

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146573.2 - phase: 0
(226 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
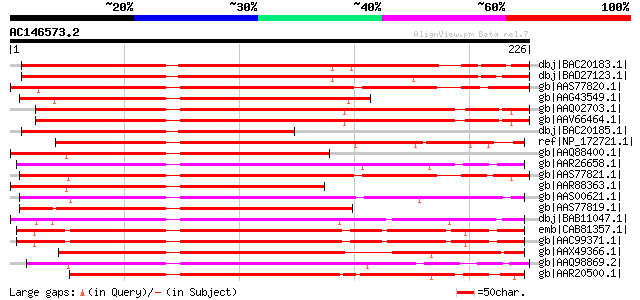
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC20183.1| dehydratiion responsive element binding protein ... 197 1e-49
dbj|BAD27123.1| dehydration responsive element binding protein 1... 194 2e-48
gb|AAS77820.1| CBF1 [Lycopersicon esculentum] gi|18535580|gb|AAK... 182 6e-45
gb|AAG43549.1| Avr9/Cf-9 rapidly elicited protein 111B [Nicotian... 179 5e-44
gb|AAQ02703.1| CBF-like protein [Glycine max] 178 9e-44
gb|AAV66464.1| drought responsive element binding protein [Glyci... 177 2e-43
dbj|BAC20185.1| dehydration responsive element binding protein 1... 175 1e-42
ref|NP_172721.1| DRE-binding protein, putative / CRT/DRE-binding... 174 1e-42
gb|AAQ88400.1| CaCBF1B [Capsicum annuum] 174 2e-42
gb|AAR26658.1| Cbcbf [Capsella bursa-pastoris] 172 5e-42
gb|AAS77821.1| CBF2 [Lycopersicon esculentum] 172 6e-42
gb|AAR88363.1| DREB-like protein 1 [Capsicum annuum] 172 6e-42
gb|AAS00621.1| DREB1 [Thellungiella salsuginea] 171 2e-41
gb|AAS77819.1| CBF3 [Lycopersicon esculentum] 169 5e-41
dbj|BAB11047.1| AP2 domain transcription factor-like protein [Ar... 167 2e-40
emb|CAB81357.1| DRE/CRT-binding protein DREB1C [Arabidopsis thal... 165 8e-40
gb|AAC99371.1| CRT/DRE binding factor 2 [Arabidopsis thaliana] 165 8e-40
gb|AAX49366.1| At1g63030 [Arabidopsis thaliana] gi|8493593|gb|AA... 164 1e-39
gb|AAQ98869.2| putative dehydration responsive element binding p... 164 2e-39
gb|AAR20500.1| DREB2-19 [Brassica napus] 163 4e-39
>dbj|BAC20183.1| dehydratiion responsive element binding protein 1 like protein
[Prunus avium]
Length = 232
Score = 197 bits (502), Expect = 1e-49
Identities = 114/230 (49%), Positives = 146/230 (62%), Gaps = 26/230 (11%)
Query: 6 SSFGDYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMR 65
SSF D S ++ + S EVILAS+ PK+RAGRRVFKETRHPVYRGVRRRNNNKWVCE+R
Sbjct: 20 SSFSDASVTARRASLSDEEVILASSCPKRRAGRRVFKETRHPVYRGVRRRNNNKWVCELR 79
Query: 66 VPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDA 125
PN N K+RIWLGTYPT EMAARAHDVA L +GK ACLNFADSAWRL +P S DA
Sbjct: 80 APN-----NKKARIWLGTYPTAEMAARAHDVAVLAFRGKLACLNFADSAWRLPVPASTDA 134
Query: 126 VEIRRAAMEAAKMF------AIEENHNQ---RSDRDAVDMNMENSKKNVLEVQVPVLSSE 176
EIRRAA EAA+ F ++E ++ ++ V M ++ + E ++ +
Sbjct: 135 AEIRRAATEAAEAFRQAEDGGVDEKESKAVVSEEKGCVGMEGSSNLFYLDEDEIFEMPRL 194
Query: 177 FEDMHHNLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNFSI 226
+DM ++L PP + Y +++D+E D + LW+FSI
Sbjct: 195 LDDMADGIMLC---------PPQCLDGY-MDWNDVETVDD--LKLWSFSI 232
>dbj|BAD27123.1| dehydration responsive element binding protein 1 [Prunus avium]
gi|23495460|dbj|BAC20184.1| dehydration responsive
element binding protein 1 like protein [Prunus avium]
Length = 240
Score = 194 bits (493), Expect = 2e-48
Identities = 119/231 (51%), Positives = 140/231 (60%), Gaps = 18/231 (7%)
Query: 6 SSFGDYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMR 65
S D S ++ + S +VILAS+RPKKRAGRRVFKETRHPVYRGVRRRNN+KWVCEMR
Sbjct: 18 SLLSDASVTTRGASCSDGDVILASSRPKKRAGRRVFKETRHPVYRGVRRRNNDKWVCEMR 77
Query: 66 VPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDA 125
PN KSRIWLGTYPT EMAARAHDVAAL +GK AC+NFADSAWRL +P S D
Sbjct: 78 EPN-----KKKSRIWLGTYPTAEMAARAHDVAALAFRGKLACINFADSAWRLPVPASMDT 132
Query: 126 VEIRRAAMEAAKMF------AIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLS----S 175
++IRRAA EAA+ F + + +R V + +N K +V + LS
Sbjct: 133 MDIRRAAAEAAEGFRPVEFGGVCSGSSDEKERMVVQVEEKNKKGSVNLERSRSLSLSYWD 192
Query: 176 EFEDMHHNLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNFSI 226
E E H LL E L +P Y N DDM V LW+FSI
Sbjct: 193 EEEVFHMPRLLHDMAEGLLLSPSQCLGGY-MNLDDMG--TDADVKLWSFSI 240
>gb|AAS77820.1| CBF1 [Lycopersicon esculentum] gi|18535580|gb|AAK57551.1| putative
transcriptional activator CBF1 [Lycopersicon esculentum]
Length = 210
Score = 182 bits (462), Expect = 6e-45
Identities = 105/232 (45%), Positives = 141/232 (60%), Gaps = 28/232 (12%)
Query: 1 MNIFLSSFGDY------SDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRR 54
MNIF + + D S SSSSS S EVILAS PKK AGR+ F+ETRHP+YRG+R+
Sbjct: 1 MNIFETYYSDSLILTESSSSSSSSSFSEEEVILASNNPKKPAGRKKFRETRHPIYRGIRK 60
Query: 55 RNNNKWVCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSA 114
RN+ KWVCE+R PN K+RIWLGT+PT EMAARAHDVAAL L+G+SACLNF+DSA
Sbjct: 61 RNSGKWVCEVREPN------KKTRIWLGTFPTAEMAARAHDVAALALRGRSACLNFSDSA 114
Query: 115 WRLTLPESNDAVEIRRAAMEAAKMFAIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLS 174
WRL +P S+++ +I++AA +A ++F EE + + N++ S V E + +
Sbjct: 115 WRLPIPASSNSKDIQKAAAQAVEIFRSEEVSGESPE---TSENVQESSDFVDEEAIFFMP 171
Query: 175 SEFEDMHHNLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNFSI 226
+M L+L PP G D D +++LWN+SI
Sbjct: 172 GLLANMAEGLML----------PPPQCAEMG---DHCVETDAYMITLWNYSI 210
>gb|AAG43549.1| Avr9/Cf-9 rapidly elicited protein 111B [Nicotiana tabacum]
Length = 219
Score = 179 bits (454), Expect = 5e-44
Identities = 93/159 (58%), Positives = 116/159 (72%), Gaps = 12/159 (7%)
Query: 5 LSSFGDYSDSSSSS----ETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKW 60
L+ + SDSSSSS S EV+LAS PKKRAGR+ F+ETRHPVYRGVR+RN++KW
Sbjct: 12 LAEYSSISDSSSSSCNRANHSDEEVMLASNNPKKRAGRKKFRETRHPVYRGVRKRNSDKW 71
Query: 61 VCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLP 120
VCE+R PN KSRIWLGT+P+ EMAARAHDVAA+ L+G+SACLNFADSAW+L +P
Sbjct: 72 VCELREPN------KKSRIWLGTFPSAEMAARAHDVAAIALRGRSACLNFADSAWKLPIP 125
Query: 121 ESNDAVEIRRAAMEAAKMFAIEENHN--QRSDRDAVDMN 157
S DA +I++AA EAA+ F E N + S D ++N
Sbjct: 126 ASTDAKDIQKAAAEAAEAFRSSEAENMPEYSGEDTKEVN 164
>gb|AAQ02703.1| CBF-like protein [Glycine max]
Length = 234
Score = 178 bits (452), Expect = 9e-44
Identities = 104/218 (47%), Positives = 137/218 (62%), Gaps = 14/218 (6%)
Query: 12 SDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMRVPNINV 71
S+ S S EV LA PKKRAGR+ F+ETRHPVYRGVRRRN++KWVCE+R PN
Sbjct: 28 SEGSRGVAFSDEEVRLAVRHPKKRAGRKKFRETRHPVYRGVRRRNSDKWVCEVREPN--- 84
Query: 72 NKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDAVEIRRA 131
K+RIWLGT+PTPEMAARAHDVAA+ L+G+ ACLNFADSAWRL +P + +A +I++A
Sbjct: 85 ---KKTRIWLGTFPTPEMAARAHDVAAMALRGRYACLNFADSAWRLPVPATAEAKDIQKA 141
Query: 132 AMEAAKMFAIEEN-HNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHHNLLLSIAN 190
A EAA+ F ++ N + ++ V+ + V E E+ ++ + N
Sbjct: 142 AAEAAQAFRPDQTLKNANTRQECVEAVAVAVAETTTATAQGVFYMEEEEQVLDMPELLRN 201
Query: 191 EPLRSNPPSPTNYYGSNYDDMEIFDTQ--IVSLWNFSI 226
L SPT+ G Y+D ++ D Q VSLWNFSI
Sbjct: 202 MVLM----SPTHCLGYEYEDADL-DAQDAEVSLWNFSI 234
>gb|AAV66464.1| drought responsive element binding protein [Glycine soja]
Length = 234
Score = 177 bits (450), Expect = 2e-43
Identities = 104/218 (47%), Positives = 136/218 (61%), Gaps = 14/218 (6%)
Query: 12 SDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMRVPNINV 71
S+ S S EV LA PKKRAGR+ F+ETRHPVYRGVRRRN++KWVCE+R PN
Sbjct: 28 SEGSRGVAFSDEEVRLAVRHPKKRAGRKKFRETRHPVYRGVRRRNSDKWVCEVREPN--- 84
Query: 72 NKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDAVEIRRA 131
K+RIWLGT+PTPEMAARAHDVAA+ L+G+ ACLNFADSAWRL +P + +A +I++A
Sbjct: 85 ---KKTRIWLGTFPTPEMAARAHDVAAMALRGRYACLNFADSAWRLPVPATAEAKDIQKA 141
Query: 132 AMEAAKMFAIEEN-HNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHHNLLLSIAN 190
A EAA+ F ++ N + ++ V+ V E E+ ++ + N
Sbjct: 142 AAEAAQAFRPDQTLKNANTRQECVEAVAVAVADTTTATAQGVFYMEEEEQVLDMPELLRN 201
Query: 191 EPLRSNPPSPTNYYGSNYDDMEIFDTQ--IVSLWNFSI 226
L SPT+ G Y+D ++ D Q VSLWNFSI
Sbjct: 202 MVLM----SPTHCLGYEYEDADL-DAQDAEVSLWNFSI 234
>dbj|BAC20185.1| dehydration responsive element binding protein 1 like protein
[Prunus avium]
Length = 130
Score = 175 bits (443), Expect = 1e-42
Identities = 88/119 (73%), Positives = 93/119 (77%), Gaps = 5/119 (4%)
Query: 6 SSFGDYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMR 65
SS D S + S EVILAS+RPKKRAGRRVFKETRHPVYRGVRRRNN+KWVCEMR
Sbjct: 16 SSLSDVSFKTRGPSWSDGEVILASSRPKKRAGRRVFKETRHPVYRGVRRRNNDKWVCEMR 75
Query: 66 VPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESND 124
PN N KSRIWLGTYPT EMAARAHDVAAL +GK ACLNFADSAWRL +P S D
Sbjct: 76 EPN-----NKKSRIWLGTYPTAEMAARAHDVAALAFRGKLACLNFADSAWRLPVPASMD 129
>ref|NP_172721.1| DRE-binding protein, putative / CRT/DRE-binding factor, putative
[Arabidopsis thaliana] gi|48479384|gb|AAT44959.1|
putative AP2/EREBP transcription factor [Arabidopsis
thaliana] gi|47605747|sp|Q9LN86|DRE1F_ARATH Dehydration
responsive element binding protein 1F (DREB1F protein)
gi|9502389|gb|AAF88096.1| T12C24.14 [Arabidopsis
thaliana]
Length = 209
Score = 174 bits (442), Expect = 1e-42
Identities = 105/221 (47%), Positives = 136/221 (61%), Gaps = 32/221 (14%)
Query: 21 SSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMRVPNINVNKNNKSRIW 80
++ ++ILA RPKKRAGRRVFKETRHPVYRG+RRRN +KWVCE+R P ++ RIW
Sbjct: 2 NNDDIILAEMRPKKRAGRRVFKETRHPVYRGIRRRNGDKWVCEVREPT------HQRRIW 55
Query: 81 LGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDAVEIRRAAMEAAKMFA 140
LGTYPT +MAARAHDVA L L+G+SACLNFADSAWRL +PESND IRR A EAA+MF
Sbjct: 56 LGTYPTADMAARAHDVAVLALRGRSACLNFADSAWRLPVPESNDPDVIRRVAAEAAEMFR 115
Query: 141 IEENHNQRS----DRDAVDMNMENSKKNVLEVQVPVLSS----EFEDMHHNLLLSIANEP 192
+ + + D VD+ + + + SS ++E++ ++ +A P
Sbjct: 116 PVDLESGITVLPCAGDDVDLGFGSGSGSGSGSEERNSSSYGFGDYEEV-STTMMRLAEGP 174
Query: 193 LRSNPPS------PTNYYGSN---YDDMEIFDTQIVSLWNF 224
L S P S PTN Y Y+DM SLW++
Sbjct: 175 LMSPPRSYMEDMTPTNVYTEEEMCYEDM--------SLWSY 207
>gb|AAQ88400.1| CaCBF1B [Capsicum annuum]
Length = 215
Score = 174 bits (441), Expect = 2e-42
Identities = 92/151 (60%), Positives = 109/151 (71%), Gaps = 18/151 (11%)
Query: 1 MNIFLSSFGDYSDSSSSSETSSS------------EVILASARPKKRAGRRVFKETRHPV 48
MNIF S + D SSSS + SS EVILAS PKK AGR+ F+ETRHPV
Sbjct: 1 MNIFRSYYSDPLTESSSSFSDSSIYSPNRAIFSDEEVILASNNPKKPAGRKKFRETRHPV 60
Query: 49 YRGVRRRNNNKWVCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACL 108
YRGVR+RN+ KWVCE+R PN KSRIWLGT+PT EMAARAHDVAA+ L+G+SACL
Sbjct: 61 YRGVRKRNSGKWVCEVREPN------KKSRIWLGTFPTAEMAARAHDVAAIALRGRSACL 114
Query: 109 NFADSAWRLTLPESNDAVEIRRAAMEAAKMF 139
NFADSAWRL +P S+D +I++AA EAA+ F
Sbjct: 115 NFADSAWRLPVPASSDTKDIQKAAAEAAEAF 145
>gb|AAR26658.1| Cbcbf [Capsella bursa-pastoris]
Length = 219
Score = 172 bits (437), Expect = 5e-42
Identities = 108/224 (48%), Positives = 134/224 (59%), Gaps = 15/224 (6%)
Query: 4 FLSSFGDYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCE 63
F FG +S SS LA++ PKK AGR+ F+ETRHPVYRGVRRRN+ KWVCE
Sbjct: 8 FSEMFGSEYESPVSSGGGDYCPTLATSCPKKPAGRKKFRETRHPVYRGVRRRNSGKWVCE 67
Query: 64 MRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESN 123
+R PN KSRIWLGT+PT +MAARAHDVAA+ L+G+SACLNFADSAWRL +PES
Sbjct: 68 VREPN------KKSRIWLGTFPTADMAARAHDVAAIALRGRSACLNFADSAWRLRIPEST 121
Query: 124 DAVEIRRAAMEAAKMFAIEENHNQRSDRD-AVDMNMENSKKNVLEVQVPVLSSEFEDMH- 181
A EI++AA EAA F E + + D DM + V Q L + EDM
Sbjct: 122 GAKEIQKAAAEAALAFQDEMMMSDTTTTDHGFDMEETFVEAIVTAEQSASLYIDEEDMFG 181
Query: 182 -HNLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNF 224
+L+ S+A L P P+ + NYD I VSLW++
Sbjct: 182 MPSLMASMAEGMLL---PLPSVQWNHNYD---IDGDDDVSLWSY 219
>gb|AAS77821.1| CBF2 [Lycopersicon esculentum]
Length = 220
Score = 172 bits (436), Expect = 6e-42
Identities = 101/225 (44%), Positives = 140/225 (61%), Gaps = 24/225 (10%)
Query: 5 LSSFGDYSDSSSSSETSSSE--VILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVC 62
LSS SD+++ + S +E +ILAS PKK AGR+ F+ETRHPVYRG+R+RN+ KWVC
Sbjct: 17 LSSSLSISDTNNLNHYSPNEEVIILASNNPKKPAGRKKFRETRHPVYRGIRKRNSGKWVC 76
Query: 63 EMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPES 122
E+R PN K+RIWLGT+PT EMAARAHDVAA+ L+G+SACLNFADS WRL +P S
Sbjct: 77 EVREPN------KKTRIWLGTFPTAEMAARAHDVAAIALRGRSACLNFADSVWRLPIPAS 130
Query: 123 NDAVEIRRAAMEAAKMFAIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHH 182
+++ +I++AA EAA++F EE + + N++ S V E + + +M
Sbjct: 131 SNSKDIQKAAAEAAEIFRSEEVSGESPE---TSENVQESSDFVDEEALFSMPGLLANMAE 187
Query: 183 NLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQ-IVSLWNFSI 226
L+L PP G +Y +E+ D + LWN+SI
Sbjct: 188 GLML----------PPPQCLEIGDHY--VELADVHAYMPLWNYSI 220
>gb|AAR88363.1| DREB-like protein 1 [Capsicum annuum]
Length = 215
Score = 172 bits (436), Expect = 6e-42
Identities = 91/149 (61%), Positives = 108/149 (72%), Gaps = 18/149 (12%)
Query: 1 MNIFLSSFGDYSDSSSSSETSSS------------EVILASARPKKRAGRRVFKETRHPV 48
MNIF S + D SSSS + SS EVILAS PKK AGR+ F+ETRHPV
Sbjct: 1 MNIFRSYYSDPLTESSSSFSDSSIYSPNRAIFSDEEVILASNNPKKPAGRKKFRETRHPV 60
Query: 49 YRGVRRRNNNKWVCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACL 108
YRGVR+RN+ KWVCE+R PN KSRIWLGT+PT EMAARAHDVAA+ L+G+SACL
Sbjct: 61 YRGVRKRNSGKWVCEVREPN------KKSRIWLGTFPTAEMAARAHDVAAIALRGRSACL 114
Query: 109 NFADSAWRLTLPESNDAVEIRRAAMEAAK 137
NFADSAWRL +P S+D +I++AA EAA+
Sbjct: 115 NFADSAWRLPVPASSDTKDIQKAAAEAAE 143
>gb|AAS00621.1| DREB1 [Thellungiella salsuginea]
Length = 216
Score = 171 bits (432), Expect = 2e-41
Identities = 102/228 (44%), Positives = 136/228 (58%), Gaps = 20/228 (8%)
Query: 5 LSSFGDYSDSSSSSETSSSEV------ILASARPKKRAGRRVFKETRHPVYRGVRRRNNN 58
++SF +++ S S V LA++ PKK AGR+ F+ETRHP+YRGVRRRN+
Sbjct: 1 MNSFSAFAEMFGSEYESPVTVGGDYCPTLATSCPKKPAGRKKFRETRHPIYRGVRRRNSG 60
Query: 59 KWVCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLT 118
KWVCE+R PN KSRIWLGT+PT EMAARAHDVAA+ L+G+SACLNFADSAWRL
Sbjct: 61 KWVCEVREPN------KKSRIWLGTFPTAEMAARAHDVAAIALRGRSACLNFADSAWRLR 114
Query: 119 LPESNDAVEIRRAAMEAAKMFAIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEF- 177
+PES A +I++AA EAA F E + SD ++ME + V+ + +
Sbjct: 115 IPESTCAKDIQKAAAEAAVAFQAEMSDTMTSDH---GLDMEETTVEVIVTEEEQSEGFYM 171
Query: 178 -EDMHHNLLLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNF 224
E+ + +AN P P+ +G NYD D VSLW++
Sbjct: 172 DEEAMFGMPRLLANMAEGMLLPPPSVQWGHNYDCDGDAD---VSLWSY 216
>gb|AAS77819.1| CBF3 [Lycopersicon esculentum]
Length = 205
Score = 169 bits (428), Expect = 5e-41
Identities = 84/145 (57%), Positives = 109/145 (74%), Gaps = 7/145 (4%)
Query: 5 LSSFGDYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEM 64
+ S +SDS ++ S EVILAS PKK AGR+ F+ETRHPVYRGVR+RN+ KWVCE+
Sbjct: 8 IESCSSFSDSIRANH-SDEEVILASNNPKKPAGRKKFRETRHPVYRGVRKRNSGKWVCEV 66
Query: 65 RVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESND 124
R PN K+RIWLGT+PT EMAARAHDVAA+ L+G+SACLNFADSAWRL P+S+D
Sbjct: 67 REPN------KKTRIWLGTFPTAEMAARAHDVAAIALRGRSACLNFADSAWRLPTPDSSD 120
Query: 125 AVEIRRAAMEAAKMFAIEENHNQRS 149
+I++AA +AA++F ++ + S
Sbjct: 121 TKDIQKAAAQAAEIFRPLKSEEEES 145
>dbj|BAB11047.1| AP2 domain transcription factor-like protein [Arabidopsis thaliana]
gi|15242244|ref|NP_200012.1| DRE-binding protein,
putative / CRT/DRE-binding factor, putative [Arabidopsis
thaliana] gi|48479306|gb|AAT44924.1| putative AP2/EREBP
transcription factor [Arabidopsis thaliana]
gi|47605744|sp|Q9FJ93|DRE1D_ARATH Dehydration responsive
element binding protein 1D (DREB1D protein) (C-repeat
binding factor 4) (C-repeat/dehydration responsive
element binding factor 4) (CRT/DRE binding factor 4)
Length = 224
Score = 167 bits (424), Expect = 2e-40
Identities = 107/232 (46%), Positives = 138/232 (59%), Gaps = 18/232 (7%)
Query: 1 MNIFLSSFGD----YSDSSSS-SETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRR 55
MN F S+F D SD S S++S LAS+ PKKRAGR+ F+ETRHP+YRGVR+R
Sbjct: 1 MNPFYSTFPDSFLSISDHRSPVSDSSECSPKLASSCPKKRAGRKKFRETRHPIYRGVRQR 60
Query: 56 NNNKWVCEMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAW 115
N+ KWVCE+R PN KSRIWLGT+PT EMAARAHDVAAL L+G+SACLNFADSAW
Sbjct: 61 NSGKWVCEVREPN------KKSRIWLGTFPTVEMAARAHDVAALALRGRSACLNFADSAW 114
Query: 116 RLTLPESNDAVEIRRAAMEAAKMFAIE-ENHNQRSDRDAVDMNMENSKKNVLEVQVPVLS 174
RL +PE+ EI++AA EAA F E ++ +A + E ++ E + +
Sbjct: 115 RLRIPETTCPKEIQKAASEAAMAFQNETTTEGSKTAAEAEEAAGEGVREG--ERRAEEQN 172
Query: 175 SEFEDMHHNLLLSIAN--EPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNF 224
M LL + N E + P G N++D + VSLW+F
Sbjct: 173 GGVFYMDDEALLGMPNFFENMAEGMLLPPPEVGWNHNDFD--GVGDVSLWSF 222
>emb|CAB81357.1| DRE/CRT-binding protein DREB1C [Arabidopsis thaliana]
gi|5596410|emb|CAB51470.1| DRE/CRT-binding protein
DREB1C [Arabidopsis thaliana] gi|3660552|dbj|BAA33436.1|
DREB1C [Arabidopsis thaliana]
gi|18416557|ref|NP_567719.1| DRE-binding protein
(DREB1C) / CRT/DRE-binding factor 2 (CBF2) [Arabidopsis
thaliana] gi|4322228|gb|AAD15976.1| CRT/DRE binding
factor 2 [Arabidopsis thaliana]
gi|3907541|gb|AAC78647.1| transcriptional activator CBF1
homolog [Arabidopsis thaliana]
gi|47605752|sp|Q9SYS6|DRE1C_ARATH Dehydration responsive
element binding protein 1C (DREB1C protein) (C-repeat
binding factor 2) (C-repeat/dehydration responsive
element binding factor 2) (CRT/DRE binding factor 2)
gi|3738228|dbj|BAA33793.1| DREB1C [Arabidopsis thaliana]
Length = 216
Score = 165 bits (418), Expect = 8e-40
Identities = 105/228 (46%), Positives = 137/228 (60%), Gaps = 25/228 (10%)
Query: 4 FLSSFG-DYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVC 62
F FG DY SS S + LA++ PKK AGR+ F+ETRHP+YRGVR+RN+ KWVC
Sbjct: 7 FSEMFGSDYESPVSSGGDYSPK--LATSCPKKPAGRKKFRETRHPIYRGVRQRNSGKWVC 64
Query: 63 EMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPES 122
E+R PN K+RIWLGT+ T EMAARAHDVAA+ L+G+SACLNFADSAWRL +PES
Sbjct: 65 ELREPN------KKTRIWLGTFQTAEMAARAHDVAAIALRGRSACLNFADSAWRLRIPES 118
Query: 123 NDAVEIRRAAMEAAKMFAIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHH 182
A EI++AA EAA F E H DA ++ME + V + P S + M
Sbjct: 119 TCAKEIQKAAAEAALNFQDEMCH---MTTDAHGLDMEETL--VEAIYTPEQSQDAFYMDE 173
Query: 183 NLLLSIANEPLRSNP------PSPTNYYGSNYDDMEIFDTQIVSLWNF 224
+L +++ L N PSP+ + N+D + VSLW++
Sbjct: 174 EAMLGMSS--LLDNMAEGMLLPSPSVQWNYNFD---VEGDDDVSLWSY 216
>gb|AAC99371.1| CRT/DRE binding factor 2 [Arabidopsis thaliana]
Length = 216
Score = 165 bits (418), Expect = 8e-40
Identities = 105/228 (46%), Positives = 137/228 (60%), Gaps = 25/228 (10%)
Query: 4 FLSSFG-DYSDSSSSSETSSSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVC 62
F FG DY SS S + LA++ PKK AGR+ F+ETRHP+YRGVR+RN+ KWVC
Sbjct: 7 FSEMFGSDYESPVSSGGDYSPK--LATSCPKKPAGRKKFRETRHPIYRGVRQRNSGKWVC 64
Query: 63 EMRVPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPES 122
E+R PN K+RIWLGT+ T EMAARAHDVAA+ L+G+SACLNFADSAWRL +PES
Sbjct: 65 ELREPN------KKTRIWLGTFQTAEMAARAHDVAAIALRGRSACLNFADSAWRLRIPES 118
Query: 123 NDAVEIRRAAMEAAKMFAIEENHNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHH 182
A EI++AA EAA F E H DA ++ME + V + P S + M
Sbjct: 119 TCAKEIQKAAAEAALNFQDEMCH---MTTDAHGLDMEETL--VEAIYTPEQSQDAFYMDE 173
Query: 183 NLLLSIANEPLRSNP------PSPTNYYGSNYDDMEIFDTQIVSLWNF 224
+L +++ L N PSP+ + N+D + VSLW++
Sbjct: 174 EAMLGMSS--LLDNMAEGMLLPSPSVQWNYNFD---VEGDDDVSLWSY 216
>gb|AAX49366.1| At1g63030 [Arabidopsis thaliana] gi|8493593|gb|AAF75816.1| Contains
similarity to transcriptional activator CBF1 from
Arabidopsis thaliana gb|U77378 and contains an AP2
PF|00847 domain gi|15221664|ref|NP_176491.1| AP2
domain-containing transcription factor, putative
[Arabidopsis thaliana] gi|48479274|gb|AAT44908.1|
putative AP2/EREBP transcription factor [Arabidopsis
thaliana] gi|47605751|sp|Q9SGJ6|DRE1E_ARATH Dehydration
responsive element binding protein 1E (DREB1E protein)
gi|12323254|gb|AAG51606.1| transcription factor DREB1A,
putative; 19375-18830 [Arabidopsis thaliana]
Length = 181
Score = 164 bits (416), Expect = 1e-39
Identities = 93/206 (45%), Positives = 125/206 (60%), Gaps = 30/206 (14%)
Query: 22 SSEVILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMRVPNINVNKNNKSRIWL 81
+ ++ +A +PKKRAGRR+FKETRHP+YRGVRRR+ +KWVCE+R P ++ R+WL
Sbjct: 3 NDDITVAEMKPKKRAGRRIFKETRHPIYRGVRRRDGDKWVCEVREPI------HQRRVWL 56
Query: 82 GTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDAVEIRRAAMEAAKMFAI 141
GTYPT +MAARAHDVA L L+G+SACLNF+DSAWRL +P S D IRR A EAA+MF
Sbjct: 57 GTYPTADMAARAHDVAVLALRGRSACLNFSDSAWRLPVPASTDPDTIRRTAAEAAEMFRP 116
Query: 142 EENHNQRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHH---NLLLSIANEPLRSNPP 198
E + V +SEF+ +++ +A EPL S P
Sbjct: 117 PEFST--------------------GITVLPSASEFDTSDEGVAGMMMRLAEEPLMSPPR 156
Query: 199 SPTNYYGSNYDDMEIFDTQIVSLWNF 224
S + S Y D E+ + +SLW++
Sbjct: 157 SYIDMNTSVYVDEEMC-YEDLSLWSY 181
>gb|AAQ98869.2| putative dehydration responsive element binding protein [Gossypium
hirsutum]
Length = 216
Score = 164 bits (414), Expect = 2e-39
Identities = 97/222 (43%), Positives = 133/222 (59%), Gaps = 21/222 (9%)
Query: 8 FGDYSDSSSSSETSSSE--VILASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMR 65
FG S+S + + S+ ++LAS+ PK+RAGR+ F+ETRHPVYRGVRRRN KWV E+R
Sbjct: 13 FGSVSESGTDRPVNFSDDYMMLASSYPKRRAGRKKFRETRHPVYRGVRRRNPGKWVSEVR 72
Query: 66 VPNINVNKNNKSRIWLGTYPTPEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDA 125
PN KSRIWLGT+P +MAARAHDVAA+ L+GKSACLNFADSAW+L +P S+D
Sbjct: 73 EPN------KKSRIWLGTFPKADMAARAHDVAAIALRGKSACLNFADSAWKLPVPASSDP 126
Query: 126 VEIRRAAMEAAKMFAIEENHNQRSDRDAV-DMNMENSKKNVLEVQVPVLSSEFEDMHHNL 184
+I++ E A+ F E+ + S DA N E K L+ + + F N+
Sbjct: 127 KDIQKTVAEVAETFRTAEHSSGNSRNDAKRSENTEMQKGFYLDEEALFGTQRF---WANM 183
Query: 185 LLSIANEPLRSNPPSPTNYYGSNYDDMEIFDTQIVSLWNFSI 226
+ P RS + +++ E+ D V LW++SI
Sbjct: 184 AAGMMMSPPRSG-------HDGGWEEHEVDD--YVPLWSYSI 216
>gb|AAR20500.1| DREB2-19 [Brassica napus]
Length = 214
Score = 163 bits (412), Expect = 4e-39
Identities = 102/201 (50%), Positives = 126/201 (61%), Gaps = 18/201 (8%)
Query: 27 LASARPKKRAGRRVFKETRHPVYRGVRRRNNNKWVCEMRVPNINVNKNNKSRIWLGTYPT 86
LA++ PKK AGR+ F+ETRHPVYRGVR RN+ KWVCE+R PN KSRIWLGT+ T
Sbjct: 29 LAASCPKKPAGRKKFRETRHPVYRGVRLRNSGKWVCEVREPN------KKSRIWLGTFLT 82
Query: 87 PEMAARAHDVAALTLKGKSACLNFADSAWRLTLPESNDAVEIRRAAMEAAKMFAIEENHN 146
E+AARAHDVAA+ L+GKSACLNFADSAWRL +PE+ EI++AA EAA F E N N
Sbjct: 83 AEIAARAHDVAAIALRGKSACLNFADSAWRLRIPETTCPKEIQKAAAEAALAFLAEIN-N 141
Query: 147 QRSDRDAVDMNMENSKKNVLEVQVPVLSSEFEDMHH--NLLLSIANEPLRSNPPSPTNYY 204
+D +DM + E V + E M LL S+A L P P+ ++
Sbjct: 142 TTTDH-GLDMEETIVEAIFTEENNDVFYMDEESMLEMPALLASMAEGMLL---PPPSVHF 197
Query: 205 GSNYDDMEIFDTQI-VSLWNF 224
G NYD FD VSLW++
Sbjct: 198 GHNYD----FDGDADVSLWSY 214
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.128 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 378,210,346
Number of Sequences: 2540612
Number of extensions: 14637997
Number of successful extensions: 58566
Number of sequences better than 10.0: 705
Number of HSP's better than 10.0 without gapping: 653
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 57148
Number of HSP's gapped (non-prelim): 875
length of query: 226
length of database: 863,360,394
effective HSP length: 123
effective length of query: 103
effective length of database: 550,865,118
effective search space: 56739107154
effective search space used: 56739107154
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146573.2


