

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146585.14 - phase: 0
(891 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
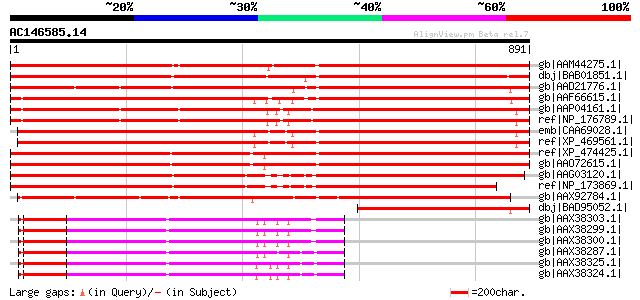
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM44275.1| receptor-like kinase RHG4 [Glycine max] gi|265185... 1168 0.0
dbj|BAB01851.1| unnamed protein product [Arabidopsis thaliana] g... 993 0.0
gb|AAD21776.1| putative receptor-like protein kinase [Arabidopsi... 858 0.0
gb|AAF66615.1| LRR receptor-like protein kinase [Nicotiana tabacum] 843 0.0
gb|AAP04161.1| putative receptor protein kinase (TMK1) [Arabidop... 841 0.0
ref|NP_176789.1| leucine-rich repeat protein kinase, putative (T... 840 0.0
emb|CAA69028.1| TMK [Oryza sativa (indica cultivar-group)] gi|74... 832 0.0
ref|XP_469561.1| gibberellin-induced receptor-like kinase TMK [O... 829 0.0
ref|XP_474425.1| OSJNBa0088H09.21 [Oryza sativa (japonica cultiv... 745 0.0
gb|AAO72615.1| receptor-like protein kinase-like protein [Oryza ... 742 0.0
gb|AAG03120.1| F5A9.23 [Arabidopsis thaliana] 739 0.0
ref|NP_173869.1| leucine-rich repeat family protein / protein ki... 738 0.0
gb|AAX92784.1| receptor-like kinase RHG4 [Oryza sativa (japonica... 730 0.0
dbj|BAD95052.1| putative receptor-like protein kinase [Arabidops... 375 e-102
gb|AAX38303.1| receptor-like protein kinase [Solanum habrochaites] 338 4e-91
gb|AAX38299.1| receptor-like protein kinase [Solanum habrochaite... 338 4e-91
gb|AAX38300.1| receptor-like protein kinase [Solanum habrochaites] 338 5e-91
gb|AAX38287.1| receptor-like protein kinase [Lycopersicon peruvi... 337 8e-91
gb|AAX38325.1| receptor-like protein kinase [Lycopersicon pimpin... 337 1e-90
gb|AAX38324.1| receptor-like protein kinase [Lycopersicon pimpin... 337 1e-90
>gb|AAM44275.1| receptor-like kinase RHG4 [Glycine max] gi|26518502|gb|AAN80746.1|
receptor-like kinase RHG4 [Glycine max]
Length = 893
Score = 1168 bits (3021), Expect = 0.0
Identities = 601/902 (66%), Positives = 695/902 (76%), Gaps = 20/902 (2%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
M + +SL P P+GWS T FCQW G++C S + VTSI+L+ Q L GTLP +LNSL+QL
Sbjct: 1 MSNFLKSLTPPPSGWSETTPFCQWKGIQCDSSSHVTSISLASQSLTGTLPSDLNSLSQLR 60
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
TL LQ+N+L+G LPSL+NLS L V NNFSSV+P AF+ L SLQTLSLG N L PW
Sbjct: 61 TLSLQDNSLTGTLPSLSNLSFLQTVYFNRNNFSSVSPTAFASLTSLQTLSLGSNPALQPW 120
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSL-AGSGI 179
+FPT+LT SSNL +D+ + G LPDIF F SL L L+YNNL+G LP+S A + +
Sbjct: 121 SFPTDLTSSSNLIDLDLATVSLTGPLPDIFDKFPSLQHLRLSYNNLTGNLPSSFSAANNL 180
Query: 180 QSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTG 239
++ W+NN GL+G++ V+SNM+ L Q WL+ N+FTG IPDLSQC ++ DLQLRDNQLTG
Sbjct: 181 ETLWLNNQAAGLSGTLLVLSNMSALNQSWLNKNQFTGSIPDLSQCTALSDLQLRDNQLTG 240
Query: 240 VVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMD 299
VVP SL S+ L+ V+L NN+LQGPVPVFGK V D I N+FC + CD RVM
Sbjct: 241 VVPASLTSLPSLKKVSLDNNELQGPVPVFGKGVNVTLDGI--NSFCLDTPG-NCDPRVMV 297
Query: 300 LLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTD 359
LL I FGYPI+ A+SW GNDPC W V+C GKI +NF KQGLQGTISPAFANLTD
Sbjct: 298 LLQIAEAFGYPIRLAESWKGNDPCDGWNYVVCAAGKIITVNFEKQGLQGTISPAFANLTD 357
Query: 360 LTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGK-- 417
L L+LNGNNL GSIP +L TL QL+TLDVS+N+LSG VPKF PKVK +T GN LGK
Sbjct: 358 LRTLFLNGNNLIGSIPDSLITLPQLQTLDVSDNNLSGLVPKFPPKVKLVTAGNALLGKPL 417
Query: 418 NHGGGAPGSAPGGSPAGSGKGASMK------KVWIIIIIVLIVVGFVVGGAWFSWKCYSR 471
+ GGG G+ P GS G G S K WI I+V IV+ F+ + SWKC+
Sbjct: 418 SPGGGPSGTTPSGSSTGGSGGESSKGNSSVSPGWIAGIVV-IVLFFIAVVLFVSWKCFVN 476
Query: 472 KGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNAT 531
K +F+RV ENG+G KLD VSNGYGG ELQSQSSGD SDLH DG T
Sbjct: 477 KLQGKFSRVKGHENGKGGFKLDAVHVSNGYGGVPVELQSQSSGDRSDLHALDG-----PT 531
Query: 532 ISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEI 591
SI VL+QVTN+FS++NILGRGGFG+VYKG+L DGTKIAVKRM SVA G+KGL EF+AEI
Sbjct: 532 FSIQVLQQVTNNFSEENILGRGGFGVVYKGQLHDGTKIAVKRMESVAMGNKGLKEFEAEI 591
Query: 592 GVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIA 651
VL+KVRHRHLVALLGYCING ERLLVYE+MPQGTLTQHLFE +E GY PLTWKQR++IA
Sbjct: 592 AVLSKVRHRHLVALLGYCINGIERLLVYEYMPQGTLTQHLFEWQEQGYVPLTWKQRVVIA 651
Query: 652 LDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAG 711
LDV RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG YSVET+LAG
Sbjct: 652 LDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAG 711
Query: 712 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNK 771
TFGYLAPEYAATGRVTTKVD+YAFG+VLMELITGRKALDD+VPDE SHLVTWFRRVL NK
Sbjct: 712 TFGYLAPEYAATGRVTTKVDIYAFGIVLMELITGRKALDDTVPDERSHLVTWFRRVLINK 771
Query: 772 ENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTT 831
ENIPKAIDQTL+PDEETM SIYKVAELAGHCT R PYQRPD+GHAVNVL PLV+QW+P++
Sbjct: 772 ENIPKAIDQTLNPDEETMESIYKVAELAGHCTAREPYQRPDMGHAVNVLVPLVEQWKPSS 831
Query: 832 HTDEST-CADDNQMSLTQALQRWQANEGTSTFFNGMT-SQTQSSSTSKPPVFADSLHSPD 889
H +E D QMSL QAL+RWQANEGTS+ FN ++ SQTQSS +SKP FAD+ S D
Sbjct: 832 HDEEEDGSGGDLQMSLPQALRRWQANEGTSSIFNDISISQTQSSISSKPVGFADTFDSMD 891
Query: 890 CR 891
CR
Sbjct: 892 CR 893
>dbj|BAB01851.1| unnamed protein product [Arabidopsis thaliana]
gi|15229508|ref|NP_189017.1| leucine-rich repeat family
protein / protein kinase family protein [Arabidopsis
thaliana]
Length = 928
Score = 993 bits (2566), Expect = 0.0
Identities = 523/909 (57%), Positives = 650/909 (70%), Gaps = 28/909 (3%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
M +L +S P P+ WSS T FC+WSGV+C+ RVT+I+L+D+ L G + +++L++L
Sbjct: 30 MLALAKSFNPPPSDWSSTTDFCKWSGVRCTG-GRVTTISLADKSLTGFIAPEISTLSELK 88
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
++ +Q N LSG +PS A LSSL ++ + NNF V GAF+GL SLQ LSL +N N++ W
Sbjct: 89 SVSIQRNKLSGTIPSFAKLSSLQEIYMDENNFVGVETGAFAGLTSLQILSLSDNNNITTW 148
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQ 180
+FP+EL S++L +I ++ I G LPDIF S +SL L L+YNN++G LP SL S IQ
Sbjct: 149 SFPSELVDSTSLTTIYLDNTNIAGVLPDIFDSLASLQNLRLSYNNITGVLPPSLGKSSIQ 208
Query: 181 SFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTGV 240
+ WINN G++G+I V+S+MT L+Q WLH N F GPIPDLS+ ++ DLQLRDN LTG+
Sbjct: 209 NLWINNQDLGMSGTIEVLSSMTSLSQAWLHKNHFFGPIPDLSKSENLFDLQLRDNDLTGI 268
Query: 241 VPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDL 300
VP +L++++ L+N++L NN+ QGP+P+F +VK D HN FC A C +VM L
Sbjct: 269 VPPTLLTLASLKNISLDNNKFQGPLPLFSPEVKVTID---HNVFCTTKAGQSCSPQVMTL 325
Query: 301 LHIVGGFGYPIQFAKSWTGNDPCKDWLCVIC--GGGKITKLNFAKQGLQGTISPAFANLT 358
L + GG GYP A+SW G+D C W V C G + LN K G G ISPA ANLT
Sbjct: 326 LAVAGGLGYPSMLAESWQGDDACSGWAYVSCDSAGKNVVTLNLGKHGFTGFISPAIANLT 385
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKF-ITDGNVWLGK 417
L +LYLNGN+LTG IP+ L ++ L+ +DVSNN+L GE+PKF VKF GN LG
Sbjct: 386 SLKSLYLNGNDLTGVIPKELTFMTSLQLIDVSNNNLRGEIPKFPATVKFSYKPGNALLGT 445
Query: 418 NHGGGAP---GSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGL 474
N G G+ G A GG SG G S KV +I+ +++ V+ F+ + +K ++
Sbjct: 446 NGGDGSSPGTGGASGGPGGSSGGGGS--KVGVIVGVIVAVLVFLAILGFVVYKFVMKRKY 503
Query: 475 RRFARVGNPENGEGNVKLDLASVSNGYGGASS--------ELQSQSSGDHSDLHGFDGGN 526
RF R + G+ V +++ +G GG ++ L S SSGD+SD +GG+
Sbjct: 504 GRFNRTDPEKVGKILVSDAVSNGGSGNGGYANGHGANNFNALNSPSSGDNSDRFLLEGGS 563
Query: 527 GGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNE 586
TI + VLRQVTN+FS+DNILGRGGFG+VY GEL DGTK AVKRM A G+KG++E
Sbjct: 564 ---VTIPMEVLRQVTNNFSEDNILGRGGFGVVYAGELHDGTKTAVKRMECAAMGNKGMSE 620
Query: 587 FQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQ 646
FQAEI VLTKVRHRHLVALLGYC+NGNERLLVYE+MPQG L QHLFE E GY+PLTWKQ
Sbjct: 621 FQAEIAVLTKVRHRHLVALLGYCVNGNERLLVYEYMPQGNLGQHLFEWSELGYSPLTWKQ 680
Query: 647 RLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVE 706
R+ IALDV RGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG YSVE
Sbjct: 681 RVSIALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVE 740
Query: 707 TKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRR 766
T+LAGTFGYLAPEYAATGRVTTKVDVYAFGVVLME++TGRKALDDS+PDE SHLVTWFRR
Sbjct: 741 TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMEILTGRKALDDSLPDERSHLVTWFRR 800
Query: 767 VLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQ 826
+L NKENIPKA+DQTL+ DEETM SIY+VAELAGHCT R P QRPD+GHAVNVL PLV++
Sbjct: 801 ILINKENIPKALDQTLEADEETMESIYRVAELAGHCTAREPQQRPDMGHAVNVLGPLVEK 860
Query: 827 WEPTTHTDESTCADDNQMSLTQALQRWQANEGT--STFFNG--MTSQTQSSSTSKPPVFA 882
W+P+ +E + D MSL QALQRWQ NEGT ST F+G SQTQSS K F
Sbjct: 861 WKPSCQEEEESFGIDVNMSLPQALQRWQ-NEGTSSSTMFHGDFSYSQTQSSIPPKASGFP 919
Query: 883 DSLHSPDCR 891
++ S D R
Sbjct: 920 NTFDSADGR 928
>gb|AAD21776.1| putative receptor-like protein kinase [Arabidopsis thaliana]
gi|15226361|ref|NP_178291.1| leucine-rich repeat protein
kinase, putative [Arabidopsis thaliana]
gi|25287709|pir||E84429 probable receptor-like protein
kinase [imported] - Arabidopsis thaliana
Length = 943
Score = 858 bits (2218), Expect = 0.0
Identities = 474/919 (51%), Positives = 596/919 (64%), Gaps = 36/919 (3%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLT 60
MQSL+ SL + + SN + C+W V+C NRVT I L + + GTLP NL SL++L
Sbjct: 33 MQSLKSSLNLTSDVDWSNPNPCKWQSVQCDGSNRVTKIQLKQKGIRGTLPTNLQSLSELV 92
Query: 61 TLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPW 120
L L N +SGP+P L+ LS L +NL N F+SV FSG++SLQ + L EN PW
Sbjct: 93 ILELFLNRISGPIPDLSGLSRLQTLNLHDNLFTSVPKNLFSGMSSLQEMYL-ENNPFDPW 151
Query: 121 TFPTELTQSSNLNSIDINQAKINGTLPDIFGSFS--SLNTLHLAYNNLSGGLPNSLAGSG 178
P + ++++L ++ ++ I G +PD FGS S SL L L+ N L G LP S AG+
Sbjct: 152 VIPDTVKEATSLQNLTLSNCSIIGKIPDFFGSQSLPSLTNLKLSQNGLEGELPMSFAGTS 211
Query: 179 IQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLT 238
IQS ++N L GSI+V+ NMT L +V L N+F+GPIPDLS S++ +R+NQLT
Sbjct: 212 IQSLFLNGQK--LNGSISVLGNMTSLVEVSLQGNQFSGPIPDLSGLVSLRVFNVRENQLT 269
Query: 239 GVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVM 298
GVVP SLVS+S L V L NN LQGP P+FGK V + + + N+FC N A CD RV
Sbjct: 270 GVVPQSLVSLSSLTTVNLTNNYLQGPTPLFGKSVGVDIVN-NMNSFCTNVAGEACDPRVD 328
Query: 299 DLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLT 358
L+ + FGYP++ A+SW GN+PC +W+ + C GG IT +N KQ L GTISP+ A LT
Sbjct: 329 TLVSVAESFGYPVKLAESWKGNNPCVNWVGITCSGGNITVVNMRKQDLSGTISPSLAKLT 388
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKN 418
L + L N L+G IP L TLS+L LDVSNND G PKF V +T+GN +GKN
Sbjct: 389 SLETINLADNKLSGHIPDELTTLSKLRLLDVSNNDFYGIPPKFRDTVTLVTEGNANMGKN 448
Query: 419 ---HGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWK--CYSRKG 473
APG++PG P+G G+ K + I++ VVG VVG C K
Sbjct: 449 GPNKTSDAPGASPGSKPSGGSDGSETSKKSSNVKIIVPVVGGVVGALCLVGLGVCLYAKK 508
Query: 474 LRRFARVGNPENG----------EGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFD 523
+R ARV +P + ++KL +A+ S GG S S S SD+H +
Sbjct: 509 RKRPARVQSPSSNMVIHPHHSGDNDDIKLTVAASSLNSGGGSDSY-SHSGSAASDIHVVE 567
Query: 524 GGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKG 583
GN ISI VLR VTN+FS++NILGRGGFG VYKGEL DGTKIAVKRM S KG
Sbjct: 568 AGN---LVISIQVLRNVTNNFSEENILGRGGFGTVYKGELHDGTKIAVKRMESSVVSDKG 624
Query: 584 LNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLT 643
L EF++EI VLTK+RHRHLVALLGYC++GNERLLVYE+MPQGTL+QHLF +E G PL
Sbjct: 625 LTEFKSEITVLTKMRHRHLVALLGYCLDGNERLLVYEYMPQGTLSQHLFHWKEEGRKPLD 684
Query: 644 WKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNY 703
W +RL IALDV RGVEYLH+LA QSFIHRDLKPSNILLGDDMRAKV+DFGLV+ APDG Y
Sbjct: 685 WTRRLAIALDVARGVEYLHTLAHQSFIHRDLKPSNILLGDDMRAKVSDFGLVRLAPDGKY 744
Query: 704 SVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTW 763
S+ET++AGTFGYLAPEYA TGRVTTKVD+++ GV+LMELITGRKALD++ P++S HLVTW
Sbjct: 745 SIETRVAGTFGYLAPEYAVTGRVTTKVDIFSLGVILMELITGRKALDETQPEDSVHLVTW 804
Query: 764 FRRVLTNKEN--IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLC 821
FRRV +K+ AID + D++T+ SI KV ELAGHC R PYQRPD+ H VNVL
Sbjct: 805 FRRVAASKDENAFKNAIDPNISLDDDTVASIEKVWELAGHCCAREPYQRPDMAHIVNVLS 864
Query: 822 PLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANE---------GTSTFFNGMTSQTQS 872
L QW+PT + D M L Q L++WQA E G+S+ G TQ+
Sbjct: 865 SLTVQWKPTETDPDDVYGIDYDMPLPQVLKKWQAFEGLSQTADDSGSSSSAYGSKDNTQT 924
Query: 873 SSTSKPPVFADSLHSPDCR 891
S ++P FADS S D R
Sbjct: 925 SIPTRPSGFADSFTSVDGR 943
>gb|AAF66615.1| LRR receptor-like protein kinase [Nicotiana tabacum]
Length = 945
Score = 843 bits (2177), Expect = 0.0
Identities = 470/923 (50%), Positives = 590/923 (63%), Gaps = 48/923 (5%)
Query: 1 MQSLRQSLKP-SPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
MQ L++ + P S GW+ C+W V+C+ D RVT I + +Q L G+LP NLN+LT+L
Sbjct: 39 MQELKKRINPPSSLGWNDPDP-CKWGKVQCTKDGRVTRIQIGNQGLKGSLPPNLNNLTEL 97
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
+QNN L+G LPS + L SL + L +N F+S+ F GL SLQ++ L +N SP
Sbjct: 98 LVFEVQNNGLTGSLPSFSGLDSLQSLLLNNNGFTSIPTDFFDGLTSLQSVYLDKN-QFSP 156
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGI 179
W+ P L ++++ + A I GT+PD F +F+SL LHL++NNL G LP+S +GS I
Sbjct: 157 WSIPESLKSATSIQTFSAVSANITGTIPDFFDAFASLTNLHLSFNNLGGSLPSSFSGSQI 216
Query: 180 QSFWINNNLPGLTGSITVISNMTLLTQV-WLHVNKFTGPIPDLSQCNSIKDLQLRDNQLT 238
QS W+N L GSI VI NMT LT+ N F+ P+PD S + +++ LRDN LT
Sbjct: 217 QSLWLNGLKGRLNGSIAVIQNMTQLTRTSGCKANAFSSPLPDFSGLSQLQNCSLRDNSLT 276
Query: 239 GVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVM 298
G VP+SLV++ L+ V L NN LQGP P F V+ + N+FC + VPCD+RV
Sbjct: 277 GPVPNSLVNLPSLKVVVLTNNFLQGPTPKFPSSVQVDML-ADTNSFCLSQPGVPCDSRVN 335
Query: 299 DLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLT 358
LL + GYP +FA++W GNDPC W+ + C GG IT LNF K GL GTISP ++++T
Sbjct: 336 TLLAVAKDVGYPREFAENWKGNDPCSPWMGITCDGGNITVLNFQKMGLTGTISPNYSSIT 395
Query: 359 DLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKN 418
L L L NNL G+IP LA L L LDVSNN L G++P F V T GNV +GK+
Sbjct: 396 SLQKLILANNNLIGTIPNELALLPNLRELDVSNNQLYGKIPPFKSNVLLKTQGNVNIGKD 455
Query: 419 H----GGGAPGSAPGGSPAGSGKGA----SMKKVWIIIIIVLIVVGFVVGG-------AW 463
+ G P + GS GSG G S KK +VVG V+GG A
Sbjct: 456 NPPPPAPGTPSGSTPGSSDGSGGGQTHANSGKKS-----STGVVVGSVIGGVCAAVVLAG 510
Query: 464 FSWKCYSRKGLRRFARVGNPE----------NGEGNVKLDLASVSNGYGGASSELQSQSS 513
C R +R RV +P + + VK+ +A S G + SS
Sbjct: 511 LFVFCLYRTKRKRSGRVQSPHTVVIHPHHSGSDQDAVKITIAGSSVNGGDSCG-----SS 565
Query: 514 GDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKR 573
DLH + GN ISI VLR VTN+FS+ NILGRGGFG VYKGEL DGTK+AVKR
Sbjct: 566 SAPGDLHIVEAGN---MVISIQVLRDVTNNFSEVNILGRGGFGTVYKGELHDGTKMAVKR 622
Query: 574 MISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFE 633
M S KGL+EF++EI VLTKVRHRHLV LLGYC++GNERLLVYE+MPQGTL+++LF
Sbjct: 623 MESGVMSEKGLDEFKSEIAVLTKVRHRHLVTLLGYCLDGNERLLVYEYMPQGTLSRYLFN 682
Query: 634 CREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFG 693
+E G PL W +RL IALDV RGVEYLH LAQQSFIHRDLKPSNILLGDDMRAKVADFG
Sbjct: 683 WKEEGLKPLEWTRRLTIALDVARGVEYLHGLAQQSFIHRDLKPSNILLGDDMRAKVADFG 742
Query: 694 LVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSV 753
LV+ APD SV T+LAGTFGYLAPEYA TGRVTTK+DV++FGV+LMELITGRKALD+S
Sbjct: 743 LVRLAPDPKASVVTRLAGTFGYLAPEYAVTGRVTTKIDVFSFGVILMELITGRKALDESQ 802
Query: 754 PDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDI 813
P+ES HLV WFRR+ NKE KAID T+D DEET+ S+ VAELAGH R P+QRPD+
Sbjct: 803 PEESMHLVPWFRRMHINKETFRKAIDPTVDLDEETLSSVSTVAELAGHSCAREPHQRPDM 862
Query: 814 GHAVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGT-----STFFNGMTS 868
GHAVNVL L + W+P ++ D MSL QA+++WQA EG S+ + +
Sbjct: 863 GHAVNVLSSLAELWKPAEVDEDEIYGIDYDMSLPQAVKKWQALEGMSGIDGSSSYLASSD 922
Query: 869 QTQSSSTSKPPVFADSLHSPDCR 891
TQ+S ++P FADS S D R
Sbjct: 923 NTQTSIPTRPSGFADSFTSADGR 945
>gb|AAP04161.1| putative receptor protein kinase (TMK1) [Arabidopsis thaliana]
Length = 942
Score = 841 bits (2172), Expect = 0.0
Identities = 476/922 (51%), Positives = 592/922 (63%), Gaps = 42/922 (4%)
Query: 1 MQSLRQSLKP-SPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M SL++SL P S GWS C+W+ + C+ RVT I + L GTL +L +L++L
Sbjct: 32 MLSLKKSLNPPSSFGWSDPDP-CKWTHIVCTGTKRVTRIQIGHSGLQGTLSPDLRNLSEL 90
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
L LQ N +SGP+PSL+ L+SL + L +NNF S+ F GL SLQ++ + N
Sbjct: 91 ERLELQWNNISGPVPSLSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDNN-PFKS 149
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIFG--SFSSLNTLHLAYNNLSGGLPNSLAGS 177
W P L +S L + N A ++G+LP G F L+ LHLA+NNL G LP SLAGS
Sbjct: 150 WEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPMSLAGS 209
Query: 178 GIQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQL 237
+QS W+N LTG ITV+ NMT L +VWLH NKF+GP+PD S ++ L LRDN
Sbjct: 210 QVQSLWLNGQK--LTGDITVLQNMTGLKEVWLHSNKFSGPLPDFSGLKELESLSLRDNSF 267
Query: 238 TGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARV 297
TG VP SL+S+ L+ V L NN LQGPVPVF V + D S N+FC ++ CD RV
Sbjct: 268 TGPVPASLLSLESLKVVNLTNNHLQGPVPVFKSSVSVDLDKDS-NSFCLSSPG-ECDPRV 325
Query: 298 MDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANL 357
LL I F YP + A+SW GNDPC +W+ + C G IT ++ K L GTISP F +
Sbjct: 326 KSLLLIASSFDYPPRLAESWKGNDPCTNWIGIACSNGNITVISLEKMELTGTISPEFGAI 385
Query: 358 TDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGK 417
L + L NNLTG IPQ L TL L+TLDVS+N L G+VP F V T+GN +GK
Sbjct: 386 KSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVPGFRSNVVVNTNGNPDIGK 445
Query: 418 NHGG-GAPGSAPGGSPAGSGKGAS-----MKKVWIIIIIVLIVVG-----FVVGGAWFSW 466
+ +PGS+ +GSG MK I IIV V+G F++G F
Sbjct: 446 DKSSLSSPGSSSPSGGSGSGINGDKDRRGMKSSTFIGIIVGSVLGGLLSIFLIGLLVF-- 503
Query: 467 KCYSRKGLRRF-------ARVGNPEN-GEGN--VKLDLASVSNGYGGASSELQSQSSGDH 516
C+ +K +RF A V +P + G N VK+ +A S GG S + +
Sbjct: 504 -CWYKKRQKRFSGSESSNAVVVHPRHSGSDNESVKITVAGSSVSVGGISDTYTLPGTSEV 562
Query: 517 SDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMIS 576
D GN ISI VLR VTN+FS DNILG GGFG+VYKGEL DGTKIAVKRM +
Sbjct: 563 GD--NIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMEN 620
Query: 577 VAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECRE 636
KG EF++EI VLTKVRHRHLV LLGYC++GNE+LLVYE+MPQGTL++HLFE E
Sbjct: 621 GVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSE 680
Query: 637 HGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK 696
G PL WKQRL +ALDV RGVEYLH LA QSFIHRDLKPSNILLGDDMRAKVADFGLV+
Sbjct: 681 EGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVR 740
Query: 697 NAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDE 756
AP+G S+ET++AGTFGYLAPEYA TGRVTTKVDVY+FGV+LMELITGRK+LD+S P+E
Sbjct: 741 LAPEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEE 800
Query: 757 SSHLVTWFRRVLTNKE-NIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGH 815
S HLV+WF+R+ NKE + KAID T+D DEET+ S++ VAELAGHC +R PYQRPD+GH
Sbjct: 801 SIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCSREPYQRPDMGH 860
Query: 816 AVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS------Q 869
AVN+L LV+ W+P+ E D MSL QAL++WQA EG S + +S
Sbjct: 861 AVNILSSLVELWKPSDQNPEDIYGIDLDMSLPQALKKWQAYEGRSDLESSTSSLLPSLDN 920
Query: 870 TQSSSTSKPPVFADSLHSPDCR 891
TQ S ++P FA+S S D R
Sbjct: 921 TQMSIPTRPYGFAESFTSVDGR 942
>ref|NP_176789.1| leucine-rich repeat protein kinase, putative (TMK1) [Arabidopsis
thaliana] gi|1174718|sp|P43298|TMK1_ARATH Putative
receptor protein kinase TMK1 precursor
gi|12322608|gb|AAG51302.1| receptor protein kinase
(TMK1), putative [Arabidopsis thaliana]
gi|166888|gb|AAA32876.1| protein kinase
Length = 942
Score = 840 bits (2171), Expect = 0.0
Identities = 476/922 (51%), Positives = 591/922 (63%), Gaps = 42/922 (4%)
Query: 1 MQSLRQSLKP-SPNGWSSNTSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M SL++SL P S GWS C+W+ + C+ RVT I + L GTL +L +L++L
Sbjct: 32 MLSLKKSLNPPSSFGWSDPDP-CKWTHIVCTGTKRVTRIQIGHSGLQGTLSPDLRNLSEL 90
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
L LQ N +SGP+PSL+ L+SL + L +NNF S+ F GL SLQ++ + N
Sbjct: 91 ERLELQWNNISGPVPSLSGLASLQVLMLSNNNFDSIPSDVFQGLTSLQSVEIDNN-PFKS 149
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIFG--SFSSLNTLHLAYNNLSGGLPNSLAGS 177
W P L +S L + N A ++G+LP G F L+ LHLA+NNL G LP SLAGS
Sbjct: 150 WEIPESLRNASALQNFSANSANVSGSLPGFLGPDEFPGLSILHLAFNNLEGELPMSLAGS 209
Query: 178 GIQSFWINNNLPGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQL 237
+QS W+N LTG ITV+ NMT L +VWLH NKF+GP+PD S ++ L LRDN
Sbjct: 210 QVQSLWLNGQK--LTGDITVLQNMTGLKEVWLHSNKFSGPLPDFSGLKELESLSLRDNSF 267
Query: 238 TGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARV 297
TG VP SL+S+ L+ V L NN LQGPVPVF V + D S N+FC ++ CD RV
Sbjct: 268 TGPVPASLLSLESLKVVNLTNNHLQGPVPVFKSSVSVDLDKDS-NSFCLSSPG-ECDPRV 325
Query: 298 MDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANL 357
LL I F YP + A+SW GNDPC +W+ + C G IT ++ K L GTISP F +
Sbjct: 326 KSLLLIASSFDYPPRLAESWKGNDPCTNWIGIACSNGNITVISLEKMELTGTISPEFGAI 385
Query: 358 TDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGK 417
L + L NNLTG IPQ L TL L+TLDVS+N L G+VP F V T+GN +GK
Sbjct: 386 KSLQRIILGINNLTGMIPQELTTLPNLKTLDVSSNKLFGKVPGFRSNVVVNTNGNPDIGK 445
Query: 418 NHGG-GAPGSAPGGSPAGSGKGAS-----MKKVWIIIIIVLIVVG-----FVVGGAWFSW 466
+ +PGS+ +GSG MK I IIV V+G F++G F
Sbjct: 446 DKSSLSSPGSSSPSGGSGSGINGDKDRRGMKSSTFIGIIVGSVLGGLLSIFLIGLLVF-- 503
Query: 467 KCYSRKGLRRF-------ARVGNPEN-GEGN--VKLDLASVSNGYGGASSELQSQSSGDH 516
C+ +K +RF A V +P + G N VK+ +A S GG S + +
Sbjct: 504 -CWYKKRQKRFSGSESSNAVVVHPRHSGSDNESVKITVAGSSVSVGGISDTYTLPGTSEV 562
Query: 517 SDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMIS 576
D GN ISI VLR VTN+FS DNILG GGFG+VYKGEL DGTKIAVKRM +
Sbjct: 563 GD--NIQMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMEN 620
Query: 577 VAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECRE 636
KG EF++EI VLTKVRHRHLV LLGYC++GNE+LLVYE+MPQGTL++HLFE E
Sbjct: 621 GVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSE 680
Query: 637 HGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVK 696
G PL WKQRL +ALDV RGVEYLH LA QSFIHRDLKPSNILLGDDMRAKVADFGLV+
Sbjct: 681 EGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVR 740
Query: 697 NAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDE 756
AP+G S+ET++AGTFGYLAPEYA TGRVTTKVDVY+FGV+LMELITGRK+LD+S P+E
Sbjct: 741 LAPEGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEE 800
Query: 757 SSHLVTWFRRVLTNKE-NIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGH 815
S HLV+WF+R+ NKE + KAID T+D DEET+ S++ VAELAGHC R PYQRPD+GH
Sbjct: 801 SIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGH 860
Query: 816 AVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS------Q 869
AVN+L LV+ W+P+ E D MSL QAL++WQA EG S + +S
Sbjct: 861 AVNILSSLVELWKPSDQNPEDIYGIDLDMSLPQALKKWQAYEGRSDLESSTSSLLPSLDN 920
Query: 870 TQSSSTSKPPVFADSLHSPDCR 891
TQ S ++P FA+S S D R
Sbjct: 921 TQMSIPTRPYGFAESFTSVDGR 942
>emb|CAA69028.1| TMK [Oryza sativa (indica cultivar-group)] gi|7434420|pir||T04124
receptor-like protein kinase (EC 2.7.1.-) - rice
Length = 962
Score = 832 bits (2150), Expect = 0.0
Identities = 464/913 (50%), Positives = 593/913 (64%), Gaps = 43/913 (4%)
Query: 14 GWSSN---TSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALS 70
GWS+ +S W+GV C S RVT++ + ++ L G L + +LT L L L +N++S
Sbjct: 58 GWSTGDPCSSPRAWAGVTCDSAGRVTAVQVGNRSLTGRLAPEVRNLTALARLELFDNSIS 117
Query: 71 GPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSS 130
G LPSLA LSSL + + +N F+ + P F GL +L +SL N PW P +L +
Sbjct: 118 GELPSLAGLSSLQYLLVHNNGFTRIPPDFFKGLTALAAVSLDNN-PFDPWPLPADLADCT 176
Query: 131 NLNSIDINQAKINGTLPDIFGS-FSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLP 189
+L + N A + G LPD FG+ SL L LA+N +SG +P SLA + +Q+ W+NN +
Sbjct: 177 SLTNFSANTANVTGALPDFFGTALPSLQRLSLAFNKMSGPVPASLATAPLQALWLNNQIG 236
Query: 190 G--LTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTGVVPDSLVS 247
GSI+ ISNMT L ++WLH N FTGP+PD S S+ DL+LRDNQLTG VPDSL+
Sbjct: 237 ENQFNGSISFISNMTSLQELWLHSNDFTGPLPDFSGLASLSDLELRDNQLTGPVPDSLLK 296
Query: 248 MSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGF 307
+ L VTL NN LQGP P F VK + + FC + PCD RV LL + GF
Sbjct: 297 LGSLTKVTLTNNLLQGPTPKFADKVKADVVPTTER-FCLSTPGQPCDPRVSLLLEVAAGF 355
Query: 308 GYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNG 367
YP + A +W GNDPC ++ V C G IT LNFA+ G G+ISPA +T L L L
Sbjct: 356 QYPAKLADNWKGNDPCDGYIGVGCDAGNITVLNFARMGFSGSISPAIGKITTLQKLILAD 415
Query: 368 NNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFI-TDGNVWLGKN------HG 420
NN+TG++P+ +A L L +D+SNN+L G++P F+ K + +GN +GK+ G
Sbjct: 416 NNITGTVPKEVAALPALTEVDLSNNNLYGKLPTFAAKNVLVKANGNPNIGKDAPAPSGSG 475
Query: 421 GGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGG---AWFSWKCYSRKGLRRF 477
G +AP G G G S II VVG + G A + CY RK + F
Sbjct: 476 GSGGSNAPDGGNGGDGSNGSPSPS-SAGIIAGSVVGAIAGVGLLAALGFYCYKRKQ-KPF 533
Query: 478 ARVGNPE----------NGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNG 527
RV +P + VK+ +A + G A+SE SQ+S D+H + GN
Sbjct: 534 GRVQSPHAMVVHPRHSGSDPDMVKITVAGGNVNGGAAASETYSQASSGPRDIHVVETGN- 592
Query: 528 GNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEF 587
ISI VLR VTN+FSD+N+LGRGGFG VYKGEL DGTKIAVKRM + G+KGLNEF
Sbjct: 593 --MVISIQVLRNVTNNFSDENVLGRGGFGTVYKGELHDGTKIAVKRMEAGVMGNKGLNEF 650
Query: 588 QAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQR 647
++EI VLTKVRHR+LV+LLGYC++GNER+LVYE+MPQGTL+QHLFE +EH PL WK+R
Sbjct: 651 KSEIAVLTKVRHRNLVSLLGYCLDGNERILVYEYMPQGTLSQHLFEWKEHNLRPLEWKKR 710
Query: 648 LIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGN-YSV 705
L IALDV RGVEYLHSLAQQ+FIHRDLKPSNILLGDDM+AKVADFGLV+ AP DG SV
Sbjct: 711 LSIALDVARGVEYLHSLAQQTFIHRDLKPSNILLGDDMKAKVADFGLVRLAPADGKCVSV 770
Query: 706 ETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFR 765
ET+LAGTFGYLAPEYA TGRVTTK DV++FGV+LMELITGRKALD++ P++S HLVTWFR
Sbjct: 771 ETRLAGTFGYLAPEYAVTGRVTTKADVFSFGVILMELITGRKALDETQPEDSMHLVTWFR 830
Query: 766 RVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQ 825
R+ +K+ KAID T+D EET+ S+ VAELAGHC R P+QRPD+GHAVNVL L
Sbjct: 831 RMQLSKDTFQKAIDPTIDLTEETLASVSTVAELAGHCCAREPHQRPDMGHAVNVLSTLSD 890
Query: 826 QWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS-------QTQSSSTSKP 878
W+P+ + + D M+L QAL++WQA E S+ F+G TS TQ+S ++P
Sbjct: 891 VWKPSDPDSDDSYGIDLDMTLPQALKKWQAFE-DSSHFDGATSSFLASLDNTQTSIPTRP 949
Query: 879 PVFADSLHSPDCR 891
P FA+S S D R
Sbjct: 950 PGFAESFTSADGR 962
>ref|XP_469561.1| gibberellin-induced receptor-like kinase TMK [Oryza sativa
(japonica cultivar-group)] gi|28301932|gb|AAO38825.1|
gibberellin-induced receptor-like kinase TMK [Oryza
sativa (japonica cultivar-group)]
Length = 962
Score = 829 bits (2142), Expect = 0.0
Identities = 463/913 (50%), Positives = 592/913 (64%), Gaps = 43/913 (4%)
Query: 14 GWSSN---TSFCQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALS 70
GWS+ +S W+GV C S RVT++ + ++ L G L + +LT L L L +N++S
Sbjct: 58 GWSTGDPCSSPRAWAGVTCDSAGRVTAVQVGNRSLTGRLAPEVRNLTALARLELFDNSIS 117
Query: 71 GPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSS 130
G LPSLA LSSL + + +N F+ + P F GL +L +SL N PW P +L +
Sbjct: 118 GELPSLAGLSSLQYLLVHNNGFTRIPPDFFKGLTALAAVSLDNN-PFDPWPLPADLADCT 176
Query: 131 NLNSIDINQAKINGTLPDIFGS-FSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNLP 189
+L + N A + G LPD FG+ SL L LA+N +SG +P SLA + +Q+ W+NN +
Sbjct: 177 SLTNFSANTANVTGALPDFFGTALPSLQRLSLAFNKMSGPVPASLATAPLQALWLNNQIG 236
Query: 190 G--LTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTGVVPDSLVS 247
GSI+ ISNMT L ++WLH N FTGP+PD S S+ DL+LRDNQLTG VPDSL+
Sbjct: 237 ENQFNGSISFISNMTSLQELWLHSNDFTGPLPDFSGLASLSDLELRDNQLTGPVPDSLLK 296
Query: 248 MSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGF 307
+ L VTL NN LQGP P F VK + + FC + PCD RV LL + F
Sbjct: 297 LGSLTKVTLTNNLLQGPTPKFADKVKADVVPTTER-FCLSTPGQPCDPRVNLLLEVAAEF 355
Query: 308 GYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNG 367
YP + A +W GNDPC ++ V C G IT LNFA+ G G+ISPA +T L L L
Sbjct: 356 QYPAKLADNWKGNDPCDGYIGVGCDAGNITVLNFARMGFSGSISPAIGKITTLQKLILAD 415
Query: 368 NNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFI-TDGNVWLGKN------HG 420
NN+TG++P+ +A L L +D+SNN+L G++P F+ K + +GN +GK+ G
Sbjct: 416 NNITGTVPKEVAALPALTEVDLSNNNLYGKLPTFAAKNVLVKANGNPNIGKDAPAPSGSG 475
Query: 421 GGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGG---AWFSWKCYSRKGLRRF 477
G +AP G G G S II VVG + G A + CY RK + F
Sbjct: 476 GSGGSNAPDGGNGGDGSNGSPSSS-SAGIIAGSVVGAIAGVGLLAALGFYCYKRKQ-KPF 533
Query: 478 ARVGNPE----------NGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNG 527
RV +P + VK+ +A + G A+SE SQ+S D+H + GN
Sbjct: 534 GRVQSPHAMVVHPRHSGSDPDMVKITVAGGNVNGGAAASETYSQASSGPRDIHVVETGN- 592
Query: 528 GNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEF 587
ISI VLR VTN+FSD+N+LGRGGFG VYKGEL DGTKIAVKRM + G+KGLNEF
Sbjct: 593 --MVISIQVLRNVTNNFSDENVLGRGGFGTVYKGELHDGTKIAVKRMEAGVMGNKGLNEF 650
Query: 588 QAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQR 647
++EI VLTKVRHR+LV+LLGYC++GNER+LVYE+MPQGTL+QHLFE +EH PL WK+R
Sbjct: 651 KSEIAVLTKVRHRNLVSLLGYCLDGNERILVYEYMPQGTLSQHLFEWKEHNLRPLEWKKR 710
Query: 648 LIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGN-YSV 705
L IALDV RGVEYLHSLAQQ+FIHRDLKPSNILLGDDM+AKVADFGLV+ AP DG SV
Sbjct: 711 LSIALDVARGVEYLHSLAQQTFIHRDLKPSNILLGDDMKAKVADFGLVRLAPADGKCVSV 770
Query: 706 ETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFR 765
ET+LAGTFGYLAPEYA TGRVTTK DV++FGV+LMELITGRKALD++ P++S HLVTWFR
Sbjct: 771 ETRLAGTFGYLAPEYAVTGRVTTKADVFSFGVILMELITGRKALDETQPEDSMHLVTWFR 830
Query: 766 RVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQ 825
R+ +K+ KAID T+D EET+ S+ VAELAGHC R P+QRPD+GHAVNVL L
Sbjct: 831 RMQLSKDTFQKAIDPTIDLTEETLASVSTVAELAGHCCAREPHQRPDMGHAVNVLSTLSD 890
Query: 826 QWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMTS-------QTQSSSTSKP 878
W+P+ + + D M+L QAL++WQA E S+ F+G TS TQ+S ++P
Sbjct: 891 VWKPSDPDSDDSYGIDLDMTLPQALKKWQAFE-DSSHFDGATSSFLASLDNTQTSIPTRP 949
Query: 879 PVFADSLHSPDCR 891
P FA+S S D R
Sbjct: 950 PGFAESFTSADGR 962
>ref|XP_474425.1| OSJNBa0088H09.21 [Oryza sativa (japonica cultivar-group)]
gi|21741125|emb|CAD41925.1| OSJNBa0070M12.3 [Oryza
sativa (japonica cultivar-group)]
gi|32488720|emb|CAE03463.1| OSJNBa0088H09.21 [Oryza
sativa (japonica cultivar-group)]
Length = 938
Score = 745 bits (1924), Expect = 0.0
Identities = 417/925 (45%), Positives = 568/925 (61%), Gaps = 45/925 (4%)
Query: 1 MQSLRQSLKPSPN--GWSSNTSF---CQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNS 55
+ LR+SL + GW + W + C RV +I+L + LAGTLP +
Sbjct: 25 LHDLRRSLTNADAVLGWGDPNAADPCAAWPHISCDRAGRVNNIDLKNAGLAGTLPSTFAA 84
Query: 56 LTQLTTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN- 114
L L L LQNN LSG LPS ++SL L +N+F S+ FSGL SL +SL +N
Sbjct: 85 LDALQDLSLQNNNLSGDLPSFRGMASLRHAFLNNNSFRSIPADFFSGLTSLLVISLDQNP 144
Query: 115 INLSP--WTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPN 172
+N+S WT P ++ + L S+ +N + G +PD G+ +SL L LAYN LSG +P+
Sbjct: 145 LNVSSGGWTIPADVAAAQQLQSLSLNGCNLTGAIPDFLGAMNSLQELKLAYNALSGPIPS 204
Query: 173 SLAGSGIQSFWINNN--LPGLTGSITVISNMTLLTQVWLHVNKFTGPIPD-LSQCNSIKD 229
+ SG+Q+ W+NN +P L+G++ +I+ M L Q WLH N F+GPIPD ++ C + D
Sbjct: 205 TFNASGLQTLWLNNQHGVPKLSGTLDLIATMPNLEQAWLHGNDFSGPIPDSIADCKRLSD 264
Query: 230 LQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNA 289
L L NQL G+VP +L SM+GL++V L NN L GPVP K KY S N FC +
Sbjct: 265 LCLNSNQLVGLVPPALESMAGLKSVQLDNNNLLGPVPAI-KAPKYT---YSQNGFCADKP 320
Query: 290 SVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGT 349
V C +VM LLH + YP + SW+GN+ C DWL + C G +T LN + GL GT
Sbjct: 321 GVACSPQVMALLHFLAEVDYPKRLVASWSGNNSCVDWLGISCVAGNVTMLNLPEYGLNGT 380
Query: 350 ISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFIT 409
IS + NL++L+ + L GNNLTG +P +L +L L+ LD+S NDL+G +P FSP VK
Sbjct: 381 ISDSLGNLSELSDINLIGNNLTGHVPDSLTSLRLLQKLDLSGNDLTGPLPTFSPSVKVNV 440
Query: 410 DGNVWLGKNHGGGAPGSAPGGSPAGSG-------------------KGASMKKVWIIIII 450
GN+ N G APGSAP GS + A + I + +
Sbjct: 441 TGNL----NFNGTAPGSAPSKDTPGSSSSRAPTLPGQGVLPENKKKRSAVVLATTIPVAV 496
Query: 451 VLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSE--L 508
++ + V F K S V EN + + + + V+N +S++
Sbjct: 497 SVVALASVCAVLIFRKKRGSVPPNAASVVVHPRENSDPDNLVKIVMVNNDGNSSSTQGNT 556
Query: 509 QSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTK 568
S SS SD+H D GN I++ VLR T +F+ DN+LGRGGFG+VYKGEL DGT
Sbjct: 557 LSGSSSRASDVHMIDTGN---FVIAVQVLRGATKNFTQDNVLGRGGFGVVYKGELHDGTM 613
Query: 569 IAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLT 628
IAVKRM + +K L+EFQAEI +LTKVRHR+LV++LGY I GNERLLVYE+M G L+
Sbjct: 614 IAVKRMEAAVISNKALDEFQAEITILTKVRHRNLVSILGYSIEGNERLLVYEYMSNGALS 673
Query: 629 QHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAK 688
+HLF+ ++ PL+WK+RL IALDV RG+EYLH+LA Q +IHRDLK +NILLGDD RAK
Sbjct: 674 KHLFQWKQFELEPLSWKKRLNIALDVARGMEYLHNLAHQCYIHRDLKSANILLGDDFRAK 733
Query: 689 VADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKA 748
V+DFGLVK+APDGN+SV T+LAGTFGYLAPEYA TG++TTK DV++FGVVLMELITG A
Sbjct: 734 VSDFGLVKHAPDGNFSVATRLAGTFGYLAPEYAVTGKITTKADVFSFGVVLMELITGMTA 793
Query: 749 LDDS-VPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSP 807
+D+S + +E+ +L +WF ++ +++ + AID TLD +ET SI +AELAGHCT+R P
Sbjct: 794 IDESRLEEETRYLASWFCQIRKDEDRLRAAIDPTLDQSDETFESISVIAELAGHCTSREP 853
Query: 808 YQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMT 867
QRPD+GHAVNVL P+V++W+P E D L Q ++ WQ E + T + ++
Sbjct: 854 TQRPDMGHAVNVLVPMVEKWKPVNDETEDYMGIDLHQPLLQMVKGWQDAEASMTDGSILS 913
Query: 868 -SQTQSSSTSKPPVFADSLHSPDCR 891
++ S ++P FA+S S D R
Sbjct: 914 LEDSKGSIPARPAGFAESFTSADGR 938
>gb|AAO72615.1| receptor-like protein kinase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 938
Score = 742 bits (1916), Expect = 0.0
Identities = 416/925 (44%), Positives = 568/925 (60%), Gaps = 45/925 (4%)
Query: 1 MQSLRQSLKPSPN--GWSSNTSF---CQWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNS 55
+ LR+SL + GW + W + C RV +I+L + LAGTLP +
Sbjct: 25 LHDLRRSLTXAEAVLGWGDPNAADPCAAWPHISCDRAGRVNNIDLKNAGLAGTLPFTFAA 84
Query: 56 LTQLTTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN- 114
L L L LQN+ LSG LPS ++SL L +N+F S+ FSGL SL +SL +N
Sbjct: 85 LDALQDLSLQNHNLSGDLPSFRGMASLRHAFLNNNSFRSIPADFFSGLTSLLVISLDQNP 144
Query: 115 INLSP--WTFPTELTQSSNLNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPN 172
+N+S WT P ++ + L S+ +N + G +PD G+ +SL L LAYN LSG +P+
Sbjct: 145 LNVSSGGWTIPADVAAAQQLQSLSLNGCNLTGAIPDFLGAMNSLQELKLAYNALSGPIPS 204
Query: 173 SLAGSGIQSFWINNN--LPGLTGSITVISNMTLLTQVWLHVNKFTGPIPD-LSQCNSIKD 229
+ SG+Q+ W+NN +P L+G++ +I+ M L Q WLH N F+GPIPD ++ C + D
Sbjct: 205 TFNASGLQTLWLNNQHGVPKLSGTLDLIATMPNLEQAWLHGNDFSGPIPDSIADCKRLSD 264
Query: 230 LQLRDNQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNA 289
L L NQL G+VP +L SM+GL++V L NN L GPVP K KY S N FC +
Sbjct: 265 LCLNSNQLVGLVPPALESMAGLKSVQLDNNNLLGPVPAI-KAPKYT---YSQNGFCADKP 320
Query: 290 SVPCDARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGT 349
V C +VM LLH + YP + SW+GN+ C DWL + C G +T LN + GL GT
Sbjct: 321 GVACSPQVMALLHFLAEVDYPKRLVASWSGNNSCVDWLGISCVAGNVTMLNLPEYGLNGT 380
Query: 350 ISPAFANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFIT 409
IS + NL++L+ + L GNNLTG +P +L +L L+ LD+S NDL+G +P FSP VK
Sbjct: 381 ISDSLGNLSELSDINLIGNNLTGHVPDSLTSLRLLQKLDLSGNDLTGPLPTFSPSVKVNV 440
Query: 410 DGNVWLGKNHGGGAPGSAPGGSPAGSG-------------------KGASMKKVWIIIII 450
GN+ N G APGSAP GS + A + I + +
Sbjct: 441 TGNL----NFNGTAPGSAPSKDTPGSSSSRAPTLPGQGVLPENKKKRSAVVLATTIPVAV 496
Query: 451 VLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNPENGEGNVKLDLASVSNGYGGASSE--L 508
++ + V F K S V EN + + + + V+N +S++
Sbjct: 497 SVVALASVCAVLIFRKKRGSVPPNAASVVVHPRENSDPDNLVKIVMVNNDGNSSSTQGNT 556
Query: 509 QSQSSGDHSDLHGFDGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTK 568
S SS SD+H D GN I++ VLR T +F+ DN+LGRGGFG+VYKGEL DGT
Sbjct: 557 LSGSSSRASDVHMIDTGN---FVIAVQVLRGATKNFTQDNVLGRGGFGVVYKGELHDGTM 613
Query: 569 IAVKRMISVAKGSKGLNEFQAEIGVLTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLT 628
IAVKRM + +K L+EFQAEI +LTKVRHR+LV++LGY I GNERLLVYE+M G L+
Sbjct: 614 IAVKRMEAAVISNKALDEFQAEITILTKVRHRNLVSILGYSIEGNERLLVYEYMSNGALS 673
Query: 629 QHLFECREHGYTPLTWKQRLIIALDVGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAK 688
+HLF+ ++ PL+WK+RL IALDV RG+EYLH+LA Q +IHRDLK +NILLGDD RAK
Sbjct: 674 KHLFQWKQFELEPLSWKKRLNIALDVARGMEYLHNLAHQCYIHRDLKSANILLGDDFRAK 733
Query: 689 VADFGLVKNAPDGNYSVETKLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKA 748
V+DFGLVK+APDGN+SV T+LAGTFGYLAPEYA TG++TTK DV++FGVVLMELITG A
Sbjct: 734 VSDFGLVKHAPDGNFSVATRLAGTFGYLAPEYAVTGKITTKADVFSFGVVLMELITGMTA 793
Query: 749 LDDS-VPDESSHLVTWFRRVLTNKENIPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSP 807
+D+S + +E+ +L +WF ++ +++ + AID TLD +ET SI +AELAGHCT+R P
Sbjct: 794 IDESRLEEETRYLASWFCQIRKDEDRLRAAIDPTLDQSDETFESISVIAELAGHCTSREP 853
Query: 808 YQRPDIGHAVNVLCPLVQQWEPTTHTDESTCADDNQMSLTQALQRWQANEGTSTFFNGMT 867
QRPD+GHAVNVL P+V++W+P E D L Q ++ WQ E + T + ++
Sbjct: 854 TQRPDMGHAVNVLVPMVEKWKPVNDETEDYMGIDLHQPLLQMVKGWQDAEASMTDGSILS 913
Query: 868 -SQTQSSSTSKPPVFADSLHSPDCR 891
++ S ++P FA+S S D R
Sbjct: 914 LEDSKGSIPARPAGFAESFTSADGR 938
>gb|AAG03120.1| F5A9.23 [Arabidopsis thaliana]
Length = 924
Score = 739 bits (1909), Expect = 0.0
Identities = 427/894 (47%), Positives = 549/894 (60%), Gaps = 40/894 (4%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSG-VKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M +LR SLK S N S + C+WS +KC + NRVT+I + D+ ++G LP +L LT L
Sbjct: 27 MIALRDSLKLSGNPNWSGSDPCKWSMFIKCDASNRVTAIQIGDRGISGKLPPDLGKLTSL 86
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
T + N L+GP+PSLA L SL V N+F+SV FSGL+SLQ +SL N
Sbjct: 87 TKFEVMRNRLTGPIPSLAGLKSLVTVYANDNDFTSVPEDFFSGLSSLQHVSLDNN-PFDS 145
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIF---GSFSSLNTLHLAYNNLSGGLPNSLAG 176
W P L +++L ++G +PD FSSL TL L+YN+L P + +
Sbjct: 146 WVIPPSLENATSLVDFSAVNCNLSGKIPDYLFEGKDFSSLTTLKLSYNSLVCEFPMNFSD 205
Query: 177 SGIQSFWINNNL--PGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRD 234
S +Q +N L GSI+ + MT LT V L N F+GP+PD S S+K +R+
Sbjct: 206 SRVQVLMLNGQKGREKLHGSISFLQKMTSLTNVTLQGNSFSGPLPDFSGLVSLKSFNVRE 265
Query: 235 NQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVF-GKDVKYNSDDISHNNFCNNNASVPC 293
NQL+G+VP SL + L +V L NN LQGP P F D+K + + + N+FC + C
Sbjct: 266 NQLSGLVPSSLFELQSLSDVALGNNLLQGPTPNFTAPDIKPDLNGL--NSFCLDTPGTSC 323
Query: 294 DARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPA 353
D RV LL IV FGYP+ FA+ W GNDPC W+ + C G IT +NF GL GTISP
Sbjct: 324 DPRVNTLLSIVEAFGYPVNFAEKWKGNDPCSGWVGITCTGTDITVINFKNLGLNGTISPR 383
Query: 354 FANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNV 413
FA+ L + L+ NNL G+IPQ LA LS L+TLDVS N L GEVP+F+ + T GN
Sbjct: 384 FADFASLRVINLSQNNLNGTIPQELAKLSNLKTLDVSKNRLCGEVPRFNTTI-VNTTGNF 442
Query: 414 WLGKNHGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKG 473
N G S+ G GS G I++ L+++G + + K
Sbjct: 443 EDCPNGNAGKKASSNAGKIVGSVIG---------ILLALLLIGVAI--------FFLVKK 485
Query: 474 LRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATIS 533
++ ++ +P+ + ++ N G S +S SG+ + L G GN IS
Sbjct: 486 KMQYHKM-HPQQQSSDQDAFKITIENLCTGVS---ESGFSGNDAHL-----GEAGNIVIS 536
Query: 534 IHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGV 593
I VLR T +F + NILGRGGFGIVYKGEL DGTKIAVKRM S KGL+EF++EI V
Sbjct: 537 IQVLRDATYNFDEKNILGRGGFGIVYKGELHDGTKIAVKRMESSIISGKGLDEFKSEIAV 596
Query: 594 LTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALD 653
LT+VRHR+LV L GYC+ GNERLLVY++MPQGTL++H+F +E G PL W +RLIIALD
Sbjct: 597 LTRVRHRNLVVLHGYCLEGNERLLVYQYMPQGTLSRHIFYWKEEGLRPLEWTRRLIIALD 656
Query: 654 VGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTF 713
V RGVEYLH+LA QSFIHRDLKPSNILLGDDM AKVADFGLV+ AP+G S+ETK+AGTF
Sbjct: 657 VARGVEYLHTLAHQSFIHRDLKPSNILLGDDMHAKVADFGLVRLAPEGTQSIETKIAGTF 716
Query: 714 GYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKEN 773
GYLAPEYA TGRVTTKVDVY+FGV+LMEL+TGRKALD + +E HL TWFRR+ NK +
Sbjct: 717 GYLAPEYAVTGRVTTKVDVYSFGVILMELLTGRKALDVARSEEEVHLATWFRRMFINKGS 776
Query: 774 IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHT 833
PKAID+ ++ +EET+ SI VAELA C++R P RPD+ H VNVL LV QW+PT +
Sbjct: 777 FPKAIDEAMEVNEETLRSINIVAELANQCSSREPRDRPDMNHVVNVLVSLVVQWKPTERS 836
Query: 834 DES--TCADDNQMSLTQ-ALQRWQANEGTSTFFNGMTSQTQSSSTSKPPVFADS 884
+S D L Q L + T T S+ +S+ S FADS
Sbjct: 837 SDSEDIYGIDYDTPLPQLILDSCFFGDNTLTSIPSRPSELESTFKSGQGRFADS 890
>ref|NP_173869.1| leucine-rich repeat family protein / protein kinase family protein
[Arabidopsis thaliana] gi|9743346|gb|AAF97970.1|
F21J9.31 [Arabidopsis thaliana]
Length = 886
Score = 738 bits (1906), Expect = 0.0
Identities = 414/843 (49%), Positives = 532/843 (62%), Gaps = 37/843 (4%)
Query: 1 MQSLRQSLKPSPNGWSSNTSFCQWSG-VKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQL 59
M +LR SLK S N S + C+WS +KC + NRVT+I + D+ ++G LP +L LT L
Sbjct: 27 MIALRDSLKLSGNPNWSGSDPCKWSMFIKCDASNRVTAIQIGDRGISGKLPPDLGKLTSL 86
Query: 60 TTLYLQNNALSGPLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSP 119
T + N L+GP+PSLA L SL V N+F+SV FSGL+SLQ +SL N
Sbjct: 87 TKFEVMRNRLTGPIPSLAGLKSLVTVYANDNDFTSVPEDFFSGLSSLQHVSLDNN-PFDS 145
Query: 120 WTFPTELTQSSNLNSIDINQAKINGTLPDIF---GSFSSLNTLHLAYNNLSGGLPNSLAG 176
W P L +++L ++G +PD FSSL TL L+YN+L P + +
Sbjct: 146 WVIPPSLENATSLVDFSAVNCNLSGKIPDYLFEGKDFSSLTTLKLSYNSLVCEFPMNFSD 205
Query: 177 SGIQSFWINNNL--PGLTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRD 234
S +Q +N L GSI+ + MT LT V L N F+GP+PD S S+K +R+
Sbjct: 206 SRVQVLMLNGQKGREKLHGSISFLQKMTSLTNVTLQGNSFSGPLPDFSGLVSLKSFNVRE 265
Query: 235 NQLTGVVPDSLVSMSGLQNVTLRNNQLQGPVPVF-GKDVKYNSDDISHNNFCNNNASVPC 293
NQL+G+VP SL + L +V L NN LQGP P F D+K + + + N+FC + C
Sbjct: 266 NQLSGLVPSSLFELQSLSDVALGNNLLQGPTPNFTAPDIKPDLNGL--NSFCLDTPGTSC 323
Query: 294 DARVMDLLHIVGGFGYPIQFAKSWTGNDPCKDWLCVICGGGKITKLNFAKQGLQGTISPA 353
D RV LL IV FGYP+ FA+ W GNDPC W+ + C G IT +NF GL GTISP
Sbjct: 324 DPRVNTLLSIVEAFGYPVNFAEKWKGNDPCSGWVGITCTGTDITVINFKNLGLNGTISPR 383
Query: 354 FANLTDLTALYLNGNNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNV 413
FA+ L + L+ NNL G+IPQ LA LS L+TLDVS N L GEVP+F+ + T GN
Sbjct: 384 FADFASLRVINLSQNNLNGTIPQELAKLSNLKTLDVSKNRLCGEVPRFNTTI-VNTTGNF 442
Query: 414 WLGKNHGGGAPGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKG 473
N G S+ G GS G I++ L+++G + + K
Sbjct: 443 EDCPNGNAGKKASSNAGKIVGSVIG---------ILLALLLIGVAI--------FFLVKK 485
Query: 474 LRRFARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHSDLHGFDGGNGGNATIS 533
++ ++ +P+ + ++ N G S +S SG+ + L G GN IS
Sbjct: 486 KMQYHKM-HPQQQSSDQDAFKITIENLCTGVS---ESGFSGNDAHL-----GEAGNIVIS 536
Query: 534 IHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGV 593
I VLR T +F + NILGRGGFGIVYKGEL DGTKIAVKRM S KGL+EF++EI V
Sbjct: 537 IQVLRDATYNFDEKNILGRGGFGIVYKGELHDGTKIAVKRMESSIISGKGLDEFKSEIAV 596
Query: 594 LTKVRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALD 653
LT+VRHR+LV L GYC+ GNERLLVY++MPQGTL++H+F +E G PL W +RLIIALD
Sbjct: 597 LTRVRHRNLVVLHGYCLEGNERLLVYQYMPQGTLSRHIFYWKEEGLRPLEWTRRLIIALD 656
Query: 654 VGRGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTF 713
V RGVEYLH+LA QSFIHRDLKPSNILLGDDM AKVADFGLV+ AP+G S+ETK+AGTF
Sbjct: 657 VARGVEYLHTLAHQSFIHRDLKPSNILLGDDMHAKVADFGLVRLAPEGTQSIETKIAGTF 716
Query: 714 GYLAPEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKEN 773
GYLAPEYA TGRVTTKVDVY+FGV+LMEL+TGRKALD + +E HL TWFRR+ NK +
Sbjct: 717 GYLAPEYAVTGRVTTKVDVYSFGVILMELLTGRKALDVARSEEEVHLATWFRRMFINKGS 776
Query: 774 IPKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHT 833
PKAID+ ++ +EET+ SI VAELA C++R P RPD+ H VNVL LV QW+PT +
Sbjct: 777 FPKAIDEAMEVNEETLRSINIVAELANQCSSREPRDRPDMNHVVNVLVSLVVQWKPTERS 836
Query: 834 DES 836
+S
Sbjct: 837 SDS 839
>gb|AAX92784.1| receptor-like kinase RHG4 [Oryza sativa (japonica cultivar-group)]
Length = 912
Score = 730 bits (1884), Expect = 0.0
Identities = 404/860 (46%), Positives = 544/860 (62%), Gaps = 24/860 (2%)
Query: 14 GWSSNTSFCQWSGVKCSSDN--RVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSG 71
GW + + C + GV C +VT +NL+D+ L+GTLPD+L+SLT LT L LQ NAL+G
Sbjct: 45 GWDGD-NVCGFEGVTCERGGAGKVTELNLADRGLSGTLPDSLSSLTSLTALQLQGNALTG 103
Query: 72 PLPSLANLSSLTDVNLGSNNFSSVTPGAFSGLNSLQTLSLGENINLSPWTFPTELTQSSN 131
+PSLA + SL + L N F+S+ P GL SLQ L++ EN+ L PW P + S+
Sbjct: 104 AVPSLARMGSLARLALDGNAFTSLPPDFLHGLTSLQYLTM-ENLPLPPWPVPDAIANCSS 162
Query: 132 LNSIDINQAKINGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGS-GIQSFWINNNLPG 190
L++ + A I+G P + + SL L L+YNNL+GGLP L+ ++S +NN
Sbjct: 163 LDTFSASNASISGPFPAVLATLVSLRNLRLSYNNLTGGLPPELSSLIAMESLQLNNQRSD 222
Query: 191 --LTGSITVISNMTLLTQVWLHVNKFTGPIPDLSQCNSIKDLQLRDNQLTGVVPDSLVSM 248
L+G I VI++M L +W+ NKFTGPIPDL+ ++ +RDN LTGVVP SL +
Sbjct: 223 DKLSGPIDVIASMKSLKLLWIQSNKFTGPIPDLNG-TQLEAFNVRDNMLTGVVPPSLTGL 281
Query: 249 SGLQNVTLRNNQLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFG 308
L+NV+L NN QGP P F D+ S N FC N PC LL + GFG
Sbjct: 282 MSLKNVSLSNNNFQGPKPAFAAIP--GQDEDSGNGFCLNTPG-PCSPLTTTLLQVAEGFG 338
Query: 309 YPIQFAKSWTGNDPCKD-WLCVICGGGKITKLNFAKQGLQGTISPAFANLTDLTALYLNG 367
YP + AK+W GNDPC W+ ++C ++ +N +++ L G ISPA ANLT L L L+
Sbjct: 339 YPYELAKTWKGNDPCSPAWVGIVCTSSDVSMINLSRKNLSGRISPALANLTRLARLDLSN 398
Query: 368 NNLTGSIPQNLATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWL----GKNHGGGA 423
NNLTG IP L TL L L+V+NN L+GEVPKF P V + GN++ GGG+
Sbjct: 399 NNLTGVIPDVLTTLPSLTVLNVANNRLTGEVPKFKPSVNVLAQGNLFGQSSGSSGGGGGS 458
Query: 424 PGSAPGGSPAGSGKGASMKKVWIIIIIVLIVVGFVVGGAWFSWKCYSRKGLRRFARVGNP 483
G + AG GK + I II+ +I++ + K + + R + +P
Sbjct: 459 DGDSSSSDSAGGGKSKPNTGMIIGIIVAVIILFACIALLVHHRKKKNVEKFRPVSTKTSP 518
Query: 484 ENGEGNVKLDLAS---VSNGYGGASSELQSQ-SSGDHSDLHGFDGGNGGNATISIHVLRQ 539
E +K+ + +SNG +EL S S+ + S++ +G +S+ VL +
Sbjct: 519 AESE-MMKIQVVGANGISNGSSAFPTELYSHVSAANSSNISELFESHG--MQLSVEVLLK 575
Query: 540 VTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRMISVAKGSKGLNEFQAEIGVLTKVRH 599
TN+FS+D ILGRGGFG+V+KG L +G +AVKR S G+KG EF AEI VL KVRH
Sbjct: 576 ATNNFSEDCILGRGGFGVVFKGNL-NGKLVAVKRCDSGTMGTKGQEEFLAEIDVLRKVRH 634
Query: 600 RHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGRGVE 659
RHLVALLGYC +GNERLLVYE+M GTL +HL + ++ G+ PLTW QR+ IALDV RG+E
Sbjct: 635 RHLVALLGYCTHGNERLLVYEYMSGGTLREHLCDLQQSGFIPLTWTQRMTIALDVARGIE 694
Query: 660 YLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYLAPE 719
YLH LAQ++FIHRDLKPSNILL D+RAKV+DFGLVK A D + S+ T++AGTFGYLAPE
Sbjct: 695 YLHGLAQETFIHRDLKPSNILLDQDLRAKVSDFGLVKLAKDTDKSLMTRIAGTFGYLAPE 754
Query: 720 YAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKENIPKAID 779
YA TG+VTTKVDVYA+GV+LME+ITGRK LDDS+PD+ +HLVT FRR + +KE K +D
Sbjct: 755 YATTGKVTTKVDVYAYGVILMEMITGRKVLDDSLPDDETHLVTIFRRNILDKEKFRKFVD 814
Query: 780 QTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTDESTCA 839
TL+ E S+ +VA+LA HCT R PYQRPD+ H VN L LV QW+PT ++
Sbjct: 815 PTLELSAEGWTSLLEVADLARHCTAREPYQRPDMCHCVNRLSSLVDQWKPTNIDEDDYEG 874
Query: 840 DDNQMSLTQALQRWQANEGT 859
+ ++M L Q L++W+ ++ T
Sbjct: 875 ETSEMGLHQQLEKWRCDDFT 894
>dbj|BAD95052.1| putative receptor-like protein kinase [Arabidopsis thaliana]
Length = 306
Score = 375 bits (962), Expect = e-102
Identities = 189/306 (61%), Positives = 227/306 (73%), Gaps = 11/306 (3%)
Query: 597 VRHRHLVALLGYCINGNERLLVYEHMPQGTLTQHLFECREHGYTPLTWKQRLIIALDVGR 656
+RHRHLVALLGYC++GNERLLVYE+MP+GTL+QHLF +E G PL W +RL IALDV R
Sbjct: 1 MRHRHLVALLGYCLDGNERLLVYEYMPRGTLSQHLFHWKEEGRKPLDWTRRLAIALDVAR 60
Query: 657 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGNYSVETKLAGTFGYL 716
GVEYLH+LA QSFIHRDLKPSNILLGDDMRAKV+DFGLV+ APDG YS+ET++AGTFGYL
Sbjct: 61 GVEYLHTLAHQSFIHRDLKPSNILLGDDMRAKVSDFGLVRLAPDGKYSIETRVAGTFGYL 120
Query: 717 APEYAATGRVTTKVDVYAFGVVLMELITGRKALDDSVPDESSHLVTWFRRVLTNKEN--I 774
APEYA TGRVTTKVD+++ GV+LMELITGRKALD++ P++S HLVTWFRRV +K+
Sbjct: 121 APEYAVTGRVTTKVDIFSLGVILMELITGRKALDETQPEDSVHLVTWFRRVAASKDENAF 180
Query: 775 PKAIDQTLDPDEETMLSIYKVAELAGHCTTRSPYQRPDIGHAVNVLCPLVQQWEPTTHTD 834
AID + D++T+ SI KV ELAGHC R PYQRPD+ H VNVL L QW+PT
Sbjct: 181 KNAIDPNISLDDDTVASIEKVWELAGHCCAREPYQRPDMAHIVNVLSSLTVQWKPTETDP 240
Query: 835 ESTCADDNQMSLTQALQRWQANE---------GTSTFFNGMTSQTQSSSTSKPPVFADSL 885
+ D M L Q L++WQA E G+S+ G TQ+S ++P FADS
Sbjct: 241 DDVYGIDYDMPLPQVLKKWQAFEGLSQTADDSGSSSSAYGSKDNTQTSIPTRPSGFADSF 300
Query: 886 HSPDCR 891
S D R
Sbjct: 301 TSVDGR 306
>gb|AAX38303.1| receptor-like protein kinase [Solanum habrochaites]
Length = 628
Score = 338 bits (868), Expect = 4e-91
Identities = 215/591 (36%), Positives = 310/591 (52%), Gaps = 53/591 (8%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F S+ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDSIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHLWLHGNQFSGKIPVEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAP----GSAPGGSPA 433
+L L LD+SNN++S +PKF+ +K + +GN L N G P + P SP
Sbjct: 400 WTSLKSLSVLDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 434 GS----------------GKGASMKK--VWIIIIIVLIVVGFV----VGGAWFSWKCYSR 471
S G+ + KK I I+V+ + GF+ + + + C
Sbjct: 460 SSVPPSRPNSSSSVIFKPGEQSPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYVCKKS 519
Query: 472 KGLRR-----FARVGNPENGEGNVKLDLASVSNG---YGGASSELQSQSSGDHSDLHGFD 523
K + +P + + VK+ +A+ +NG AS QS H
Sbjct: 520 KDKHQAPTALVVHPRDPSDSDNVVKIAIANQTNGSLSTVNASGSASIQSGESHMI----- 574
Query: 524 GGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 575 --EAGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 623
Score = 59.3 bits (142), Expect = 5e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSVLDLSNNNISPPLP 421
>gb|AAX38299.1| receptor-like protein kinase [Solanum habrochaites]
gi|61105037|gb|AAX38298.1| receptor-like protein kinase
[Solanum habrochaites]
Length = 628
Score = 338 bits (868), Expect = 4e-91
Identities = 215/591 (36%), Positives = 310/591 (52%), Gaps = 53/591 (8%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F S+ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDSIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHLWLHGNQFSGKIPVEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVQSWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAP----GSAPGGSPA 433
+L L LD+SNN++S +PKF+ +K + +GN L N G P + P SP
Sbjct: 400 WTSLKSLSVLDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 434 GS----------------GKGASMKK--VWIIIIIVLIVVGFV----VGGAWFSWKCYSR 471
S G+ + KK I I+V+ + GF+ + + + C
Sbjct: 460 SSVPPSRPNSSSSVIFKPGEQSPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYVCKKS 519
Query: 472 KGLRR-----FARVGNPENGEGNVKLDLASVSNG---YGGASSELQSQSSGDHSDLHGFD 523
K + +P + + VK+ +A+ +NG AS QS H
Sbjct: 520 KDKHQAPTALVVHPRDPSDSDNVVKIAIANQTNGSLSTVNASGSASIQSGESHMI----- 574
Query: 524 GGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 575 --EAGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 623
Score = 59.3 bits (142), Expect = 5e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSVLDLSNNNISPPLP 421
>gb|AAX38300.1| receptor-like protein kinase [Solanum habrochaites]
Length = 628
Score = 338 bits (867), Expect = 5e-91
Identities = 214/591 (36%), Positives = 310/591 (52%), Gaps = 53/591 (8%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F ++ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDTIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHIWLHGNQFSGKIPVEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAP----GSAPGGSPA 433
+L L LD+SNN++S +PKF+ +K + +GN L N G P + P SP
Sbjct: 400 WTSLKSLSVLDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 434 GS----------------GKGASMKK--VWIIIIIVLIVVGFV----VGGAWFSWKCYSR 471
S G+ + KK I I+V+ + GF+ + + + C
Sbjct: 460 SSVPPSRPNSSSSVIFKPGEQSPEKKDSKSKIAIVVVPIAGFLLLVFLAIPLYIYVCKKS 519
Query: 472 KGLRR-----FARVGNPENGEGNVKLDLASVSNG---YGGASSELQSQSSGDHSDLHGFD 523
K + +P + + VK+ +A+ +NG AS QS H
Sbjct: 520 KDKHQAPTALVVHPRDPSDSDNVVKIAIANQTNGSLSTVNASGSASIQSGESHMI----- 574
Query: 524 GGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 575 --EAGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 623
Score = 59.3 bits (142), Expect = 5e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLESVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSVLDLSNNNISPPLP 421
>gb|AAX38287.1| receptor-like protein kinase [Lycopersicon peruvianum]
Length = 629
Score = 337 bits (865), Expect = 8e-91
Identities = 213/592 (35%), Positives = 314/592 (52%), Gaps = 54/592 (9%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F ++ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDTIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSTNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHIWLHGNQFSGKIPVEIGNITNLKDLSVNTNNLVGLIPESLANMR-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLVTVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGGAP----GSAPGGSPA 433
+L L LD+SNN++S +PKF+ +K + +GN L N G P + P SP
Sbjct: 400 WTSLKSLSILDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 434 GS----------------------GKGASMKKVWIIIIIV---LIVVGFVVGGAWFSWKC 468
S K S K+ I+++ + L++V + + + C
Sbjct: 460 SSVVPSSRPNSSSSVIFKPGEQSPEKKDSKSKIAIVVVPIAGSLLLVFLAI--PLYIYVC 517
Query: 469 YSRKGLRR-----FARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHS-DLHGF 522
K + +P + + VK+ +A+ +N G S+ S S+ HS + H
Sbjct: 518 KKSKDKHQAPTALVVHPRDPSDSDNVVKIAIANQTN--GSLSTVNASGSASIHSGESHMI 575
Query: 523 DGGNGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
+ GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 576 E---AGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 624
Score = 59.3 bits (142), Expect = 5e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLVTVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSILDLSNNNISPPLP 421
>gb|AAX38325.1| receptor-like protein kinase [Lycopersicon pimpinellifolium]
Length = 628
Score = 337 bits (863), Expect = 1e-90
Identities = 212/589 (35%), Positives = 313/589 (52%), Gaps = 49/589 (8%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F ++ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDTIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSSNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHLWLHGNQFSGKIPVEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLETVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGG--------APGSAPG 429
+L L LD+SNN++S +PKF+ +K + +GN L N G P +P
Sbjct: 400 WTSLKSLSILDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 430 GSPAGSGKGASMKKVW--------------IIIIIVLIVVGFV----VGGAWFSWKCYSR 471
S S +S ++ I I+V+ + GF+ + + + C
Sbjct: 460 SSVPSSRPNSSSSVIFKPSEQSPEKKDSKSKIAIVVVPIAGFLLLVCLAIPLYIYVCKKS 519
Query: 472 KGLRR-----FARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHS-DLHGFDGG 525
K + +P + + VK+ +A+ +N G S+ S S+ HS + H +
Sbjct: 520 KDKHQAPTALVVHPRDPSDSDNVVKIAIANQTN--GSLSTVNASGSASIHSGESHMIE-- 575
Query: 526 NGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 576 -AGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 623
Score = 58.9 bits (141), Expect = 7e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLETVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSILDLSNNNISPPLP 421
>gb|AAX38324.1| receptor-like protein kinase [Lycopersicon pimpinellifolium]
gi|61105081|gb|AAX38320.1| receptor-like protein kinase
[Lycopersicon pimpinellifolium]
gi|61105079|gb|AAX38319.1| receptor-like protein kinase
[Lycopersicon pimpinellifolium]
gi|61105077|gb|AAX38318.1| receptor-like protein kinase
[Lycopersicon pimpinellifolium]
gi|61105075|gb|AAX38317.1| receptor-like protein kinase
[Lycopersicon pimpinellifolium]
gi|61105073|gb|AAX38316.1| receptor-like protein kinase
[Lycopersicon pimpinellifolium]
Length = 628
Score = 337 bits (863), Expect = 1e-90
Identities = 212/589 (35%), Positives = 313/589 (52%), Gaps = 49/589 (8%)
Query: 24 WSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGPLPSLANLSSLT 83
W + CS +R+ I + L G LP NLN L++LT L LQ N SG LPS + LS L+
Sbjct: 46 WPHIVCSG-SRIQQIQVMGLGLKGPLPQNLNKLSRLTHLGLQKNQFSGKLPSFSGLSELS 104
Query: 84 DVNLGSNNFSSVTPGAFSGLNSLQTLSLGEN-INL-SPWTFPTELTQSSNLNSIDINQAK 141
L N F ++ F GL +LQ L+L EN +N S W+ P L S+ L ++ +
Sbjct: 105 FAYLDFNQFDTIPLDFFDGLVNLQVLALDENPLNATSGWSLPNGLQDSAQLINLTMINCN 164
Query: 142 INGTLPDIFGSFSSLNTLHLAYNNLSGGLPNSLAGSGIQSFWINNNL-PGLTGSITVISN 200
+ G LP+ G+ SSL L L+ N LSG +P + + ++ W+N+ G++GSI V++
Sbjct: 165 LAGPLPEFLGTMSSLEVLLLSSNRLSGPIPGTFKDAVLKMLWLNDQSGDGMSGSIDVVAT 224
Query: 201 MTLLTQVWLHVNKFTGPIP-DLSQCNSIKDLQLRDNQLTGVVPDSLVSMSGLQNVTLRNN 259
M LT +WLH N+F+G IP ++ ++KDL + N L G++P+SL +M L N+ L NN
Sbjct: 225 MVSLTHLWLHGNQFSGKIPVEIGNLTNLKDLSVNTNNLVGLIPESLANMP-LDNLDLNNN 283
Query: 260 QLQGPVPVFGKDVKYNSDDISHNNFCNNNASVPCDARVMDLLHIVGGFGYPIQFAKSWTG 319
GPVP F K + N+FC C VM LL + G YP + +SW+G
Sbjct: 284 HFMGPVPKF----KATNVSFMSNSFCQTKQGAVCAPEVMALLEFLDGVNYPSRLVESWSG 339
Query: 320 NDPCKD-WLCVICGGG-KITKLNFAKQGLQGTISPAFANLTDLTALYLNGNNLTGSIPQN 377
N+PC W + C K++ +N K L GT+SP+ ANL +T +YL NNL+G +P +
Sbjct: 340 NNPCDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLETVTRIYLESNNLSGFVPSS 399
Query: 378 LATLSQLETLDVSNNDLSGEVPKFSPKVKFITDGNVWLGKNHGGG--------APGSAPG 429
+L L LD+SNN++S +PKF+ +K + +GN L N G P +P
Sbjct: 400 WTSLKSLSILDLSNNNISPPLPKFTTPLKLVLNGNPKLTSNPPGANPSPNNSTTPADSPT 459
Query: 430 GSPAGSGKGASMKKVW--------------IIIIIVLIVVGFV----VGGAWFSWKCYSR 471
S S +S ++ I I+V+ + GF+ + + + C
Sbjct: 460 SSVPSSRPNSSSSVIFKPSEQSPEKKDSKSKIAIVVVPIAGFLLLVCLAIPLYIYVCKKS 519
Query: 472 KGLRR-----FARVGNPENGEGNVKLDLASVSNGYGGASSELQSQSSGDHS-DLHGFDGG 525
K + +P + + VK+ +A+ +N G S+ S S+ HS + H +
Sbjct: 520 KDKHQAPTALVVHPRDPSDSDNVVKIAIANQTN--GSLSTVNASGSASIHSGESHMIE-- 575
Query: 526 NGGNATISIHVLRQVTNDFSDDNILGRGGFGIVYKGELPDGTKIAVKRM 574
GN IS+ VLR VT +FS +N LGRGGFG+VYKGEL DGT+IAVKRM
Sbjct: 576 -AGNLLISVQVLRNVTKNFSPENELGRGGFGVVYKGELDDGTQIAVKRM 623
Score = 58.9 bits (141), Expect = 7e-07
Identities = 33/86 (38%), Positives = 52/86 (60%), Gaps = 4/86 (4%)
Query: 15 WSSNTSFC--QWSGVKCSSDNRVTSINLSDQKLAGTLPDNLNSLTQLTTLYLQNNALSGP 72
WS N C +W G+ C + +V+ INL L+GTL ++ +L +T +YL++N LSG
Sbjct: 337 WSGNNP-CDGRWWGISCDDNQKVSVINLPKSNLSGTLSPSIANLETVTRIYLESNNLSGF 395
Query: 73 LP-SLANLSSLTDVNLGSNNFSSVTP 97
+P S +L SL+ ++L +NN S P
Sbjct: 396 VPSSWTSLKSLSILDLSNNNISPPLP 421
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,639,663,320
Number of Sequences: 2540612
Number of extensions: 76403629
Number of successful extensions: 377839
Number of sequences better than 10.0: 23857
Number of HSP's better than 10.0 without gapping: 10387
Number of HSP's successfully gapped in prelim test: 13518
Number of HSP's that attempted gapping in prelim test: 261514
Number of HSP's gapped (non-prelim): 55794
length of query: 891
length of database: 863,360,394
effective HSP length: 137
effective length of query: 754
effective length of database: 515,296,550
effective search space: 388533598700
effective search space used: 388533598700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146585.14


