

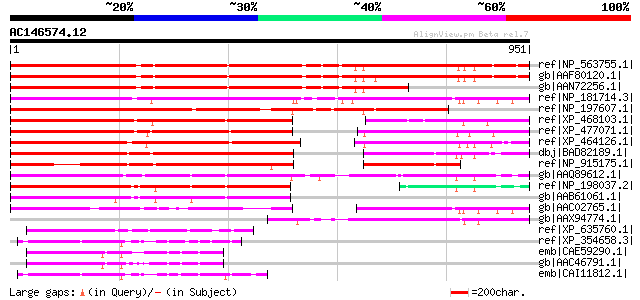
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146574.12 - phase: 0 /pseudo
(951 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_563755.1| expressed protein [Arabidopsis thaliana] 833 0.0
gb|AAF80120.1| Contains similarity to an unknown protein T11A7.7... 827 0.0
gb|AAN72256.1| At1g05960/T21E18_20 [Arabidopsis thaliana] gi|141... 719 0.0
ref|NP_181714.3| cyclin-related [Arabidopsis thaliana] 677 0.0
ref|NP_197607.1| expressed protein [Arabidopsis thaliana] 599 e-169
ref|XP_468103.1| cyclin-like protein [Oryza sativa (japonica cul... 567 e-160
ref|XP_477071.1| cyclin-like protein [Oryza sativa (japonica cul... 561 e-158
ref|XP_464126.1| cyclin-like protein [Oryza sativa (japonica cul... 558 e-157
dbj|BAD82189.1| cyclin-like [Oryza sativa (japonica cultivar-gro... 553 e-155
ref|NP_915175.1| P0674H09.27 [Oryza sativa (japonica cultivar-gr... 453 e-125
gb|AAQ89612.1| At5g26850 [Arabidopsis thaliana] gi|26449532|dbj|... 438 e-121
ref|NP_198037.2| expressed protein [Arabidopsis thaliana] 388 e-106
gb|AAB61061.1| Hypothetical protein F2P16.24 [Arabidopsis thalia... 372 e-101
gb|AAC02765.1| hypothetical protein [Arabidopsis thaliana] gi|25... 344 9e-93
gb|AAX94774.1| cyclin-related protein 1 [Cucumis sativus] 342 3e-92
ref|XP_635760.1| hypothetical protein DDB0219543 [Dictyostelium ... 87 3e-15
ref|XP_354658.3| PREDICTED: similar to KIAA0953 protein [Mus mus... 83 4e-14
emb|CAE59290.1| Hypothetical protein CBG02625 [Caenorhabditis br... 82 8e-14
gb|AAC46791.1| Hypothetical protein C32D5.3 [Caenorhabditis eleg... 81 2e-13
emb|CAI11812.1| novel protein [Danio rerio] 81 2e-13
>ref|NP_563755.1| expressed protein [Arabidopsis thaliana]
Length = 982
Score = 833 bits (2152), Expect = 0.0
Identities = 485/1007 (48%), Positives = 658/1007 (65%), Gaps = 82/1007 (8%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PACGNLC FCPSLRARSR PVKRYKK++AEI PRN+ AE NDRKIGKLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
AS+NPLRIPKITE LEQ+CYK+LRN + GSVKV+LCIY+KLLSSC+EQ+PLF+ SLL I+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
RTLLEQT+ +EV+ILGCNTLVDFI QT ++MFNLEG IPKLCQLAQE+GDDER+L LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQ-SKSNLAKVEKLNSQSQNQLV 239
SAG+Q L+ MV F+GEHS LSMD D IIS ILENY+DL+ + + +V++++
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILENYMDLEKGQEDTKEVDQISD------- 233
Query: 240 QEFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPL 299
+ P VS N T +++E+ +D +K+P+YWS VCL NIAKLAKE TTVRRVLEPL
Sbjct: 234 TKIPNMTKKVSFKPNPVTDYKLEN-MDISKSPSYWSMVCLCNIAKLAKETTTVRRVLEPL 292
Query: 300 FHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQID 359
FD+ ++WS +KGVA VL++LQ L ESG N H+++S L+KHLDHKNV KQ LQI+
Sbjct: 293 LTAFDSGDYWSPQKGVASSVLLFLQSRLEESGENCHVLVSSLIKHLDHKNVIKQQGLQIN 352
Query: 360 IINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIE 419
++N+ T +A + KQQAS A+ I+DLIKHLR+CLQN+AE +D+ D N+ LQ ++E
Sbjct: 353 MVNVATCLALHAKQQASGAMTAVIADLIKHLRKCLQNAAE-SDVSVDKTKQNSDLQHALE 411
Query: 420 MCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKA 479
CI +LSNKVGDAGPI D+ AVVLE +S++ +++RTT SA+ + A +++ VPNV YH K
Sbjct: 412 NCIAELSNKVGDAGPILDMFAVVLETISTNVVLSRTTASAILRAAHIVSVVPNVSYHKKV 471
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKISKKVESDGLSIQHE 539
FPDALFHQLLLAM+H D T++ AH+I S+VL+ ++ PW DQ K + + S LS+
Sbjct: 472 FPDALFHQLLLAMSHADCTTRVEAHNIFSVVLLGTLRLPWSDQHKETSEAVSGSLSV--- 528
Query: 540 SLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI 599
G + + E++ + SL S + ++ S V Q L+ + + +
Sbjct: 529 --DGICTVRNQEEEKEKVEKSLNSELCKDVN---HISRPSVSGQTSQQLSCQSLDSLKDL 583
Query: 600 *DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMR-------QWHTLIALLYYLLVL----- 647
D + K + S + ++L +L I + +A Y + +L
Sbjct: 584 DDGI-----KSLCSLRLSSHQVNMLLSSLWIQATSTDNTPENFEAMASTYQITLLFSLAK 638
Query: 648 --SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKV 705
++MALV+CFQLAFSLR++SL+Q+GG+ SRRRS+ T AS+MLIF A+ ++ +L+P +
Sbjct: 639 RSNHMALVQCFQLAFSLRNLSLNQDGGMQHSRRRSIFTFASYMLIFGAKISNILELVPII 698
Query: 706 KASLTEAPVDPFLELVDDNLLRAVCIK-SDKVVFGSVEDEVAAMKSLSAVQLDDRQLKET 764
K SLT VDP+L L D LRAVC + +GS +D+ AA+ S S + DDR+LKE
Sbjct: 699 KESLTAQMVDPYLVLEGDIRLRAVCSGFPQEETYGSDKDDSAALNS-SVIVTDDRRLKEI 757
Query: 765 VISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQIEFP--- 821
VI++F +K L E+E +++ ++ FS DDA+ G LF +TP P SPL Q E P
Sbjct: 758 VITHFTSKLQTLSEEEQLNLRKEIQSDFSLDDAHSLGGQLFTDTPGPSSPLNQTELPAFE 817
Query: 822 -----DVDEMKGVGP--------------------N*VEASQIVEHHFQPTAQM------ 850
D+ +G+ P + + ++++E + Q+
Sbjct: 818 EVELSDIAAFEGISPGASGSQSGHRTSLSTNTNPVDVLSVNELLESVSETARQVASLPVS 877
Query: 851 -----F*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEVSRQPVKALEYS- 904
+ ++N L TGKQQKM +RSFK Q TKAI S +NE+ + +K E +
Sbjct: 878 SIPVPYDQMMNQCEALVTGKQQKMSVLRSFK-PQATKAIT-SEDNEKDEQYLLKETEEAG 935
Query: 905 KGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
+ D K + Q Q Q+ F SQ+ Q+S RLPPSSPYDKFLKAAG
Sbjct: 936 EDDEKAIIVADVQPQGQLGFFSQEV-PQNSFRLPPSSPYDKFLKAAG 981
>gb|AAF80120.1| Contains similarity to an unknown protein T11A7.7 gi|2335096 from
Arabidopsis thaliana BAC T11A7 gb|AC002339 and contains
a tropomyosin PF|00261 domain. ESTs gb|AI995205,
gb|N37925, gb|F13889, gb|AV523107, gb|AV535948,
gb|AV558461, gb|F13888 come from this gene
gi|25406886|pir||F86194 hypothetical protein [imported]
- Arabidopsis thaliana
Length = 1628
Score = 827 bits (2137), Expect = 0.0
Identities = 486/1014 (47%), Positives = 658/1014 (63%), Gaps = 89/1014 (8%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PACGNLC FCPSLRARSR PVKRYKK++AEI PRN+ AE NDRKIGKLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
AS+NPLRIPKITE LEQ+CYK+LRN + GSVKV+LCIY+KLLSSC+EQ+PLF+ SLL I+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
RTLLEQT+ +EV+ILGCNTLVDFI QT ++MFNLEG IPKLCQLAQE+GDDER+L LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQ-SKSNLAKVEKLNSQSQNQLV 239
SAG+Q L+ MV F+GEHS LSMD D IIS ILENY+DL+ + + +V++++
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILENYMDLEKGQEDTKEVDQISD------- 233
Query: 240 QEFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPL 299
+ P VS N T +++E+ +D +K+P+YWS VCL NIAKLAKE TTVRRVLEPL
Sbjct: 234 TKIPNMTKKVSFKPNPVTDYKLEN-MDISKSPSYWSMVCLCNIAKLAKETTTVRRVLEPL 292
Query: 300 FHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQID 359
FD+ ++WS +KGVA VL++LQ L ESG N H+++S L+KHLDHKNV KQ LQI+
Sbjct: 293 LTAFDSGDYWSPQKGVASSVLLFLQSRLEESGENCHVLVSSLIKHLDHKNVIKQQGLQIN 352
Query: 360 IINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIE 419
++N+ T +A + KQQAS A+ I+DLIKHLR+CLQN+AE +D+ D N+ LQ ++E
Sbjct: 353 MVNVATCLALHAKQQASGAMTAVIADLIKHLRKCLQNAAE-SDVSVDKTKQNSDLQHALE 411
Query: 420 MCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKA 479
CI +LSNKVGDAGPI D+ AVVLE +S++ +++RTT SA+ + A +++ VPNV YH K
Sbjct: 412 NCIAELSNKVGDAGPILDMFAVVLETISTNVVLSRTTASAILRAAHIVSVVPNVSYHKKV 471
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKISKKVESDGLSIQHE 539
FPDALFHQLLLAM+H D T++ AH+I S+VL+ ++ PW DQ K + + S LS+
Sbjct: 472 FPDALFHQLLLAMSHADCTTRVEAHNIFSVVLLGTLRLPWSDQHKETSEAVSGSLSV--- 528
Query: 540 SLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI 599
G + + E++ + SL S + ++ S V Q L+ + + +
Sbjct: 529 --DGICTVRNQEEEKEKVEKSLNSELCKDVN---HISRPSVSGQTSQQLSCQSLDSLKDL 583
Query: 600 *DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMR-------QWHTLIALLYYLLVL----- 647
D + K + S + ++L +L I + +A Y + +L
Sbjct: 584 DDGI-----KSLCSLRLSSHQVNMLLSSLWIQATSTDNTPENFEAMASTYQITLLFSLAK 638
Query: 648 --SYMALVRCFQLAFSLRSISLDQ-------EGGLPPSRRRSLLTLASHMLIFSARAADF 698
++MALV+CFQLAFSLR++SL+Q EGG+ SRRRS+ T AS+MLIF A+ ++
Sbjct: 639 RSNHMALVQCFQLAFSLRNLSLNQDDFWYNVEGGMQHSRRRSIFTFASYMLIFGAKISNI 698
Query: 699 SDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIK-SDKVVFGSVEDEVAAMKSLSAVQLD 757
+L+P +K SLT VDP+L L D LRAVC + +GS +D+ AA+ S S + D
Sbjct: 699 LELVPIIKESLTAQMVDPYLVLEGDIRLRAVCSGFPQEETYGSDKDDSAALNS-SVIVTD 757
Query: 758 DRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQ 817
DR+LKE VI++F +K L E+E +++ ++ FS DDA+ G LF +TP P SPL Q
Sbjct: 758 DRRLKEIVITHFTSKLQTLSEEEQLNLRKEIQSDFSLDDAHSLGGQLFTDTPGPSSPLNQ 817
Query: 818 IEFP--------DVDEMKGVGP--------------------N*VEASQIVEHHFQPTAQ 849
E P D+ +G+ P + + ++++E + Q
Sbjct: 818 TELPAFEEVELSDIAAFEGISPGASGSQSGHRTSLSTNTNPVDVLSVNELLESVSETARQ 877
Query: 850 M-----------F*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEVSRQPV 898
+ + ++N L TGKQQKM +RSFK Q TKAI S +NE+ + +
Sbjct: 878 VASLPVSSIPVPYDQMMNQCEALVTGKQQKMSVLRSFK-PQATKAIT-SEDNEKDEQYLL 935
Query: 899 KALEYS-KGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
K E + + D K + Q Q Q+ F SQ+ Q+S RLPPSSPYDKFLKAAG
Sbjct: 936 KETEEAGEDDEKAIIVADVQPQGQLGFFSQEV-PQNSFRLPPSSPYDKFLKAAG 988
>gb|AAN72256.1| At1g05960/T21E18_20 [Arabidopsis thaliana]
gi|14194169|gb|AAK56279.1| At1g05960/T21E18_20
[Arabidopsis thaliana]
Length = 731
Score = 719 bits (1856), Expect = 0.0
Identities = 394/745 (52%), Positives = 524/745 (69%), Gaps = 37/745 (4%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PACGNLC FCPSLRARSR PVKRYKK++AEI PRN+ AE NDRKIGKLCEY
Sbjct: 1 MGVMSRRVLPACGNLCFFCPSLRARSRHPVKRYKKMLAEIFPRNQEAEPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
AS+NPLRIPKITE LEQ+CYK+LRN + GSVKV+LCIY+KLLSSC+EQ+PLF+ SLL I+
Sbjct: 61 ASRNPLRIPKITEYLEQKCYKELRNGNIGSVKVVLCIYKKLLSSCKEQMPLFSCSLLSIV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
RTLLEQT+ +EV+ILGCNTLVDFI QT ++MFNLEG IPKLCQLAQE+GDDER+L LR
Sbjct: 121 RTLLEQTKEEEVQILGCNTLVDFISLQTVNSHMFNLEGLIPKLCQLAQEMGDDERSLQLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQ-SKSNLAKVEKLNSQSQNQLV 239
SAG+Q L+ MV F+GEHS LSMD D IIS ILENY+DL+ + + +V++++
Sbjct: 181 SAGMQALAFMVSFIGEHSQLSMDLDMIISVILENYMDLEKGQEDTKEVDQISD------- 233
Query: 240 QEFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPL 299
+ P VS N T +++E+ +D +K+P+YWS VCL NIAKLAKE TTVRRVLEPL
Sbjct: 234 TKIPNMTKKVSFKPNPVTDYKLEN-MDISKSPSYWSMVCLCNIAKLAKETTTVRRVLEPL 292
Query: 300 FHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQID 359
FD+ ++WS +KGVA VL++LQ L ESG N H+++S L+KHLDHKNV KQ LQI+
Sbjct: 293 LTAFDSGDYWSPQKGVASSVLLFLQSRLEESGENCHVLVSSLIKHLDHKNVIKQQGLQIN 352
Query: 360 IINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIE 419
++N+ T +A + KQQAS A+ I+DLIKHLR+CLQN+AE +D+ D N+ LQ ++E
Sbjct: 353 MVNVATCLALHAKQQASGAMTAVIADLIKHLRKCLQNAAE-SDVSVDKTKQNSDLQHALE 411
Query: 420 MCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKA 479
CI +LSNKVGDAGPI D+ AVVLE +S++ +++RTT SA+ + A +++ VPNV YH K
Sbjct: 412 NCIAELSNKVGDAGPILDMFAVVLETISTNVVLSRTTASAILRAAHIVSVVPNVSYHKKV 471
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKISKKVESDGLSIQHE 539
FPDALFHQLLLAM+H D T++ AH+I S+VL+ ++ PW DQ K + + S LS+
Sbjct: 472 FPDALFHQLLLAMSHADCTTRVEAHNIFSVVLLGTLRLPWSDQHKETSEAVSGSLSV--- 528
Query: 540 SLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI 599
G + + E++ + SL S + ++ S V Q L+ + + +
Sbjct: 529 --DGICTVRNQEEEKEKVEKSLNSELCKDVN---HISRPSVSGQTSQQLSCQSLDSLKDL 583
Query: 600 *DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMR-------QWHTLIALLYYLLVL----- 647
D + K + S + ++L +L I + +A Y + +L
Sbjct: 584 DDGI-----KSLCSLRLSSHQVNMLLSSLWIQATSTDNTPENFEAMASTYQITLLFSLAK 638
Query: 648 --SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKV 705
++MALV+CFQLAFSLR++SL+Q+GG+ SRRRS+ T AS+MLIF A+ ++ +L+P +
Sbjct: 639 RSNHMALVQCFQLAFSLRNLSLNQDGGMQHSRRRSIFTFASYMLIFGAKISNILELVPII 698
Query: 706 KASLTEAPVDPFLELVDDNLLRAVC 730
K SLT VDP+L L D LRAVC
Sbjct: 699 KESLTAQMVDPYLVLEGDIRLRAVC 723
>ref|NP_181714.3| cyclin-related [Arabidopsis thaliana]
Length = 1025
Score = 677 bits (1746), Expect = 0.0
Identities = 408/1044 (39%), Positives = 610/1044 (58%), Gaps = 119/1044 (11%)
Query: 2 GVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYA 61
GV+SR+V+P CG+LCI CP+LRARSRQPVKRYKKLIAEI PRN+ +NDRKIGKLCEYA
Sbjct: 6 GVISRQVLPVCGSLCILCPALRARSRQPVKRYKKLIAEIFPRNQEEGINDRKIGKLCEYA 65
Query: 62 SKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGIIR 121
+KN +R+PKI+++LE RCYK+LRNE+F S K+ +CIYR+LL +C+EQIPLF+S L ++
Sbjct: 66 AKNAVRMPKISDSLEHRCYKELRNENFHSAKIAMCIYRRLLVTCKEQIPLFSSGFLRTVQ 125
Query: 122 TLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLRS 181
LL+QTR DE++I+GC +L +F+I Q DG+ +FNLEGF+PKLCQL E GDD+R+ LR+
Sbjct: 126 ALLDQTRQDEMQIVGCQSLFEFVINQKDGSSLFNLEGFLPKLCQLGLEGGDDDRSRSLRA 185
Query: 182 AGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQE 241
AGLQ LS+M+ MGE+SH+ +FD ++SA+LENY + +N + S + V E
Sbjct: 186 AGLQALSAMIWLMGEYSHIPSEFDNVVSAVLENYGHPKILTNA-------NDSGRKWVDE 238
Query: 242 FPKEEAHVS---SMLNVAT-------GFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATT 291
K E HV+ S++NV + E+ K++ + +P++WSKVCL+N+AKL +EATT
Sbjct: 239 VLKNEGHVAYEDSLINVPSWRTVVNDKGELNVKMEDSLDPSFWSKVCLHNMAKLGEEATT 298
Query: 292 VRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVA 351
+RR+LE LF FD WS+E +A+ VL LQFL+ SG +H +LS+L+KHLDHK+V
Sbjct: 299 MRRILESLFRNFDEGCLWSTENSIAFPVLRDLQFLMEISGQRTHFLLSMLIKHLDHKSVL 358
Query: 352 KQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLN 411
K P +Q++I+ +T+ +++ K + S ++ AISD+++HLR+C+ +S + ++G DA
Sbjct: 359 KHPSMQLNILEVTSSLSETAKVEHSATIVSAISDIMRHLRKCMHSSLDEANLGTDAANCI 418
Query: 412 TKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVP 471
+ +++ C++QL+ KVGDAGPI D MA++LEN+S+ T VARTTI+AV++TA++I S+P
Sbjct: 419 RMVSVAVDKCLVQLTKKVGDAGPILDAMALMLENISAVTDVARTTIAAVFRTAQIIASIP 478
Query: 472 NVLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSP-----WLDQKK-- 524
N+ Y NKAFP+ALFHQLL AM HPD +T+IGAH I S+VL+P+ V P D KK
Sbjct: 479 NLQYQNKAFPEALFHQLLQAMVHPDHKTRIGAHRIFSVVLVPTSVCPRPSSTTTDLKKGM 538
Query: 525 ------------------ISKKVESDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISMIL 566
+ +K++ D S L+ + NG P EE+
Sbjct: 539 GLPRSLSRTASVFSSSAALFEKLKKDKFS---SMLTSDHSQNGMPEEER-------GSST 588
Query: 567 VELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI*DLLG*AV--------TK*VSCFLQSG 618
E+L S + + Q LT + + DLL + + + L S
Sbjct: 589 GEILDRLKSSYRQAYSTWNQPLTSVVDNSV----DLLNSELDVVHIRLSSHQIGLLLSSI 644
Query: 619 FRQHLLKM----ALQIMRQWHTLIALLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLP 674
+ Q + + + ++L+ L + S+ AL+R FQ+A SLR ISL + G LP
Sbjct: 645 WAQSISPANTPDNYEAIANTYSLVLLFSRVKNSSHDALIRSFQMALSLRDISLMEGGPLP 704
Query: 675 PSRRRSLLTLASHMLIFSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSD 734
PSRRRSL TLA+ M++FS++A + L K +L +DPFL LVDD+ L+AV
Sbjct: 705 PSRRRSLFTLAASMVLFSSKAFNLFSLADFTKVTLQGPRLDPFLNLVDDHKLKAVNSDQL 764
Query: 735 KVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSP 794
KV +G +D+ +A+ +LS + L + T++ + + E+ ++ QLL F P
Sbjct: 765 KVAYGCEKDDASALDTLSNIALSTEHSRGTLVYEIVKSLEDMCNSEMDKMREQLLTEFMP 824
Query: 795 DDAYPSGPPLFMETPRPGSPLAQIEFPDV--------DEMKGVG---------------P 831
DDA P G +T + QI+ DV D+ G G P
Sbjct: 825 DDACPLGTRFLEDTHK----TYQIDSGDVKPRKEDAEDQEFGDGTETVTKNNHVTFSEIP 880
Query: 832 N*VEASQIVEHHFQPTAQMF*VLINS------------WNRLETGKQQKMLTIRSFKNQQ 879
+ + +QI+E + T Q+ + ++ L GKQQK+ ++ + + +
Sbjct: 881 DLLTVNQILESVVETTRQVGRISFHTAADASYKEMTLHCENLLMGKQQKISSLLNSQLRH 940
Query: 880 ETKAIVLSSENEE----VSRQPVKALEYSKGDLKLVTQEQFQAQD--------QIRFRSQ 927
E+ +++E S P+ + G + ++F + Q ++
Sbjct: 941 ESSVNCSPRQHDEEIKIASFHPMINSAFHTGVEVPLLSKEFDMKSPRTPVGTIQSPCYAE 1000
Query: 928 DTRKQHSLRLPPSSPYDKFLKAAG 951
+ RLP SSPYD FLKAAG
Sbjct: 1001 LQNNPQAFRLPASSPYDNFLKAAG 1024
>ref|NP_197607.1| expressed protein [Arabidopsis thaliana]
Length = 980
Score = 599 bits (1545), Expect = e-169
Identities = 367/829 (44%), Positives = 506/829 (60%), Gaps = 58/829 (6%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGV+SR V P C +LC FCP+LRARSR PVKRYK L+A+I PR++ + NDRKIGKLCEY
Sbjct: 1 MGVVSRTVFPVCESLCCFCPALRARSRHPVKRYKHLLADIFPRSQDEQPNDRKIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNPLRIPKIT +LEQRCYK+LR E F SVK+++ IY+KLL SC EQ+ LFASS LG+I
Sbjct: 61 AAKNPLRIPKITTSLEQRCYKELRMEQFHSVKIVMSIYKKLLVSCNEQMLLFASSYLGLI 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+QTR DE+RILGC L DF+ Q +GTYMFNL+G IPK+C LA E+G+++ L
Sbjct: 121 HILLDQTRYDEMRILGCEALYDFVTSQAEGTYMFNLDGLIPKICPLAHELGEEDSTTNLC 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
+AGLQ LSS+V FMGE SH+S++FD ++S +LENY S+S+ + V + N +
Sbjct: 181 AAGLQALSSLVWFMGEFSHISVEFDNVVSVVLENYGG-HSQSSTSAVNQDNKVASIDKEL 239
Query: 241 EFPKEEAHVSSMLNVAT--GFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEP 298
+ E ++S + G I + + AKNP +WS+VCL+N+AKLAKEATTVRRVLE
Sbjct: 240 SPAEAETRIASWTRIVDDRGKAIGNNREDAKNPKFWSRVCLHNLAKLAKEATTVRRVLES 299
Query: 299 LFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQI 358
LF YFD WS+E G+A VL +Q L+ SG+ L HLDHKNV K+P +Q+
Sbjct: 300 LFRYFDFNEVWSTENGLAVYVLQDVQLLIERSGSYC------LRLHLDHKNVLKKPRMQL 353
Query: 359 DIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSI 418
+I+ + T +AQ K SVA+IGA+SD+I+HLR+ + S + +++GN+ N K ++ +
Sbjct: 354 EIVYVATALAQQTKVLPSVAIIGALSDMIRHLRKSIHCSLDDSNLGNEMIQYNLKFEAVV 413
Query: 419 EMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNK 478
E C+LQLS KVGDAGPI D+MAV+LE++S+ T++ART I+A
Sbjct: 414 EQCLLQLSQKVGDAGPILDIMAVMLESMSNITVMARTLIAA------------------- 454
Query: 479 AFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVS---------PWLDQKKISKKV 529
AFPDALFHQLL AM D E+++GAH I S+VL+PS VS P Q+ +S+ V
Sbjct: 455 AFPDALFHQLLQAMVCADHESRMGAHRIFSVVLVPSSVSPSSVLNSRRPADMQRTLSRTV 514
Query: 530 E---SDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQLVKCCYKQ 586
S + L ++ ++ E+ S +S + + +F ++ K
Sbjct: 515 SVFSSSAALFRKLKLESDNSVDDTAKMERV---STLSRSTSKFIRGESFDDEEPKNNTSS 571
Query: 587 FLTKYWISEIFRI*DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMRQWHTLIALLYYLLV 646
L++ + + + + V+ SG ++ M Q + IA + L++
Sbjct: 572 VLSR--LKSSYSRSQSVKRNPSSMVADQNSSGSSPEKPSLSPHNMPQNYEAIANTFSLVL 629
Query: 647 L-------SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFS 699
L S LV FQLAFSLR++SL G L PSRRRSL TLA+ M+IFSA+A +
Sbjct: 630 LFGRTKHSSNEVLVWSFQLAFSLRNLSLG--GPLQPSRRRSLFTLATSMIIFSAKAFNIP 687
Query: 700 DLIPKVKASLTEAPVDPFLELVDDNLLRAVCI-KSDKVV--FGSVEDEVAAMKSLSAV-Q 755
L+ K SL E VDPFL+LV+D L AV ++D+ +GS ED+ A +SL + +
Sbjct: 688 PLVNSAKTSLQEKTVDPFLQLVEDCKLDAVFYGQADQPAKNYGSKEDDDDASRSLVTIEE 747
Query: 756 LDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPL 804
Q +E S M KL + E S+IK QL+ F P D P G L
Sbjct: 748 ASQNQSREHYASMIMKFLGKLSDQESSAIKEQLVSDFIPIDGCPVGTQL 796
>ref|XP_468103.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|51964448|ref|XP_507009.1| PREDICTED OJ1293_A01.36
gene product [Oryza sativa (japonica cultivar-group)]
gi|47497474|dbj|BAD19529.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 997
Score = 567 bits (1461), Expect = e-160
Identities = 287/531 (54%), Positives = 388/531 (73%), Gaps = 19/531 (3%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGV+SR V+PAC LC CPSLR RSR PVKRYKKL++EI P+++ E NDRKIGKLCEY
Sbjct: 3 MGVVSREVLPACERLCFLCPSLRTRSRHPVKRYKKLLSEIFPKSQDEEPNDRKIGKLCEY 62
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
S+NPLR+PKIT LEQ+ YK+LR E FGSVKV++ IYRK++ SC+EQ+PLFA+SLL I+
Sbjct: 63 ISRNPLRVPKITVYLEQKFYKELRVEHFGSVKVVMAIYRKVICSCQEQLPLFANSLLNIV 122
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LLEQ R D++R + C TL F+ Q D TYMFNLE IPKLCQLAQE+G+ E+ ++
Sbjct: 123 EALLEQNRQDDLRTIACRTLFYFVNNQVDSTYMFNLESQIPKLCQLAQEMGEKEKISIVH 182
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKS-NLAKVEKLNSQSQNQLV 239
+AGLQ LSSMV FMGEHSH+S + D ++SA+LENY + S N A +E + Q V
Sbjct: 183 AAGLQALSSMVWFMGEHSHISAELDNVVSAVLENYESPYANSDNDAAIE----DRRTQWV 238
Query: 240 QEFPKEEAHVSSMLNVATGF-----------EIESKLDTAKNPAYWSKVCLYNIAKLAKE 288
E K E H S + + T E+ + +++P +WS +CL+N+A++++E
Sbjct: 239 SEVLKAEDHEPSGITILTRVPSWKAIRAPRGELSLTTEESESPNFWSGICLHNLARISRE 298
Query: 289 ATTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHK 348
ATTVRRVLE +F YFD N WS KG+A CVL+ +Q ++ +SG NSH++LS+LVKHL+HK
Sbjct: 299 ATTVRRVLEAIFRYFDNNNLWSPSKGLALCVLLDMQIVIEKSGQNSHILLSMLVKHLEHK 358
Query: 349 NVAKQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIG-NDA 407
NV KQ +DII +TT++A++ K Q+S A++ AISD+++HL + +Q +D+G D
Sbjct: 359 NVLKQTDKILDIIEVTTRLAEHSKAQSSTALMAAISDMVRHLSKNMQ--LLVSDVGPGDG 416
Query: 408 HTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLI 467
+N + + + C++QLS KVGDAGPI D +AVVLEN+SS+T VAR+TI+A Y+TA++I
Sbjct: 417 MVMNDRYGKATDECLVQLSRKVGDAGPILDALAVVLENISSTTPVARSTIAATYRTAQII 476
Query: 468 TSVPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSP 518
S+PN+LY +KAFP+ALFHQLLLAM +PD ET +GAH I S+VL+PS VSP
Sbjct: 477 ASLPNLLYQSKAFPEALFHQLLLAMVYPDCETHLGAHRIFSVVLVPSSVSP 527
Score = 125 bits (315), Expect = 5e-27
Identities = 111/316 (35%), Positives = 164/316 (51%), Gaps = 22/316 (6%)
Query: 652 LVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKASLTE 711
LV FQLAFSL+SISL Q G LPPSRRRSL T+A+ ML+F ++A LIP VK LT
Sbjct: 687 LVGSFQLAFSLQSISL-QAGFLPPSRRRSLFTMATSMLVFFSKAFGIPSLIPLVKDLLTT 745
Query: 712 APVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMT 771
+ VDPFL LV+D L+ V +S V+GS +D+ A+KSLS + ++D Q K+ +S +
Sbjct: 746 SIVDPFLRLVEDCKLQVV--ESCLTVYGSKDDDDLALKSLSNININD-QSKQASVSLILD 802
Query: 772 KFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQIEFPDVDEMKGVG- 830
L E ELS+I+ QLL+ FS DDA P G T + + A++ + E+ VG
Sbjct: 803 SLKDLSEAELSTIRKQLLEEFSADDACPLGSHSNESTSQSPAYNAKLHQKSL-EVIPVGF 861
Query: 831 ----PN*VE-ASQIVEHHFQPTAQMF*VLINSW--NRLETGKQQKMLTIRS-----FKN- 877
VE A+ + E Q + +N + +ET + L++ + FK
Sbjct: 862 IFEDDTLVEPANSLAEPQLQQPLDNGLIDVNQLLESVVETSRHVGRLSVSTNLDLPFKEV 921
Query: 878 QQETKAIVLSSENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHS--L 935
+A+++ + + V + + + + Q Q F S + + HS
Sbjct: 922 ANRCEALLIGKQQKLSVCMSVHQKQDGESPMDKLGSPQ-QISPTAGFVSTNDEQCHSDFC 980
Query: 936 RLPPSSPYDKFLKAAG 951
+LP SPYDKFL +G
Sbjct: 981 KLPVLSPYDKFLAGSG 996
>ref|XP_477071.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|34393302|dbj|BAC83231.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 1066
Score = 561 bits (1445), Expect = e-158
Identities = 275/538 (51%), Positives = 386/538 (71%), Gaps = 28/538 (5%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGV+SR+V+PACG+LC FCP LRARSRQPVKRYK ++AEI P+ + E N+R+IGKLCEY
Sbjct: 1 MGVISRKVLPACGSLCYFCPGLRARSRQPVKRYKSILAEIFPKTQDEEPNERRIGKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
S+NPLR+PKIT +LEQR YK+LR+E +G KV++ IYR+LL SC+EQ+PLFASSLL I+
Sbjct: 61 CSRNPLRVPKITVSLEQRIYKELRSEQYGFAKVVMLIYRRLLVSCKEQMPLFASSLLSIV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
TLL+Q R D++RI+GC TL DF + Q DGTY FNLEG +P+LC+L+QEVG+DE+ + LR
Sbjct: 121 HTLLDQKRQDDMRIIGCETLFDFAVNQVDGTYQFNLEGLVPRLCELSQEVGEDEQTIALR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
+A LQ LS+M+ FMGE SH+S +FD ++ +LENY Q N +V K S +QL Q
Sbjct: 181 AAALQALSAMIWFMGELSHISSEFDNVVQVVLENYRP-QKMQNDGQVTK---DSSDQLEQ 236
Query: 241 EFPKEEAHVS--------------------SMLNVATGFEIESKLDTAKNPAYWSKVCLY 280
E PK E + +++NV G + ++ AK+P +WS++C++
Sbjct: 237 EAPKTEDSKAEDSKTEDSSPFVISAVPLWENIVNVKGG--VNLTVEEAKDPKFWSRICVH 294
Query: 281 NIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSI 340
N+A+L++EATT RR+LE LF YF + WS E G+A CVL+ +Q L+ +G N HLMLS+
Sbjct: 295 NMARLSREATTFRRILESLFRYFGNNSSWSPENGLALCVLLDMQLLVENAGQNMHLMLSL 354
Query: 341 LVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEA 400
L+KH++HK + KQ +Q+ I+ + +A+ QAS A IGAISDL++HL+R + +
Sbjct: 355 LIKHIEHKTMVKQQEMQLSIVEVAATLAEQSIAQASAATIGAISDLVRHLKRTFHITLGS 414
Query: 401 TDIGNDAHTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAV 460
D ++ N K + +I+ C+ QL+ KV DAGP+ D+MAV+LEN++S+ +VAR+T +AV
Sbjct: 415 KD--SELVKWNEKFRKAIDDCLGQLAKKVTDAGPVLDMMAVMLENIASTPVVARSTAAAV 472
Query: 461 YQTAKLITSVPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSP 518
Y+TA++I SVPN+ Y NK FP+ALFHQLLL M HPD E ++ AH I ++VL+PS V+P
Sbjct: 473 YRTAQIIASVPNITYQNKVFPEALFHQLLLTMIHPDHEARVAAHRIFAIVLVPSSVAP 530
Score = 144 bits (362), Expect = 2e-32
Identities = 118/366 (32%), Positives = 163/366 (44%), Gaps = 52/366 (14%)
Query: 638 IALLYYLLVLS-------YMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLI 690
IA Y LL+L + AL + FQ+AFSLR SL + LPP RRRSL TLA+ M+I
Sbjct: 700 IAHTYSLLLLFSGAKASVFEALTQSFQVAFSLRGYSLTEPDSLPPCRRRSLFTLATAMII 759
Query: 691 FSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAV--CIKSDKVVFGSVEDEVAAM 748
FS+R + LIP K L E DPFL LVD+ L+AV + ++GS ED A+
Sbjct: 760 FSSRTFNVLPLIPICKQMLNERTGDPFLRLVDECKLQAVKDSVDDPSKIYGSPEDNTNAL 819
Query: 749 KSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMET 808
KSLSA++L + Q +E ++S M + + E EL+S+KNQLL FSPDD P+ F T
Sbjct: 820 KSLSAIELSESQSRECIVSTIMNNITNMLEAELNSVKNQLLSDFSPDDMCPTSTHFFEAT 879
Query: 809 PRPGSPLAQIE-----------------FPDVDEMKGVGPN*VEASQIV----------- 840
SP + F + E N V + ++
Sbjct: 880 GDNSSPGSHDNDHHPEAVLIDLGNDHDIFGEASESTAASANAVPVTDLLSIDQLLETVVT 939
Query: 841 ------EHHFQPTAQMF*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIV--------L 886
E T F + + L K QKM + SF + L
Sbjct: 940 DPAPHTERVSVSTDMPFKEMSSQCEALTVRKHQKMASFMSFSQDMTMDPMATNQPFQTDL 999
Query: 887 SSENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFR-SQDTRKQHSLRLPPSSPYDK 945
S ++ Q D L Q+ ++ + + + +QH LRLP SSPYD
Sbjct: 1000 SLFHDPYPPQVGVPNTNPFVDDNLYGYPQYMNMNEANPQPTYEQAQQHFLRLPASSPYDN 1059
Query: 946 FLKAAG 951
F +AAG
Sbjct: 1060 FRRAAG 1065
>ref|XP_464126.1| cyclin-like protein [Oryza sativa (japonica cultivar-group)]
gi|45736188|dbj|BAD13233.1| cyclin-like protein [Oryza
sativa (japonica cultivar-group)]
Length = 1035
Score = 558 bits (1438), Expect = e-157
Identities = 282/540 (52%), Positives = 388/540 (71%), Gaps = 18/540 (3%)
Query: 2 GVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYA 61
GV+SR+V+PACG LC FCP LRARSRQPVKRYKK+IA+I P + E N+R+IGKLCEY
Sbjct: 12 GVVSRKVLPACGGLCYFCPGLRARSRQPVKRYKKIIADIFPATQDEEPNERRIGKLCEYV 71
Query: 62 SKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGIIR 121
++N R+PKIT LEQRCYK+LRNE +G VKV++ IYRKLL SC++Q+PL ASS L II
Sbjct: 72 ARNHHRVPKITAYLEQRCYKELRNEQYGFVKVVVLIYRKLLVSCKKQMPLLASSALSIIC 131
Query: 122 TLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLRS 181
TLL+QTR D++RI+GC TL DF + Q DGTY FNLE +PKLC+LAQ V +E+ +LR+
Sbjct: 132 TLLDQTRRDDMRIIGCETLFDFTVSQVDGTYQFNLEELVPKLCELAQIVKAEEKDNMLRA 191
Query: 182 AGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQ---NQL 238
+ LQ LS+M+ FMGE SH+S FD +I +LE+Y NL K++ N S+ N+
Sbjct: 192 STLQALSAMIWFMGEFSHISSAFDNVIQVVLESY-------NLQKMQNDNIDSEAPGNRW 244
Query: 239 VQEFPKEEAHV------SSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTV 292
V++ K E + S V E+ + AK+P +WS+VC++N+AKL++EATT
Sbjct: 245 VEQVLKAEGNATISRIPSWKSIVDDKGELHLPAEDAKDPNFWSRVCVHNMAKLSREATTF 304
Query: 293 RRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAK 352
RRVLE LF +FD N WSS+ +A+CVL+ +Q L+ G N LM+SILVKHL+HK+V K
Sbjct: 305 RRVLESLFRHFDNNNSWSSQNTLAFCVLLDMQILMENQGQNIDLMISILVKHLEHKSVLK 364
Query: 353 QPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNT 412
QP +Q+ ++ + +A+ + +AS A IGAISDLI+H+++ L + + D+ + N
Sbjct: 365 QPEMQLSVVEVIASLAEQSRAEASAATIGAISDLIRHMKKTLHVALGSRDL--EVIKWND 422
Query: 413 KLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPN 472
KL+++++ CILQLS KVGDAGP+ D+M+V+LEN+S + +VA T SAVY+TA++ITS+PN
Sbjct: 423 KLRNAVDECILQLSKKVGDAGPVLDMMSVMLENISRTPLVAIATTSAVYRTAQIITSIPN 482
Query: 473 VLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKISKKVESD 532
+ Y NK FP+ALFHQLLLAM HPD ET++ AH I S+VL+PS VSP+ ++ V+ D
Sbjct: 483 LSYRNKVFPEALFHQLLLAMVHPDHETRVSAHRIFSVVLVPSSVSPFSKSTSPNQLVKHD 542
Score = 146 bits (369), Expect = 3e-33
Identities = 130/372 (34%), Positives = 181/372 (47%), Gaps = 63/372 (16%)
Query: 633 QWHTLIALLYYLLVLS-------YMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLA 685
Q + IA Y LL+L + AL FQ+AFSL S SL+ L PSRRRSL TLA
Sbjct: 673 QNYEAIAHTYSLLLLFSGSKASIFEALAPSFQVAFSLMSYSLEGTDSLLPSRRRSLFTLA 732
Query: 686 SHMLIFSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEV 745
+ M++F +RA + + LIP K+ L E +DPFL LV D L+AV S++ +GS ED+
Sbjct: 733 TSMIMFFSRAFNVAPLIPICKSMLNERTMDPFLHLVQDTKLQAVKDCSEE-TYGSPEDDN 791
Query: 746 AAMKSLSAVQLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLF 805
A+KSLSAV+L Q +E++ S M LP+ EL +I++QLL FSPDD P+ F
Sbjct: 792 NALKSLSAVELTQSQSRESMASTIMNNIRDLPDSELQTIRSQLLSDFSPDDMCPTSALFF 851
Query: 806 METPR-PGSPLAQIEFPDV--------DEMKGVGPN*VEAS-----------------QI 839
T R PG DV D V N EA+ +
Sbjct: 852 ELTVRNPGCDEDSSNQEDVLINMANDNDTFGEVYEN-TEATTASVPTANLLGIDELLESV 910
Query: 840 VEHHFQPTAQM---------F*VLINSWNRLETGKQQKMLTIRSFKNQQE---------- 880
V TA+ F + N L KQQKM + SFK++ +
Sbjct: 911 VTDAPSQTARCSVSTAPNIPFKEMTNQCEVLSMEKQQKMSVLLSFKHKNQSNVLPINQAD 970
Query: 881 -TKAIVLSSENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPP 939
T A+ +SS+++ + ++L+ G K V + ++ + D +Q L+LP
Sbjct: 971 NTGAVHISSDDQNTNPFLQQSLD---GYPKYVAD-----GEALQVAADDVFQQQFLKLPA 1022
Query: 940 SSPYDKFLKAAG 951
SSPYD FLKAAG
Sbjct: 1023 SSPYDTFLKAAG 1034
>dbj|BAD82189.1| cyclin-like [Oryza sativa (japonica cultivar-group)]
gi|56784331|dbj|BAD82352.1| cyclin-like [Oryza sativa
(japonica cultivar-group)]
Length = 980
Score = 553 bits (1425), Expect = e-155
Identities = 267/519 (51%), Positives = 373/519 (71%), Gaps = 6/519 (1%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PAC +LC FCPSLRARSRQPVKRYKK+IAEI E NDR+IGKLC+Y
Sbjct: 1 MGVMSRRVLPACSSLCYFCPSLRARSRQPVKRYKKIIAEIYQLPPDGEPNDRRIGKLCDY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
S+NP RIPKITE LE+RCYKDLR+E+F KV+ CIYRKLL SC++ PL A+S L II
Sbjct: 61 VSRNPTRIPKITEYLEERCYKDLRHENFTLAKVVPCIYRKLLCSCKDHTPLLATSTLSII 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
RTLL+Q D++R+LGC LVDF+ Q D T+MFNLEG IPKLCQ++QE+ +D++ LR
Sbjct: 121 RTLLDQRMNDDLRVLGCLMLVDFLNGQVDSTHMFNLEGLIPKLCQISQELREDDKGFRLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
A LQ L+SMV++MG+HSH+SM+ D+++S I+ Y + L+ E + Q + LV
Sbjct: 181 CAALQALASMVQYMGDHSHISMELDEVVSVIVSCY---EVNQTLSIKEVVRLQDDDDLVI 237
Query: 241 EFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLF 300
VS + + ++NPA+W++VCL N+A +AKEATTV RVL+PLF
Sbjct: 238 NGSLTGLPVSGQNSAKVA---SDTMSASENPAHWARVCLRNMASIAKEATTVWRVLDPLF 294
Query: 301 HYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDI 360
FD+ N+WS E G+A+ +L +Q L+ +SG N HL+LS +KH+DHK+VAK+P Q I
Sbjct: 295 RLFDSHNYWSPENGIAFSILQEMQALMDKSGQNGHLLLSFTIKHIDHKSVAKKPAKQTSI 354
Query: 361 INITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEM 420
+ + + +A++ K +ASV + A SDLIKHLR+C+ + E+ + ND N+ L ++E
Sbjct: 355 LKVASLLAKHAKLKASVTIASATSDLIKHLRKCMHCAVESPNAQNDVDKWNSALYVALEE 414
Query: 421 CILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNKAF 480
C++QL+ KVGD GP+ D++ V+LEN+S + +ARTTIS+V++T ++ S+ LY+ KAF
Sbjct: 415 CLVQLTEKVGDVGPVLDMVGVMLENLSCTATIARTTISSVFRTVQIAASIHKSLYNQKAF 474
Query: 481 PDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPW 519
P+ALFHQLLLAM HPD++T++G+H +LS ++ PS++ PW
Sbjct: 475 PEALFHQLLLAMMHPDKKTRVGSHRVLSTIIAPSLLCPW 513
Score = 177 bits (450), Expect = 1e-42
Identities = 131/345 (37%), Positives = 180/345 (51%), Gaps = 52/345 (15%)
Query: 648 SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKA 707
S+ ALVRCFQLAFSLR +SL+QE GL PSRRR L T+AS MLIFSA+ AD IP VKA
Sbjct: 646 SHAALVRCFQLAFSLRRMSLNQENGLQPSRRRCLYTMASAMLIFSAKVADIPQTIPLVKA 705
Query: 708 SLTEAPVDPFLELVDDNLLRAVCIKSDK--VVFGSVEDEVAAMKSLSAVQLDDRQLKETV 765
++ E VDP L L+DD L +S +V+GS EDE A LS V +D QLKE V
Sbjct: 706 AVPEKMVDPHLCLIDDCRLVISSPQSSNSGIVYGSEEDESDARNFLSCVNKNDTQLKEIV 765
Query: 766 ISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQ-------- 817
IS+F KF L E + + I+ QLLQ FS DD++P PLFMETP S A+
Sbjct: 766 ISHFKEKFENLSE-KFNGIEEQLLQEFSLDDSFPLSAPLFMETPHSCSMYAEKDDHCFDE 824
Query: 818 ----IEFPDVDEM---------------KGVGPN*VEASQIVEHHFQPTAQM-------- 850
E D D++ + + +Q++E + Q+
Sbjct: 825 EVIPCEMDDDDDIVFEHSGSQSDRKTSGSMASSDVLNVNQLIESVHETARQVANAPVSAN 884
Query: 851 ---F*VLINSWNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEVSRQPVKALEYSKGD 907
+ + + L KQQKM + SFK+ + T + ++EN LE ++
Sbjct: 885 LVPYDQMKSQCEALVMEKQQKMSVLLSFKHSR-TDSRGSTAEN---------GLETNESS 934
Query: 908 LKLVTQEQFQAQDQI-RFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
+ + Q ++++ R S + S RLPP+SPYDKF++AAG
Sbjct: 935 ARSEPETQSTRKERMRRSDSASSESDRSFRLPPASPYDKFMRAAG 979
>ref|NP_915175.1| P0674H09.27 [Oryza sativa (japonica cultivar-group)]
Length = 894
Score = 453 bits (1166), Expect = e-125
Identities = 233/523 (44%), Positives = 332/523 (62%), Gaps = 56/523 (10%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MGVMSRRV+PAC +LC FCPSLRARSRQPVKRYKK+IAEI E NDR+IGKLC+Y
Sbjct: 1 MGVMSRRVLPACSSLCYFCPSLRARSRQPVKRYKKIIAEIYQLPPDGEPNDRRIGKLCDY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
S+NP RIPK Q Y+
Sbjct: 61 VSRNPTRIPKEASVFMQGSYR--------------------------------------- 81
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
D++R+LGC LVDF+ Q D T+MFNLEG IPKLCQ++QE+ +D++ LR
Sbjct: 82 -------MNDDLRVLGCLMLVDFLNGQVDSTHMFNLEGLIPKLCQISQELREDDKGFRLR 134
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
A LQ L+SMV++MG+HSH+SM+ D+++S I+ Y + L+ E + Q + LV
Sbjct: 135 CAALQALASMVQYMGDHSHISMELDEVVSVIVSCY---EVNQTLSIKEVVRLQDDDDLVI 191
Query: 241 EFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLF 300
VS + + S ++NPA+W++VCL N+A +AKEATTV RVL+PLF
Sbjct: 192 NGSLTGLPVSGQNSAKVASDTMS---ASENPAHWARVCLRNMASIAKEATTVWRVLDPLF 248
Query: 301 HYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDI 360
FD+ N+WS E G+A+ +L +Q L+ +SG N HL+LS +KH+DHK+VAK+P Q I
Sbjct: 249 RLFDSHNYWSPENGIAFSILQEMQALMDKSGQNGHLLLSFTIKHIDHKSVAKKPAKQTSI 308
Query: 361 INITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEM 420
+ + + +A++ K +ASV + A SDLIKHLR+C+ + E+ + ND N+ L ++E
Sbjct: 309 LKVASLLAKHAKLKASVTIASATSDLIKHLRKCMHCAVESPNAQNDVDKWNSALYVALEE 368
Query: 421 CILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLYHNK-- 478
C++QL+ KVGD GP+ D++ V+LEN+S + +ARTTIS+V++T ++ S+ LY+ K
Sbjct: 369 CLVQLTEKVGDVGPVLDMVGVMLENLSCTATIARTTISSVFRTVQIAASIHKSLYNQKAS 428
Query: 479 --AFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPW 519
AFP+ALFHQLLLAM HPD++T++G+H +LS ++ PS++ PW
Sbjct: 429 FLAFPEALFHQLLLAMMHPDKKTRVGSHRVLSTIIAPSLLCPW 471
Score = 158 bits (399), Expect = 9e-37
Identities = 96/180 (53%), Positives = 116/180 (64%), Gaps = 3/180 (1%)
Query: 648 SYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFSDLIPKVKA 707
S+ ALVRCFQLAFSLR +SL+QE GL PSRRR L T+AS MLIFSA+ AD IP VKA
Sbjct: 604 SHAALVRCFQLAFSLRRMSLNQENGLQPSRRRCLYTMASAMLIFSAKVADIPQTIPLVKA 663
Query: 708 SLTEAPVDPFLELVDDNLLRAVCIKSDK--VVFGSVEDEVAAMKSLSAVQLDDRQLKETV 765
++ E VDP L L+DD L +S +V+GS EDE A LS V +D QLKE V
Sbjct: 664 AVPEKMVDPHLCLIDDCRLVISSPQSSNSGIVYGSEEDESDARNFLSCVNKNDTQLKEIV 723
Query: 766 ISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQIEFPDVDE 825
IS+F KF L E + + I+ QLLQ FS DD++P PLFMETP S A+ + DE
Sbjct: 724 ISHFKEKFENLSE-KFNGIEEQLLQEFSLDDSFPLSAPLFMETPHSCSMYAEKDDHCFDE 782
>gb|AAQ89612.1| At5g26850 [Arabidopsis thaliana] gi|26449532|dbj|BAC41892.1|
unknown protein [Arabidopsis thaliana]
Length = 970
Score = 438 bits (1126), Expect = e-121
Identities = 317/1029 (30%), Positives = 508/1029 (48%), Gaps = 140/1029 (13%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MG +SR V PAC ++CI CP+LR+RSRQPVKRYKKL+ EI P++ N+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNP+RIPKI + LE+RCYKDLR+E + ++ Y K+L C++Q+ FA+SLL ++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+ ++ D ILGC TL FI Q DGTY ++E F K+C LA+E G++ + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQ 240
++GLQ LS+MV +MGE SH+ D+I+ + D + ++ E + + + +
Sbjct: 181 ASGLQCLSAMVWYMGEFSHIFATVDEIV----QTNEDREEQNCNWVNEVIRCEGRGTTIC 236
Query: 241 EFPKEEAHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLF 300
P +++ A + + P W+++CL + LAKE+TT+R++L+P+F
Sbjct: 237 NSP---SYMIVRPRTARKDPTLLTKEETEMPKVWAQICLQRMVDLAKESTTLRQILDPMF 293
Query: 301 HYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDI 360
YF++ W+ G+A VL +L+ SG + L+LS +V+HLD+K+VA P L+ I
Sbjct: 294 SYFNSRRQWTPPNGLAMIVLSDAVYLMETSG-SQQLVLSTVVRHLDNKHVANDPELKAYI 352
Query: 361 INITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEM 420
I + +A+ ++ + + I ++DL +HLR+ Q A A IG++ LN +Q+SIE
Sbjct: 353 IQVAGCLAKLIRTSSYLRDISFVNDLCRHLRKSFQ--ATARSIGDEELNLNVMIQNSIED 410
Query: 421 CILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITS-VPNVLYHNKA 479
C+ +++ + + P+FD+MAV +E + SS IV+R + ++ A ++S + + +
Sbjct: 411 CLREIAKGIVNTQPLFDMMAVSVEGLPSSGIVSRAAVGSLLILAHAMSSALSPSMRSQQV 470
Query: 480 FPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPS------------------------- 514
FPD L LL AM HP+ ET++GAH I S++L+ S
Sbjct: 471 FPDTLLDALLKAMLHPNVETRVGAHEIFSVILLQSSGQSQAGLASVRASGYLNESRNWRS 530
Query: 515 -------VVSPWLDQKKISK---KVESDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISM 564
V+ LD+ + K K+E +G + HE L K + P H L S+
Sbjct: 531 DTTSAFTSVTARLDKLRKEKDGVKIEKNGYNNTHEDL--------KNYKSSPKFHKLNSI 582
Query: 565 I-----LVELLCWWAFSNQLVKCCYKQFLTKYWISEIFRI*DLLG*AVTK*VSCFLQSGF 619
I + L + + Q L+ +WI
Sbjct: 583 IDRTAGFINLADMLPSMMKFTEDQIGQLLSAFWIQSAL---------------------- 620
Query: 620 RQHLLKMALQIMRQWHTLIALLYYLLVLSYMALVRCFQLAFSLRSISLD-QEGGLPPSRR 678
+L ++ + +L+ L L +VR FQL FSLR++SLD G LP +
Sbjct: 621 -PDILPSNIEAIAHSFSLVLLSLRLKNPDDGLVVRAFQLLFSLRTLSLDLNNGTLPSVCK 679
Query: 679 RSLLTLASHMLIFSARAADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVF 738
R +L L++ ML+F+A+ + +KA L VDP+L + D+L V +++ F
Sbjct: 680 RLILALSTSMLMFAAKIYQIPHICEMLKAQL-PGDVDPYL-FIGDDLQLHVRPQANMKDF 737
Query: 739 GSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTK-FSKLPEDELSSIKNQLLQGFSPDDA 797
GS D A L ++ +L T+I+ + K KL + E + +K Q+L+ F+PDDA
Sbjct: 738 GSSSDSQMATSMLFEMR-SKVELSNTIITDIVAKNLPKLSKLEEADVKMQILEQFTPDDA 796
Query: 798 YPSGPPLFMETPRPGSPLAQ------------------------IEFPDVDEMKGVGPN* 833
+ G +E P+P +++ + FP P
Sbjct: 797 FMFGSRPNIE-PQPNQSISKESLSFDEDIPAGSMVEDEVTSELSVRFPPRGSPSPSIPQV 855
Query: 834 VEASQIVEHHFQPTAQM-----------F*VLINSWNRLETGKQQKMLTIRSFKNQQETK 882
+ Q++E + Q+ + + N TG ++K+ + +N+Q
Sbjct: 856 ISIGQLMESALEVAGQVVGSSVSTSPLPYDTMTNRCETFGTGTREKLSRWLATENRQMNG 915
Query: 883 AIVLSSENEEVSRQPVKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSP 942
S E ALE D + +E QD +RLPP+SP
Sbjct: 916 LYGNSLEES-------SALEKVVEDGNIYGRESGMLQD----------SWSMMRLPPASP 958
Query: 943 YDKFLKAAG 951
+D FLKAAG
Sbjct: 959 FDNFLKAAG 967
>ref|NP_198037.2| expressed protein [Arabidopsis thaliana]
Length = 919
Score = 388 bits (996), Expect = e-106
Identities = 211/526 (40%), Positives = 329/526 (62%), Gaps = 20/526 (3%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MG +SR V PAC ++CI CP+LR+RSRQPVKRYKKL+ EI P++ N+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNP+RIPKI + LE+RCYKDLR+E + ++ Y K+L C++Q+ FA+SLL ++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+ ++ D ILGC TL FI Q DGTY ++E F K+C LA+E G++ + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 SAGLQTLSSMVKFMGEHSHLSMDFDKIISAILENY-VDLQSKSNLAKVEKLNSQSQNQLV 239
++GLQ LS+MV +MGE SH+ D+I+ AIL+NY D+ ++N E Q+ N V
Sbjct: 181 ASGLQCLSAMVWYMGEFSHIFATVDEIVHAILDNYEADMIVQTN----EDREEQNCN-WV 235
Query: 240 QEFPKEEAHVSSMLNVATGFEIESKL----------DTAKNPAYWSKVCLYNIAKLAKEA 289
E + E +++ N + + + + + P W+++CL + LAKE+
Sbjct: 236 NEVIRCEGRGTTICNSPSYMIVRPRTARKDPTLLTKEETEMPKVWAQICLQRMVDLAKES 295
Query: 290 TTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKN 349
TT+R++L+P+F YF++ W+ G+A VL +L+ SG + L+LS +V+HLD+K+
Sbjct: 296 TTLRQILDPMFSYFNSRRQWTPPNGLAMIVLSDAVYLMETSG-SQQLVLSTVVRHLDNKH 354
Query: 350 VAKQPILQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHT 409
VA P L+ II + +A+ ++ + + I ++DL +HLR+ Q A A IG++
Sbjct: 355 VANDPELKAYIIQVAGCLAKLIRTSSYLRDISFVNDLCRHLRKSFQ--ATARSIGDEELN 412
Query: 410 LNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITS 469
LN +Q+SIE C+ +++ + + P+FD+MAV +E + SS IV+R + ++ A ++S
Sbjct: 413 LNVMIQNSIEDCLREIAKGIVNTQPLFDMMAVSVEGLPSSGIVSRAAVGSLLILAHAMSS 472
Query: 470 -VPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPS 514
+ + + FPD L LL AM HP+ ET++GAH I S++L+ S
Sbjct: 473 ALSPSMRSQQVFPDTLLDALLKAMLHPNVETRVGAHEIFSVILLQS 518
Score = 53.5 bits (127), Expect = 3e-05
Identities = 67/274 (24%), Positives = 109/274 (39%), Gaps = 56/274 (20%)
Query: 714 VDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTK- 772
VDP+L + DD L V +++ FGS D A L ++ +L T+I+ + K
Sbjct: 663 VDPYLFIGDDLQLH-VRPQANMKDFGSSSDSQMATSMLFEMR-SKVELSNTIITDIVAKN 720
Query: 773 FSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQ--------------- 817
KL + E + +K Q+L+ F+PDDA+ G +E P+P +++
Sbjct: 721 LPKLSKLEEADVKMQILEQFTPDDAFMFGSRPNIE-PQPNQSISKESLSFDEDIPAGSMV 779
Query: 818 ---------IEFPDVDEMKGVGPN*VEASQIVEHHFQPTAQM-----------F*VLINS 857
+ FP P + Q++E + Q+ + + N
Sbjct: 780 EDEVTSELSVRFPPRGSPSPSIPQVISIGQLMESALEVAGQVVGSSVSTSPLPYDTMTNR 839
Query: 858 WNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEVSRQPVKALEYSKGDLKLVTQEQFQ 917
TG ++K+ + +N+Q S E ALE D + +E
Sbjct: 840 CETFGTGTREKLSRWLATENRQMNGLYGNSLEESS-------ALEKVVEDGNIYGRESGM 892
Query: 918 AQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
QD +RLPP+SP+D FLKAAG
Sbjct: 893 LQDSWSM----------MRLPPASPFDNFLKAAG 916
>gb|AAB61061.1| Hypothetical protein F2P16.24 [Arabidopsis thaliana]
gi|7485250|pir||T01764 hypothetical protein
A_IG002P16.24 - Arabidopsis thaliana
Length = 907
Score = 372 bits (954), Expect = e-101
Identities = 210/550 (38%), Positives = 328/550 (59%), Gaps = 43/550 (7%)
Query: 1 MGVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEY 60
MG +SR V PAC ++CI CP+LR+RSRQPVKRYKKL+ EI P++ N+RKI KLCEY
Sbjct: 1 MGFISRNVFPACESMCICCPALRSRSRQPVKRYKKLLGEIFPKSPDGGPNERKIVKLCEY 60
Query: 61 ASKNPLRIPKITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLGII 120
A+KNP+RIPKI + LE+RCYKDLR+E + ++ Y K+L C++Q+ FA+SLL ++
Sbjct: 61 AAKNPIRIPKIAKFLEERCYKDLRSEQMKFINIVTEAYNKMLCHCKDQMAYFATSLLNVV 120
Query: 121 RTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALLLR 180
LL+ ++ D ILGC TL FI Q DGTY ++E F K+C LA+E G++ + LR
Sbjct: 121 TELLDNSKQDTPTILGCQTLTRFIYSQVDGTYTHSIEKFALKVCSLAREEGEEHQKQCLR 180
Query: 181 SAGLQTLSSMVK------------------FMGEHSHLSMDFDKIISAILENY-VDLQSK 221
++GLQ LS+MV +MGE SH+ D+I+ AIL+NY D+ +
Sbjct: 181 ASGLQCLSAMVLELVLLIFQIQSILLPKVWYMGEFSHIFATVDEIVHAILDNYEADMIVQ 240
Query: 222 SNLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATGFEIESKL----------DTAKNP 271
+N E Q+ N V E + E +++ N + + + + + P
Sbjct: 241 TN----EDREEQNCN-WVNEVIRCEGRGTTICNSPSYMIVRPRTARKDPTLLTKEETEMP 295
Query: 272 AYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESG 331
W+++CL + LAKE+TT+R++L+P+F YF++ W+ G+A VL +L+ S
Sbjct: 296 KVWAQICLQRMVDLAKESTTLRQILDPMFSYFNSRRQWTPPNGLAMIVLSDAVYLMETSV 355
Query: 332 ------NNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVAVIGAISD 385
+ L+LS +V+HLD+K+VA P L+ II + +A+ ++ + + I ++D
Sbjct: 356 MFLYILGSQQLVLSTVVRHLDNKHVANDPELKAYIIQVAGCLAKLIRTSSYLRDISFVND 415
Query: 386 LIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMAVVLEN 445
L +HLR+ Q A A IG++ LN +Q+SIE C+ +++ + + P+FD+MAV +E
Sbjct: 416 LCRHLRKSFQ--ATARSIGDEELNLNVMIQNSIEDCLREIAKGIVNTQPLFDMMAVSVEG 473
Query: 446 VSSSTIVARTTISAVYQTAKLITS-VPNVLYHNKAFPDALFHQLLLAMAHPDRETQIGAH 504
+ SS IV+R + ++ A ++S + + + FPD L LL AM HP+ ET++GAH
Sbjct: 474 LPSSGIVSRAAVGSLLILAHAMSSALSPSMRSQQVFPDTLLDALLKAMLHPNVETRVGAH 533
Query: 505 SILSMVLMPS 514
I S++L+ S
Sbjct: 534 EIFSVILLQS 543
Score = 40.0 bits (92), Expect = 0.37
Identities = 33/105 (31%), Positives = 56/105 (52%), Gaps = 4/105 (3%)
Query: 714 VDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAVQLDDRQLKETVISYFMTK- 772
VDP+L + DD L V +++ FGS D A L ++ +L T+I+ + K
Sbjct: 688 VDPYLFIGDDLQLH-VRPQANMKDFGSSSDSQMATSMLFEMR-SKVELSNTIITDIVAKN 745
Query: 773 FSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQ 817
KL + E + +K Q+L+ F+PDDA+ G +E P+P +++
Sbjct: 746 LPKLSKLEEADVKMQILEQFTPDDAFMFGSRPNIE-PQPNQSISK 789
>gb|AAC02765.1| hypothetical protein [Arabidopsis thaliana] gi|25408758|pir||F84846
hypothetical protein At2g41830 [imported] - Arabidopsis
thaliana
Length = 961
Score = 344 bits (882), Expect = 9e-93
Identities = 202/523 (38%), Positives = 298/523 (56%), Gaps = 105/523 (20%)
Query: 2 GVMSRRVVPACGNLCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYA 61
GV+SR+V+P CG+LCI CP+LRARSRQPVKRYKKLIAEI PRN+ +NDRKIGKLCEYA
Sbjct: 6 GVISRQVLPVCGSLCILCPALRARSRQPVKRYKKLIAEIFPRNQEEGINDRKIGKLCEYA 65
Query: 62 SKNPLRIPK---ITENLEQRCYKDLRNESFGSVKVILCIYRKLLSSCREQIPLFASSLLG 118
+KN +R+PK I+++LE RCYK+LRNE+F S K+ +CIYR+LL +C+EQIPLF+S L
Sbjct: 66 AKNAVRMPKFFQISDSLEHRCYKELRNENFHSAKIAMCIYRRLLVTCKEQIPLFSSGFLR 125
Query: 119 IIRTLLEQTRADEVRILGCNTLVDFIIFQTDGTYMFNLEGFIPKLCQLAQEVGDDERALL 178
++ LL+QTR DE++I+GC +L +F+I Q LCQL E GDD+R+
Sbjct: 126 TVQALLDQTRQDEMQIVGCQSLFEFVINQ---------------LCQLGLEGGDDDRSRS 170
Query: 179 LRSAGLQTLSSMVKFMGEHSHLSMDFDKIISAI-LENYVDLQSKSNLA-KVEKLNSQSQN 236
LR+AGLQ LS+M+ MGE+SH+ +FD ++ + Y DL S + V N QS+
Sbjct: 171 LRAAGLQALSAMIWLMGEYSHIPSEFDNVLLGFRVPIYSDLLSFIEINWFVVSNNRQSEK 230
Query: 237 QLVQEFPKEEAHVSSML-NVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRV 295
V + VS++L N + + D+ + W L N +A E + +
Sbjct: 231 HSV-----GDKVVSAVLENYGHPKILTNANDSGRK---WVDEVLKNEGHVAYEDSLI--- 279
Query: 296 LEPLFHYFDTENHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPI 355
W + ++ + G +H +LS+L+KHLDHK+V K P
Sbjct: 280 ---------NVPSWRT--------------VVNDKGQRTHFLLSMLIKHLDHKSVLKHPS 316
Query: 356 LQIDIINITTQVAQNVKQQASVAVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQ 415
+Q++I+ +T+ +++ K + S ++ AISD+++HLR+C+ +S + ++G DA +
Sbjct: 317 MQLNILEVTSSLSETAKVEHSATIVSAISDIMRHLRKCMHSSLDEANLGTDAANCIRMVS 376
Query: 416 SSIEMCILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVPNVLY 475
+++ C++QL+ K
Sbjct: 377 VAVDKCLVQLTKK----------------------------------------------- 389
Query: 476 HNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSP 518
AFP+ALFHQLL AM HPD +T+IGAH I S+VL+P+ V P
Sbjct: 390 ---AFPEALFHQLLQAMVHPDHKTRIGAHRIFSVVLVPTSVCP 429
Score = 131 bits (330), Expect = 9e-29
Identities = 107/364 (29%), Positives = 172/364 (46%), Gaps = 51/364 (14%)
Query: 635 HTLIALLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSAR 694
++L+ L + S+ AL+R FQ+A SLR ISL + G LPPSRRRSL TLA+ M++FS++
Sbjct: 601 YSLVLLFSRVKNSSHDALIRSFQMALSLRDISLMEGGPLPPSRRRSLFTLAASMVLFSSK 660
Query: 695 AADFSDLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDKVVFGSVEDEVAAMKSLSAV 754
A + L K +L +DPFL LVDD+ L+AV KV +G +D+ +A+ +LS +
Sbjct: 661 AFNLFSLADFTKVTLQGPRLDPFLNLVDDHKLKAVNSDQLKVAYGCEKDDASALDTLSNI 720
Query: 755 QLDDRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSP 814
L + T++ + + E+ ++ QLL F PDDA P G +T +
Sbjct: 721 ALSTEHSRGTLVYEIVKSLEDMCNSEMDKMREQLLTEFMPDDACPLGTRFLEDTHK---- 776
Query: 815 LAQIEFPDV--------DEMKGVG---------------PN*VEASQIVEHHFQPTAQMF 851
QI+ DV D+ G G P+ + +QI+E + T Q+
Sbjct: 777 TYQIDSGDVKPRKEDAEDQEFGDGTETVTKNNHVTFSEIPDLLTVNQILESVVETTRQVG 836
Query: 852 *VLINS------------WNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEE----VSR 895
+ ++ L GKQQK+ ++ + + + E+ +++E S
Sbjct: 837 RISFHTAADASYKEMTLHCENLLMGKQQKISSLLNSQLRHESSVNCSPRQHDEEIKIASF 896
Query: 896 QPVKALEYSKGDLKLVTQEQFQAQD--------QIRFRSQDTRKQHSLRLPPSSPYDKFL 947
P+ + G + ++F + Q ++ + RLP SSPYD FL
Sbjct: 897 HPMINSAFHTGVEVPLLSKEFDMKSPRTPVGTIQSPCYAELQNNPQAFRLPASSPYDNFL 956
Query: 948 KAAG 951
KAAG
Sbjct: 957 KAAG 960
>gb|AAX94774.1| cyclin-related protein 1 [Cucumis sativus]
Length = 489
Score = 342 bits (877), Expect = 3e-92
Identities = 229/534 (42%), Positives = 295/534 (54%), Gaps = 104/534 (19%)
Query: 473 VLYHNKAFPDALFHQLLLAMAHPDRETQIGAHSILSMVLMPSVVSPWLDQKKIS------ 526
V Y+ KAFPDALFHQLLLAMAHPD ET+IGAH I S+VLMPS+ P ++QK IS
Sbjct: 4 VSYYKKAFPDALFHQLLLAMAHPDHETRIGAHDIFSIVLMPSIKCPMMEQKTISSDTVSW 63
Query: 527 -------KKVESDGLSIQHESLSGEDPLNGKPVEEKPIHHSLISMILVELLCWWAFSNQL 579
+K+ S G S + + + +NGK ++ +S V LL
Sbjct: 64 LPFSSPTQKLTSGGFSFKDDDNHVSESINGK------LNSLRLSSHQVRLL--------- 108
Query: 580 VKCCYKQFLTKYWISEIFRI*DLLG*AVTK*VSCFLQSGFRQHLLKMALQIMRQWHTLIA 639
L+ W+ + M Q +++
Sbjct: 109 --------LSSIWVQAT-----------------------SADNTPANFEAMAQTYSIAL 137
Query: 640 LLYYLLVLSYMALVRCFQLAFSLRSISLDQEGGLPPSRRRSLLTLASHMLIFSARAADFS 699
L S+MALVRCFQLAFSLRSI++DQEGGL PSRRRS+ TLAS ML+FSAR D
Sbjct: 138 LFTRSKTSSHMALVRCFQLAFSLRSIAVDQEGGLLPSRRRSIFTLASFMLLFSARVGDLP 197
Query: 700 DLIPKVKASLTEAPVDPFLELVDDNLLRAVCIKSDK--VVFGSVEDEVAAMKSLSAVQLD 757
DL +KASL VDP L+LV+D L AV +KS+K V FGS EDEVAA+K LS ++LD
Sbjct: 198 DLTTVIKASLDNKMVDPHLQLVNDIRLLAVRVKSEKDSVPFGSEEDEVAALKFLSILELD 257
Query: 758 DRQLKETVISYFMTKFSKLPEDELSSIKNQLLQGFSPDDAYPSGPPLFMETPRPGSPLAQ 817
++QLKETV+S+F K++ L E ELSSI+ QLL GF PD+AYP G PLFMETPRP SPLA+
Sbjct: 258 EQQLKETVVSHFTIKYANLSEAELSSIREQLLHGFLPDEAYPLGAPLFMETPRPCSPLAK 317
Query: 818 IEFPDVDEMKGVGP----------------------------N*VEASQIVEHHFQPTAQ 849
+ FPD DE G+ P + + +Q++E + Q
Sbjct: 318 LAFPDYDE--GMPPAALTDDEAFLEPSGSQSDRKTSLSISNLDILNVNQLLESVLETARQ 375
Query: 850 MF*VLINS-----------WNRLETGKQQKMLTIRSFKNQQETKAIVLSSENEEV-SRQP 897
+ ++S L + KQQKM + SFK+++E KAIVLSSE E + P
Sbjct: 376 VASFPVSSAPVPYDQMKSQCEALVSCKQQKMSVLHSFKHKKEEKAIVLSSEIETLYPPLP 435
Query: 898 VKALEYSKGDLKLVTQEQFQAQDQIRFRSQDTRKQHSLRLPPSSPYDKFLKAAG 951
+ +E +GDLK E + QDQ S + + HSLRLPPSSPYDKFLKAAG
Sbjct: 436 LNTMEIVQGDLKFYNNETNRGQDQPLLCSHEYGR-HSLRLPPSSPYDKFLKAAG 488
>ref|XP_635760.1| hypothetical protein DDB0219543 [Dictyostelium discoideum]
gi|60464145|gb|EAL62306.1| hypothetical protein
DDB0219543 [Dictyostelium discoideum]
Length = 946
Score = 86.7 bits (213), Expect = 3e-15
Identities = 96/426 (22%), Positives = 189/426 (43%), Gaps = 40/426 (9%)
Query: 31 KRYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQRCYKDLRNESFGS 90
K+YK+++ I P+ + E+ I KL + +P ++ K+ +E + K+L +
Sbjct: 7 KKYKRIVKTIFPQVEGGEMQLGNISKLVYFCEMSPEQLQKVGPYIEAKAQKNLSKKRLVY 66
Query: 91 VKVILCIYRKLLSSCREQIPLFASSLLGIIRTLLEQTRADEVRILGCNTLVDFIIFQTDG 150
V + I R+L+ CR+ + LF+ + +++ LL QT ++I T + F Q D
Sbjct: 67 VSTCIIIIRELIIGCRKHLALFSGNATRLMKILLIQTEYPSLQIEATETFIRFASVQDDS 126
Query: 151 TYM-FNLEGFIPKLCQLAQEV-GDDERALLLRSAGLQTLSSMVKFMGEHSHLSMDFDKII 208
+ ++ FI +A+ + DD +R GL+ +++ V + L+ D D I
Sbjct: 127 NQIPPEIDEFIKYFIIMAKNMDNDDTMRRTIRGEGLRGVTAYVSL----ASLTDDLDTFI 182
Query: 209 SA---ILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATGFEIESKL 265
+ I+ +D N+ +++ S ++ ++ H SM N E+
Sbjct: 183 TKHTDIIYTILD-----NMQYRDQIPSPLIIINIKIVIIDKYHNRSMEN-RNQHHDETGE 236
Query: 266 DTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYCVLMYLQF 325
T N + CL +++ + TV ++ + +Y D+ W+ EK + + Q
Sbjct: 237 TTITNVKLLATECLKDLSGRV-DNMTVASLVTTIVNYLDSHQKWNFEKFAVHALTSVTQS 295
Query: 326 LLAESGNNSHLMLSILVKH--LDH-KNVAKQPIL-QIDIINITTQVAQNVKQQASVAVIG 381
+ G N +ML+ ++H LDH V K+ +L + I+N TT V
Sbjct: 296 I---KGQNYTIMLTSFLRHLELDHPPTVTKEIVLTSVAIVNDTTSSINMV---------- 342
Query: 382 AISDLIKHLRRCLQNSAE-ATDIGNDAHTLNTKLQSSIEMCILQLSNKVGDAGPIFDLMA 440
++ L+K L R ++NS++ D+ LQ+SI I + K+ G ++M
Sbjct: 343 -VTSLLKILVRSIENSSKHLPDVDQ-----QFSLQNSIVESIGLIGRKIKSTGKKVEIMN 396
Query: 441 VVLENV 446
++ ++
Sbjct: 397 QIINSL 402
>ref|XP_354658.3| PREDICTED: similar to KIAA0953 protein [Mus musculus]
Length = 817
Score = 83.2 bits (204), Expect = 4e-14
Identities = 98/422 (23%), Positives = 186/422 (43%), Gaps = 62/422 (14%)
Query: 15 LCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITEN 74
+C C +LR R YK+L+ I P + L + KL YA P ++ +I
Sbjct: 4 VCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKTNMEKLTFYALSAPEKLDRIGAY 56
Query: 75 LEQRCYKDLRNESFGSVKVILCIYRKLLSSCR-EQIPLFASSLLGIIRTLLEQTRADEVR 133
L +R +D+ +G V + + +LL +C + I LF S L ++ LLE + + ++
Sbjct: 57 LSERLIRDVGRHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVAKLLESEKPN-LQ 115
Query: 134 ILGCNTLVDFIIFQTD-GTYMFNLEGFIPKLCQLAQEVGDD-ERALLLRSAGLQTLSSMV 191
ILG N+ V F + D +Y + + F+ + ++ DD E +R +G++ L +V
Sbjct: 116 ILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSSHDDLEIKTKIRMSGIKGLQGVV 175
Query: 192 -KFMGEHSHLSM----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEE 246
K + + ++ DKI+ ++L NL VE+ S+S + L + P++E
Sbjct: 176 RKTVNDELQANIWDPQHMDKIVPSLL---------FNLQHVEEAESRSPSPL--QAPEKE 224
Query: 247 AHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTE 306
+NPA ++ CL + A ++ ++P+ + D
Sbjct: 225 ---------------------KENPAELAERCLRELLGRAAFG-NIKNAIKPVLIHLDNH 262
Query: 307 NHWSSEKGVAYC--VLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINIT 364
+ W + C ++MY + +SHL++ L+ HLD N ++ I+ +
Sbjct: 263 SLWEPKVFATRCFKIIMY-----SIQPQHSHLVIQQLLSHLD-ANSRSAATVRAGIVEVL 316
Query: 365 TQVAQNVKQQASV--AVIGAISDLIKHLRRCLQNSAEATDIGNDAHTLNTKLQSSIEMCI 422
++ A + SV V+ + L++ LR + + + G A +L +K+ E C+
Sbjct: 317 SEAA-IIAATGSVGPTVLEMFNTLLRQLRLSIDYALTGSYDG--AVSLGSKIIKEHEECM 373
Query: 423 LQ 424
Q
Sbjct: 374 FQ 375
>emb|CAE59290.1| Hypothetical protein CBG02625 [Caenorhabditis briggsae]
Length = 859
Score = 82.0 bits (201), Expect = 8e-14
Identities = 86/373 (23%), Positives = 155/373 (41%), Gaps = 40/373 (10%)
Query: 32 RYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQRCYKDLRNESFGSV 91
RY++L+ I PR L + KL YA +P ++ +I E L R +DL + V
Sbjct: 13 RYRRLVDSIYPRAVTDGLLHSNMQKLTFYAISHPEKLERIGEYLVMRMVRDLNRQRPVQV 72
Query: 92 KVILCIYRKLLSSCRE--QIPLFASSLLGIIRTLLEQTRADEVRILGCNTLVDFI-IFQT 148
K+ + +LL +C +P F+ + L +++ LLE A ++ L ++ V F I ++
Sbjct: 73 KIAVEAMDQLLQACHSSPSLPQFSENHLRMVQRLLESNNA-KMEQLATDSFVTFSNIEES 131
Query: 149 DGTYMFNLEGFIPKLCQLAQ-----EVGDDERALLLRSAGLQTLSSMV-KFMGEHSHLSM 202
+Y + FI K Q+ G+D R L R AGL+ L +V K + + H ++
Sbjct: 132 SPSYHRQYDFFIDKFSQMCHANPQAAYGEDFR--LARCAGLRGLRGVVWKSVTDDLHPNI 189
Query: 203 ----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATG 258
DKI+ +IL N + + +H+ N T
Sbjct: 190 WEQQHMDKIVPSILFNLQEPDDNGGFS--------------------SSHIPKFDNNFTD 229
Query: 259 FEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYC 318
+ D P S CL + A ++R V+EP+ + D WS ++
Sbjct: 230 STQSHRGDDEATPKVLSDRCLRELMGKASFG-SLRAVIEPVLKHMDLHKRWSPPP--SFA 286
Query: 319 VLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVA 378
+ ++ + + NS+ ++ L+ HLD + +I I + + +
Sbjct: 287 IHVFRAIIYSIQSQNSYFVIQELINHLD-SMCSADASTRIGIATVLSSIVSIAGTSIGPL 345
Query: 379 VIGAISDLIKHLR 391
++ + L+KHLR
Sbjct: 346 LLSIFNSLLKHLR 358
>gb|AAC46791.1| Hypothetical protein C32D5.3 [Caenorhabditis elegans]
gi|2496919|sp|Q09263|YQD3_CAEEL Hypothetical protein
C32D5.3 in chromosome II gi|17532235|ref|NP_495269.1|
transmembrane protein cmp44E (2G608) [Caenorhabditis
elegans]
Length = 859
Score = 80.9 bits (198), Expect = 2e-13
Identities = 91/373 (24%), Positives = 161/373 (42%), Gaps = 39/373 (10%)
Query: 32 RYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITENLEQRCYKDLRNESFGSV 91
RY++L+ I PR L + KL YA +P ++ +I E L R +DL + V
Sbjct: 13 RYRRLVDSIYPRAVTDGLLYSNMQKLTFYAISHPEKLERIGEYLVMRMVRDLSRQRPVQV 72
Query: 92 KVILCIYRKLLSSCRE--QIPLFASSLLGIIRTLLEQTRADEVRILGCNTLVDFI-IFQT 148
K+ + +LL +C +P F+ + L +++ LLE A ++ L ++ V F I ++
Sbjct: 73 KIAVEAMDQLLQACHSSPSLPQFSENHLRMVQRLLESNNA-KMEQLATDSFVTFSNIEES 131
Query: 149 DGTYMFNLEGFIPKLCQLAQ-----EVGDDERALLLRSAGLQTLSSMV-KFMGEHSHLSM 202
+Y + FI K Q+ GDD R L R AGL+ L +V K + + H ++
Sbjct: 132 SPSYHRQYDFFIDKFSQMCHANPQAAYGDDFR--LARCAGLRGLRGVVWKSVTDDLHPNI 189
Query: 203 ----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEEAHVSSMLNVATG 258
DKI+ +IL N LQ + K S +Q+ PK + N
Sbjct: 190 WEQQHMDKIVPSILFN---LQEPDDSGK-----GFSSSQI----PKFD-------NTFAD 230
Query: 259 FEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTENHWSSEKGVAYC 318
++D P S CL + A ++R V+EP+ + D W+ ++
Sbjct: 231 STQSHRVDDEATPKVLSDRCLRELMGKASFG-SLRAVIEPVLKHMDLHKRWTPPP--SFA 287
Query: 319 VLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQVAQNVKQQASVA 378
+ ++ + + NS+ ++ L+ HLD + +I I + + +
Sbjct: 288 IHVFRAIIYSIQSQNSYFVIQELINHLD-SMCSADASTRIGIATVLSSIVSIAGTSIGPL 346
Query: 379 VIGAISDLIKHLR 391
++ + L+KHLR
Sbjct: 347 LLSIFNSLLKHLR 359
>emb|CAI11812.1| novel protein [Danio rerio]
Length = 636
Score = 80.9 bits (198), Expect = 2e-13
Identities = 104/471 (22%), Positives = 198/471 (41%), Gaps = 68/471 (14%)
Query: 15 LCIFCPSLRARSRQPVKRYKKLIAEILPRNKVAELNDRKIGKLCEYASKNPLRIPKITEN 74
+C C +LR R YK+L+ I P + L + KL YA P ++ +I
Sbjct: 1 VCGCCGALRPR-------YKRLVDNIFPEDPEDGLVKANMEKLTFYALSAPEKLDRIGAY 53
Query: 75 LEQRCYKDLRNESFGSVKVILCIYRKLLSSCR-EQIPLFASSLLGIIRTLLEQTRADEVR 133
L +R +D+ +G V + + +LL +C + I LF S L ++R LLE + + ++
Sbjct: 54 LSERLSRDVARHRYGYVCIAMEALDQLLMACHCQSINLFVESFLKMVRKLLEADKPN-LQ 112
Query: 134 ILGCNTLVDFIIFQTD-GTYMFNLEGFIPKLCQLAQE-VGDDERALLLRSAGLQTLSSMV 191
ILG N+ V F + D +Y + + F+ + ++ D + +R AG++ L +V
Sbjct: 113 ILGTNSFVKFANIEEDTPSYHRSYDFFVSRFSEMCHSGYEDPDIRTKIRMAGIKGLQGVV 172
Query: 192 -KFMGEHSHLSM----DFDKIISAILENYVDLQSKSNLAKVEKLNSQSQNQLVQEFPKEE 246
K + + ++ DKI+ ++L NL E S+S + L Q KE+
Sbjct: 173 RKTVNDELQANIWDPQHMDKIVPSLL---------FNLQSGEGTESRSPSPL-QASEKEK 222
Query: 247 AHVSSMLNVATGFEIESKLDTAKNPAYWSKVCLYNIAKLAKEATTVRRVLEPLFHYFDTE 306
++PA ++ C + A ++ + P+ + D
Sbjct: 223 ----------------------ESPAELTERCFRELLGRAAYG-NIKNAVTPVLMHLDNH 259
Query: 307 NHWSSEKGVAYCVLMYLQFLLAESGNNSHLMLSILVKHLDHKNVAKQPILQIDIINITTQ 366
+ W +G + V + + + +SHL++ L+ HLD N ++ I+ + +
Sbjct: 260 SLW---EGKTFAVRCFKIIMYSIQSQHSHLVIQQLLGHLD-ANSKSSATVRAGIVEVLLE 315
Query: 367 VAQNVKQQASV--AVIGAISDLIKHLRRC----LQNSAEATDIGNDAHTLNTKLQSSIEM 420
VA + SV V+ + L++HLR L S + T+IG + + ++
Sbjct: 316 VAA-IAASGSVGPTVLEVFNTLLRHLRLSVDYELTGSYDCTNIG--TKIIKEHEERQLQE 372
Query: 421 CILQLSNKVGDAGPIFDLMAVVLENVSSSTIVARTTISAVYQTAKLITSVP 471
+++ + P + V+L I+ + I ++ T I S P
Sbjct: 373 AVIRTIGSFANTLPTYQRSEVML------FIMGKVPIPGLHPTLPSIGSGP 417
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.323 0.137 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,412,101,869
Number of Sequences: 2540612
Number of extensions: 54751677
Number of successful extensions: 194680
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 194495
Number of HSP's gapped (non-prelim): 135
length of query: 951
length of database: 863,360,394
effective HSP length: 138
effective length of query: 813
effective length of database: 512,755,938
effective search space: 416870577594
effective search space used: 416870577594
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146574.12


