVGHSMSDPGITYRTREEVNHVRQTRDPIENIRQIILDNKIATEDQLAAIEETVRD 335
Query: 350 QVDEAIAKAKESQMPDPSDLFTNVYVKGLGV 380
++++A KA + +P +LFTNVY++ + V
Sbjct: 336 EMEKASEKAIAAPLPQARELFTNVYLQEVPV 366
>emb|CAG90582.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50426983|ref|XP_462096.1| unnamed protein product
[Debaryomyces hansenii]
Length = 398
Score = 386 bits (992), Expect = e-106
Identities = 194/354 (54%), Positives = 254/354 (70%), Gaps = 6/354 (1%)
Query: 28 RSISSSSTETLTVET-SIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLY 86
R+++S+S++ ++++ F +N E P + +T L+ + DM+++RRME+A+D+LY
Sbjct: 20 RTMASASSDLVSIKLPESSFEGYNLEIPELSFETEKDTLLQMYKDMIIIRRMEMASDALY 79
Query: 87 KAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRK 146
KAK IRGFCHL GQEAVAVG+EAAIN+KD VIT+YR H RG ++ EV ELMG++
Sbjct: 80 KAKKIRGFCHLSIGQEAVAVGIEAAINKKDSVITSYRCHGFTYMRGASVKEVLGELMGKR 139
Query: 147 DGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAAN 206
G S GKGGSMH Y + GFYGG+GIVGAQ+PLG GLAF KY + N TF LYGDGA+N
Sbjct: 140 SGVSYGKGGSMHMYAQ--GFYGGNGIVGAQVPLGAGLAFAHKYRGEGNCTFNLYGDGASN 197
Query: 207 QGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVL 266
QGQ+FE+ N+A LWDLP + CENN YGMGT+ RS+ YYKRG Y PGLKV+GMD+L
Sbjct: 198 QGQVFESYNMAKLWDLPCVFACENNKYGMGTSASRSSAMTEYYKRGQYIPGLKVNGMDIL 257
Query: 267 AVKQACKFAKEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERV 325
A QA KFAK+ NGPL+LE +TYRY GHSMSDPG+TYRTR+E+ +R DPI +
Sbjct: 258 ACYQASKFAKDWCTSGNGPLVLEYETYRYGGHSMSDPGTTYRTREEVQHMRSRNDPIAGL 317
Query: 326 RKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
+ +L DI+TE+E+K +K ARK VDE +A+A+ P+ LF +VYV G
Sbjct: 318 KATLLELDIATEEEIKSYDKAARKYVDEQVAEAEADAPPEAKMDILFEDVYVPG 371
>gb|EAA75271.1| conserved hypothetical protein [Gibberella zeae PH-1]
gi|46122153|ref|XP_385630.1| conserved hypothetical
protein [Gibberella zeae PH-1]
Length = 409
Score = 379 bits (974), Expect = e-104
Identities = 195/360 (54%), Positives = 253/360 (70%), Gaps = 6/360 (1%)
Query: 22 SSAIRHRSISSSSTETLTVETSIP-FTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEI 80
+SA S+ + E +V S F ++ +PP T++ T EL + +MV+ R+ME+
Sbjct: 34 ASASLSHSVPKADDEPFSVNLSDESFETYELDPPPYTLEVTKKELKDMYREMVVTRQMEM 93
Query: 81 AADSLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFS 140
AAD LYK K IRGFCHL GQEAVAVG+E AI ++D +ITAYR H L RG ++ +
Sbjct: 94 AADRLYKEKKIRGFCHLSTGQEAVAVGIEHAITKEDDIITAYRCHGYALLRGASVRSIIG 153
Query: 141 ELMGRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLY 200
EL+GR++G S GKGGSMH + K GFYGG+GIVGAQ+P+G GLAF KYN + N + LY
Sbjct: 154 ELLGRREGISYGKGGSMHMFAK--GFYGGNGIVGAQVPVGAGLAFAHKYNNNKNASIILY 211
Query: 201 GDGAANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKV 260
GDGA+NQGQ+FEA N+A LW+LPA+ CENN YGMGTA RS+ YYKRG Y PGLKV
Sbjct: 212 GDGASNQGQVFEAFNMAKLWNLPALFGCENNKYGMGTAAARSSALTDYYKRGQYIPGLKV 271
Query: 261 DGMDVLAVKQACKFAKEH-ALKNGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQER 319
+GMDVLAVK A K+ KE+ A GPL+LE TYRY GHSMSDPG+TYRTR+EI +R
Sbjct: 272 NGMDVLAVKAAVKYGKEYTAADKGPLVLEYVTYRYGGHSMSDPGTTYRTREEIQRMRSTN 331
Query: 320 DPIERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPD--PSDLFTNVYVKG 377
DPI +++ +L +I+TE+ELK I+KEAR V+E +A A+ P+ P LF ++YV+G
Sbjct: 332 DPIAGLKQKILDWEITTEEELKKIDKEARAHVNEEVAAAEAMAAPEAKPEILFEDIYVRG 391
>gb|EAK96452.1| hypothetical protein CaO19.10609 [Candida albicans SC5314]
gi|46437028|gb|EAK96381.1| hypothetical protein
CaO19.3097 [Candida albicans SC5314]
Length = 401
Score = 377 bits (967), Expect = e-103
Identities = 189/357 (52%), Positives = 252/357 (69%), Gaps = 6/357 (1%)
Query: 25 IRHRSISSSSTETLTVET-SIPFTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAAD 83
+ RS++ ++++ +T+E + + +N E P+ + +T L+ + DM+++RRME+AAD
Sbjct: 20 VAKRSMAKAASDLVTIELPASSYEGYNLEVPALSFETEKETLLKMYKDMIIIRRMEMAAD 79
Query: 84 SLYKAKLIRGFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELM 143
+LYK+K IRGFCHL GQEA+AVG+E AI D VIT+YR H RG ++ V +ELM
Sbjct: 80 ALYKSKKIRGFCHLSVGQEAIAVGIENAITPTDTVITSYRCHGFAFMRGASVKSVLAELM 139
Query: 144 GRKDGCSKGKGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDG 203
GR+ G + GKGGSMH + GFYGG+GIVGAQ+PLG GLAF KY D VTF LYGDG
Sbjct: 140 GRRSGIANGKGGSMHMFTN--GFYGGNGIVGAQVPLGAGLAFSHKYKNDKAVTFDLYGDG 197
Query: 204 AANQGQLFEALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGM 263
A+NQGQ+FEA N+A LW+LP I CENN YGMGT+ RS+ YYKRG Y PGLK++GM
Sbjct: 198 ASNQGQVFEAYNMAKLWNLPVIFACENNKYGMGTSAARSSAMTEYYKRGQYIPGLKINGM 257
Query: 264 DVLAVKQACKFAKEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPI 322
DVLA QA KFAK+ A + NGPL+LE +TYRY GHSMSDPG+TYRTR+E+ +R DPI
Sbjct: 258 DVLATYQASKFAKDWASQGNGPLVLEYETYRYGGHSMSDPGTTYRTREEVQHMRSRNDPI 317
Query: 323 ERVRKLVLAHDISTEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
++ ++L +I++E E+K +K ARK VDE +A A+ P+ LF +VYV G
Sbjct: 318 AGLKAVLLEKEIASEDEIKSYDKAARKYVDEQVAAAEADAPPEAKMDILFEDVYVPG 374
>emb|CAG62267.1| unnamed protein product [Candida glabrata CBS138]
gi|50293763|ref|XP_449293.1| unnamed protein product
[Candida glabrata]
Length = 408
Score = 375 bits (963), Expect = e-102
Identities = 195/348 (56%), Positives = 245/348 (70%), Gaps = 8/348 (2%)
Query: 36 ETLTVETSIPFTS---HNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIR 92
E VE S+P TS + EPPS T L+ F DMV++RRME+A D+LYKAK IR
Sbjct: 37 ENENVEISLPETSFEGYMLEPPSLNYSATKGSLLQMFKDMVIIRRMEMACDALYKAKKIR 96
Query: 93 GFCHLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKG 152
GFCHL GQEA+AVG+E AI ++D VIT+YR H RG ++ V +ELMG++ G S G
Sbjct: 97 GFCHLSVGQEAIAVGIENAITKRDSVITSYRCHGFTYMRGASVQAVLAELMGKRSGVSFG 156
Query: 153 KGGSMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFE 212
KGGSMH + GFYGG+GIVGAQ+P+G GLAF +Y + +FTLYGDGA+NQGQ+FE
Sbjct: 157 KGGSMHLFAP--GFYGGNGIVGAQVPVGAGLAFAHQYKNEDACSFTLYGDGASNQGQVFE 214
Query: 213 ALNIAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQAC 272
+ N+A LW+LP + CENN YGMGTA RS+ Y+KRG Y PGLKV+GMD+LAV QA
Sbjct: 215 SFNMAKLWNLPVVFCCENNKYGMGTAAARSSAMTEYFKRGQYIPGLKVNGMDILAVYQAS 274
Query: 273 KFAKEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLA 331
KFAKE L GPL+LE +TYRY GHSMSDPG+TYRTRDEI +R + DPI ++ +L
Sbjct: 275 KFAKEWCLSGKGPLVLEYETYRYGGHSMSDPGTTYRTRDEIQHMRSKNDPIAGLKMHLLE 334
Query: 332 HDISTEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
I+TE+E+K +K ARK VDE + A +S P+ S LF +VYVKG
Sbjct: 335 LGIATEEEVKAYDKAARKYVDEQVELADKSAPPEAKLSILFEDVYVKG 382
>emb|CAA50657.1| PDA1 [Saccharomyces cerevisiae] gi|37362644|ref|NP_011105.2| E1
alpha subunit of the pyruvate dehydrogenase (PDH)
complex, catalyzes the direct oxidative decarboxylation
of pyruvate to acetyl-CoA, regulated by glucose; Pda1p
[Saccharomyces cerevisiae]
gi|730222|sp|P16387|ODPA_YEAST Pyruvate dehydrogenase E1
component alpha subunit, mitochondrial precursor
(PDHE1-A)
Length = 420
Score = 371 bits (952), Expect = e-101
Identities = 190/345 (55%), Positives = 244/345 (70%), Gaps = 8/345 (2%)
Query: 39 TVETSIP---FTSHNCEPPSTTVQTTASELMSFFNDMVLMRRMEIAADSLYKAKLIRGFC 95
TV+ +P F S+ EPP + +T+ + L+ + DMV++RRME+A D+LYKAK IRGFC
Sbjct: 52 TVQIELPESSFESYMLEPPDLSYETSKATLLQMYKDMVIIRRMEMACDALYKAKKIRGFC 111
Query: 96 HLYDGQEAVAVGMEAAINRKDCVITAYRDHCTFLCRGGTLVEVFSELMGRKDGCSKGKGG 155
HL GQEA+AVG+E AI + D +IT+YR H RG ++ V +ELMGR+ G S GKGG
Sbjct: 112 HLSVGQEAIAVGIENAITKLDSIITSYRCHGFTFMRGASVKAVLAELMGRRAGVSYGKGG 171
Query: 156 SMHFYRKEGGFYGGHGIVGAQIPLGVGLAFGQKYNKDPNVTFTLYGDGAANQGQLFEALN 215
SMH Y GFYGG+GIVGAQ+PLG GLAF +Y + +FTLYGDGA+NQGQ+FE+ N
Sbjct: 172 SMHLYAP--GFYGGNGIVGAQVPLGAGLAFAHQYKNEDACSFTLYGDGASNQGQVFESFN 229
Query: 216 IAALWDLPAILVCENNHYGMGTAEWRSAKSPAYYKRGDYAPGLKVDGMDVLAVKQACKFA 275
+A LW+LP + CENN YGMGTA RS+ Y+KRG Y PGLKV+GMD+LAV QA KFA
Sbjct: 230 MAKLWNLPVVFCCENNKYGMGTAASRSSAMTEYFKRGQYIPGLKVNGMDILAVYQASKFA 289
Query: 276 KEHALK-NGPLILEMDTYRYHGHSMSDPGSTYRTRDEISGVRQERDPIERVRKLVLAHDI 334
K+ L GPL+LE +TYRY GHSMSDPG+TYRTRDEI +R + DPI ++ ++ I
Sbjct: 290 KDWCLSGKGPLVLEYETYRYGGHSMSDPGTTYRTRDEIQHMRSKNDPIAGLKMHLIDLGI 349
Query: 335 STEKELKDIEKEARKQVDEAIAKAKESQMPDP--SDLFTNVYVKG 377
+TE E+K +K ARK VDE + A + P+ S LF +VYVKG
Sbjct: 350 ATEAEVKAYDKSARKYVDEQVELADAAPPPEAKLSILFEDVYVKG 394
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 679,340,883
Number of Sequences: 2540612
Number of extensions: 29693019
Number of successful extensions: 83332
Number of sequences better than 10.0: 885
Number of HSP's better than 10.0 without gapping: 639
Number of HSP's successfully gapped in prelim test: 246
Number of HSP's that attempted gapping in prelim test: 81381
Number of HSP's gapped (non-prelim): 958
length of query: 395
length of database: 863,360,394
effective HSP length: 130
effective length of query: 265
effective length of database: 533,080,834
effective search space: 141266421010
effective search space used: 141266421010
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146588.4


