

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146569.7 + phase: 0
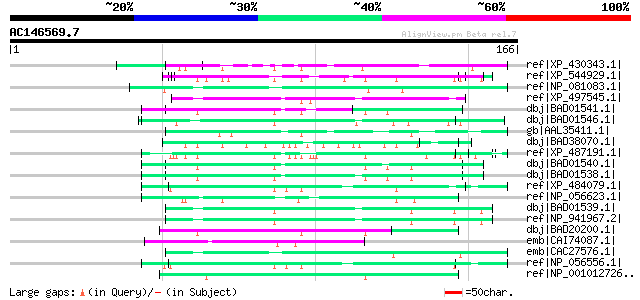
(166 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_430343.1| PREDICTED: hypothetical protein XP_430343 [Gall... 60 2e-08
ref|XP_544929.1| PREDICTED: similar to keratin associated protei... 57 2e-07
ref|NP_081083.1| hypothetical protein LOC68673 [Mus musculus] gi... 55 9e-07
ref|XP_497545.1| PREDICTED: keratin associated protein 4-3 [Homo... 53 3e-06
dbj|BAD01541.1| keratin associated protein [Homo sapiens] gi|384... 52 5e-06
dbj|BAD01546.1| keratin associated protein [Homo sapiens] gi|425... 52 8e-06
gb|AAL35411.1| PFTAIRE-interacting factor 2 [Drosophila melanoga... 51 1e-05
dbj|BAD38070.1| putative keratin associated protein [Oryza sativ... 51 1e-05
ref|XP_487191.1| PREDICTED: similar to keratin associated protei... 51 1e-05
dbj|BAD01540.1| keratin associated protein [Homo sapiens] gi|425... 51 1e-05
dbj|BAD01538.1| keratin associated protein [Homo sapiens] gi|384... 51 1e-05
ref|XP_484079.1| PREDICTED: similar to keratin associated protei... 51 1e-05
ref|NP_056623.1| keratin associated protein 5-1 [Mus musculus] g... 50 2e-05
dbj|BAD01539.1| keratin associated protein [Homo sapiens] gi|425... 50 2e-05
ref|NP_941967.2| keratin associated protein 10-5 [Homo sapiens] 50 2e-05
dbj|BAD20200.1| keratin associated protein [Homo sapiens] gi|605... 50 3e-05
emb|CAI74087.1| hypothetical protein [Theileria annulata] 50 3e-05
emb|CAC27576.1| keratin associated protein 4.5 [Homo sapiens] gi... 50 3e-05
ref|NP_056556.1| keratin associated protein 9-1 [Mus musculus] g... 49 4e-05
ref|NP_001012726.1| keratin associated protein 5-3 [Homo sapiens... 49 4e-05
>ref|XP_430343.1| PREDICTED: hypothetical protein XP_430343 [Gallus gallus]
Length = 287
Score = 60.5 bits (145), Expect = 2e-08
Identities = 44/141 (31%), Positives = 56/141 (39%), Gaps = 24/141 (17%)
Query: 36 EKKKMMEACRTVLFGSCNSCC---KTNCNGKCEKCT--CGSAKCDGIQCVTICFKCENPK 90
EK + C+ C +CC + C C+ C C CD C T C C+ P
Sbjct: 41 EKTVCCDPCQKPCCDPCQTCCDPCQKPCCDPCQTCCDPCQKPCCD--PCQTCCDPCQKP- 97
Query: 91 SCCE-------CKPIIKSCCNNKKCDDGCTP-KKPPSPKVQQCPQWCTCSRCYGYAPYQP 142
CC+ C P K CC+ C C P +KP Q C C + C P Q
Sbjct: 98 -CCDPCQQSVCCDPCQKPCCD--PCQTCCDPCQKPCCDPCQTCCDPCQKTVCCD--PCQT 152
Query: 143 YCNPCPPPNYMVCYDPNPEPC 163
C+PC VC DP +PC
Sbjct: 153 CCDPCQKT---VCCDPCQKPC 170
Score = 53.1 bits (126), Expect = 3e-06
Identities = 45/124 (36%), Positives = 53/124 (42%), Gaps = 27/124 (21%)
Query: 52 CNS-CC-----KTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNN 105
C+S CC KT C C+K C CD Q T+C CE K+ C C P K CC+
Sbjct: 7 CSSGCCSVVKSKTVCCSPCQKTVC----CDPCQ-KTVC--CE--KTVC-CDPCQKPCCD- 55
Query: 106 KKCDDGCTP-KKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPP-----NYMVCYDPN 159
C C P +KP Q C C C P Q C+PC P VC DP
Sbjct: 56 -PCQTCCDPCQKPCCDPCQTCCDPCQKPCC---DPCQTCCDPCQKPCCDPCQQSVCCDPC 111
Query: 160 PEPC 163
+PC
Sbjct: 112 QKPC 115
Score = 49.3 bits (116), Expect = 4e-05
Identities = 31/104 (29%), Positives = 40/104 (37%), Gaps = 11/104 (10%)
Query: 64 CEKCTCGSAKCDGIQCVTICFK-CENPKSCCECKPIIKSCCNNKKCDDGCTPKKPPSPKV 122
C C S C ++ T+C C+ C C+ + CC C D C +KP
Sbjct: 2 CPNVCCSSGCCSVVKSKTVCCSPCQKTVCCDPCQKTV--CCEKTVCCDPC--QKPCCDPC 57
Query: 123 QQCPQWCTCSRCYGYAPYQPYCNPCPPP---NYMVCYDPNPEPC 163
Q C C C P Q C+PC P C DP +PC
Sbjct: 58 QTCCDPCQKPCC---DPCQTCCDPCQKPCCDPCQTCCDPCQKPC 98
Score = 38.5 bits (88), Expect = 0.069
Identities = 36/132 (27%), Positives = 45/132 (33%), Gaps = 25/132 (18%)
Query: 45 RTVLFGSCNSCC----KTNCNGKCEKCTCGSAK--CDGIQCVTICFKCEN---------- 88
+TV C +CC KT C C+K C + CD Q C +
Sbjct: 143 KTVCCDPCQTCCDPCQKTVCCDPCQKPCCDPCQTCCDPCQKPCFPLTCSHWSTGFAYTPT 202
Query: 89 ---PKSCCECKPIIKSCC-NNKKCDDGCTPKKPPSPKVQQCPQWCTC-SRCYGYAPYQPY 143
P CC CC ++K C C + P P C C S+C P Q
Sbjct: 203 NTMPNGCCGGGSNCTVCCYSSKSCKTPCCKTQSCCPSQCCYPSQCCCPSQC--CCPSQCC 260
Query: 144 CNPCPPPNYMVC 155
PC P M C
Sbjct: 261 AMPCCMP--MTC 270
>ref|XP_544929.1| PREDICTED: similar to keratin associated protein 10-8 [Canis
familiaris]
Length = 729
Score = 57.0 bits (136), Expect = 2e-07
Identities = 33/104 (31%), Positives = 43/104 (40%), Gaps = 7/104 (6%)
Query: 53 NSCCKTNCNGK-CEKCTCGSAKCDGIQCVTICFK--CENPKSCCECKPIIKSCCNNKKCD 109
+SC + CN C++ C S C + C +C K C CC+ SCCN+ C
Sbjct: 128 SSCQPSCCNSSPCQEGCCVSVCCKPVCCTPVCCKPLCCESSPCCQQSSCQPSCCNSSPCQ 187
Query: 110 DGCTPK---KPPSPKVQQC-PQWCTCSRCYGYAPYQPYCNPCPP 149
+GC KP C P C S C + QP C C P
Sbjct: 188 EGCCVSVCCKPVCCTPVCCKPLCCEASPCCQQSSCQPSCGSCSP 231
Score = 52.8 bits (125), Expect = 4e-06
Identities = 35/109 (32%), Positives = 45/109 (41%), Gaps = 13/109 (11%)
Query: 51 SCNSCCKTNCNGKCEKCT--CG-SAKCDGIQCVTICFK--CENPKSCCECKPIIKSCCNN 105
+C C+ +C G C C CG S C + C +C K C CC+ SCCN+
Sbjct: 79 ACPGSCQPSC-GSCSPCQEGCGVSVCCKPVCCTPVCCKPLCCESSPCCQQSSCQPSCCNS 137
Query: 106 KKCDDGCTPK---KPPSPKVQQC-PQWCTCSRCYGYAPYQPYC---NPC 147
C +GC KP C P C S C + QP C +PC
Sbjct: 138 SPCQEGCCVSVCCKPVCCTPVCCKPLCCESSPCCQQSSCQPSCCNSSPC 186
Score = 49.7 bits (117), Expect = 3e-05
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 13/107 (12%)
Query: 54 SCCKTNC--NGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKPIIKSCCNNKKC 108
S C+++C + C++ +CGS C + C IC K CC CKP+ CC C
Sbjct: 616 SSCQSSCCSSSPCQEDSCGSVCCKPMCCTPICCK----PVCCTPICCKPV---CCQASPC 668
Query: 109 DDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPPNYMVC 155
+P +P S C C + C +P QP C P ++C
Sbjct: 669 CQP-SPCRPSSCVSLLCRPLCRPACCASPSPCQPSCRPQASSVSLLC 714
Score = 46.2 bits (108), Expect = 3e-04
Identities = 34/116 (29%), Positives = 46/116 (39%), Gaps = 12/116 (10%)
Query: 55 CCKTNCNGK--CEKCTCGSAKCDGIQCVTICFK--CENPKSCCECKPIIKSCCNNKKC-D 109
CCK C C+ C C + C +C+K C CC+ SCC++ C +
Sbjct: 413 CCKPVCCTPVCCKPVCCTPVCCKPVCCTPVCYKPVCCESSPCCQQSSCQPSCCSSSPCQE 472
Query: 110 DGCTPK--KPPSPKVQQC-PQWCTCSRCYGYAPYQPYC---NPCPPPNYM-VCYDP 158
D C KP C P C S C + QP C +PC + + VC P
Sbjct: 473 DSCVSVCCKPVCCTPVCCKPLCCESSPCCQQSSCQPSCCSSSPCQEDSCVSVCCKP 528
Score = 45.4 bits (106), Expect = 6e-04
Identities = 33/117 (28%), Positives = 41/117 (34%), Gaps = 24/117 (20%)
Query: 55 CCKTNC--------NGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKPIIKSCC 103
CCK C C+ C S+ C CV++C K CC CKP+ CC
Sbjct: 489 CCKPLCCESSPCCQQSSCQPSCCSSSPCQEDSCVSVCCK----PVCCTPVCCKPV---CC 541
Query: 104 NNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNP--CPPPNYMVCYDP 158
C C P + P C+ S C + C P C P VC P
Sbjct: 542 TPVCCKPVCCKASPCCQQSSCQPSCCSSSPCQEDSCVSVCCKPVCCTP----VCCKP 594
Score = 43.9 bits (102), Expect = 0.002
Identities = 38/134 (28%), Positives = 52/134 (38%), Gaps = 33/134 (24%)
Query: 55 CCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--CENP----KSCCECKPIIK------SC 102
CC+ + C+ C S+ C CV++C K C P CCE P + SC
Sbjct: 454 CCQQS---SCQPSCCSSSPCQEDSCVSVCCKPVCCTPVCCKPLCCESSPCCQQSSCQPSC 510
Query: 103 CNNKKCDDG-----------CTPK--KPPSPKVQQC-PQWCTCSRCYGYAPYQPYC---N 145
C++ C + CTP KP C P C S C + QP C +
Sbjct: 511 CSSSPCQEDSCVSVCCKPVCCTPVCCKPVCCTPVCCKPVCCKASPCCQQSSCQPSCCSSS 570
Query: 146 PCPPPNYM-VCYDP 158
PC + + VC P
Sbjct: 571 PCQEDSCVSVCCKP 584
Score = 43.1 bits (100), Expect = 0.003
Identities = 37/127 (29%), Positives = 50/127 (39%), Gaps = 21/127 (16%)
Query: 53 NSCCKTNCNGK-CEKCTCGSAKCDGIQCVTICFK--CENPKSCCE----------CKPII 99
+SC + CN C++ C S C + C +C K C CC+ C P
Sbjct: 174 SSCQPSCCNSSPCQEGCCVSVCCKPVCCTPVCCKPLCCEASPCCQQSSCQPSCGSCSPCQ 233
Query: 100 KSCCNNKKCDD-GCTPK--KPPSPKVQQC-PQWCTCSRCYGYAPYQPYC---NPCPPPNY 152
+ CC + C CTP KP C P C S C + QP C +PC +
Sbjct: 234 EGCCMSVCCKPVCCTPVCCKPVCCTPVCCKPLCCESSLCCQQSSCQPSCCSSSPCQEDSC 293
Query: 153 M-VCYDP 158
+ VC P
Sbjct: 294 VSVCCKP 300
Score = 41.2 bits (95), Expect = 0.011
Identities = 39/132 (29%), Positives = 48/132 (35%), Gaps = 31/132 (23%)
Query: 51 SCNS-CCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKCD 109
SC S CCK C C C C + C +C CE CC+ SCC++ C
Sbjct: 576 SCVSVCCKPVC---CTPVCCKPVCCTPVCCTPVC--CET-SPCCQQSSCQSSCCSSSPCQ 629
Query: 110 DG-----------CTP--KKPPSPKVQQC-PQWCTCSRCYGYAPYQP------YCNP-CP 148
+ CTP KP C P C S C +P +P C P C
Sbjct: 630 EDSCGSVCCKPMCCTPICCKPVCCTPICCKPVCCQASPCCQPSPCRPSSCVSLLCRPLCR 689
Query: 149 PPNYMVCYDPNP 160
P C P+P
Sbjct: 690 P---ACCASPSP 698
Score = 40.8 bits (94), Expect = 0.014
Identities = 30/112 (26%), Positives = 39/112 (34%), Gaps = 16/112 (14%)
Query: 50 GSCNSCCKTNCNGKCEK-CTCGSAKCDGIQCVTICFK--CENPKSCCECKPIIKSCCNNK 106
GSC+ C + C C K C C + C +C K C CC+ SCC++
Sbjct: 227 GSCSPCQEGCCMSVCCKPVCCTPVCCKPVCCTPVCCKPLCCESSLCCQQSSCQPSCCSSS 286
Query: 107 KCDDG-----------CTPK--KPPSPKVQQCPQWCTCSRCYGYAPYQPYCN 145
C + CTP KP K C C C P C+
Sbjct: 287 PCQEDSCVSVCCKPMCCTPVCCKPVCCKPVCCKPVCCTPVCCKPVCCTPVCS 338
Score = 35.0 bits (79), Expect = 0.77
Identities = 34/112 (30%), Positives = 41/112 (36%), Gaps = 18/112 (16%)
Query: 51 SCNS-CCKTNCNGKCEKCTCGSAKCDGIQCVTICFK----CENPK---SCCECKPIIKSC 102
SC S CCK C C C C + C +C K C+ SCC P +
Sbjct: 520 SCVSVCCKPVC---CTPVCCKPVCCTPVCCKPVCCKASPCCQQSSCQPSCCSSSPCQEDS 576
Query: 103 CNNKKCDD-GCTPK--KPPSPKVQQC-PQWCTCSRCYGYAPYQPYC---NPC 147
C + C CTP KP C P C S C + Q C +PC
Sbjct: 577 CVSVCCKPVCCTPVCCKPVCCTPVCCTPVCCETSPCCQQSSCQSSCCSSSPC 628
>ref|NP_081083.1| hypothetical protein LOC68673 [Mus musculus]
gi|12835082|dbj|BAB23145.1| unnamed protein product [Mus
musculus]
Length = 167
Score = 54.7 bits (130), Expect = 9e-07
Identities = 38/129 (29%), Positives = 51/129 (39%), Gaps = 12/129 (9%)
Query: 40 MMEACRTVLF--GSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKP 97
M+ +C +V G CC+ +C C+ C + C CV+ C + SCC
Sbjct: 1 MVSSCGSVCSEKGCGQGCCQPSC---CQTTCCRTTCCRPSCCVSSCCR----PSCCVSSC 53
Query: 98 IIKSCCNNKKCDDGCTPKK--PPSPKVQQCPQ-WCTCSRCYGYAPYQPYCNPCPPPNYMV 154
SCC + C C PS + C Q C S C G + +P C PC P
Sbjct: 54 CRPSCCRPQCCQSVCCQPTCCRPSCCISSCCQPSCGSSSCCGSSCCRPCCRPCCSPCCSP 113
Query: 155 CYDPNPEPC 163
C P PC
Sbjct: 114 CCRPCCRPC 122
Score = 43.9 bits (102), Expect = 0.002
Identities = 34/113 (30%), Positives = 48/113 (42%), Gaps = 27/113 (23%)
Query: 54 SCCKTNCN-GKCEKCTCGSAKCDGIQCVTICFK-CENP--KSCCE--CKPIIKSCCNNKK 107
+CC+ +C C + +CGS+ C G C C + C +P CC C+P + CC
Sbjct: 72 TCCRPSCCISSCCQPSCGSSSCCGSSCCRPCCRPCCSPCCSPCCRPCCRPCCRPCCR--- 128
Query: 108 CDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC--NPCPPPNYMVCYDP 158
C +P C Q C + C Y+P C + CP P M C P
Sbjct: 129 ---PCCCLRP------VCGQVCCQTTC-----YRPTCVISTCPRP--MCCAIP 165
Score = 35.4 bits (80), Expect = 0.59
Identities = 14/48 (29%), Positives = 18/48 (37%), Gaps = 7/48 (14%)
Query: 52 CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-------CENPKSC 92
C CC+ C C C C C + C T C++ C P C
Sbjct: 115 CRPCCRPCCRPCCRPCCCLRPVCGQVCCQTTCYRPTCVISTCPRPMCC 162
>ref|XP_497545.1| PREDICTED: keratin associated protein 4-3 [Homo sapiens]
Length = 158
Score = 53.1 bits (126), Expect = 3e-06
Identities = 34/101 (33%), Positives = 42/101 (40%), Gaps = 13/101 (12%)
Query: 54 SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKP--IIKSCCNNKKC 108
SCC+ +C C+ C + C C++ C C P SCC C+P I SCC C
Sbjct: 23 SCCRPSC---CQTTCCRTTCCRPSCCISSCCSCCKP-SCCRTTCCRPSCCISSCCRPSCC 78
Query: 109 DDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPP 149
C KP + C C S CY QP C C P
Sbjct: 79 ISSCC--KPSCCQTTCCRPSCCISSCYRPQCCQPSC--CRP 115
Score = 44.3 bits (103), Expect = 0.001
Identities = 31/99 (31%), Positives = 45/99 (45%), Gaps = 15/99 (15%)
Query: 44 CRTVLFG-SC--NSCCKTNC--NGKCEKCTCGSAKCDGIQCVTICFK--CENPKSCCECK 96
CRT SC +SCC+ +C + C+ C + C C++ C++ C P SCC
Sbjct: 58 CRTTCCRPSCCISSCCRPSCCISSCCKPSCCQTTCCRPSCCISSCYRPQCCQP-SCCRPA 116
Query: 97 PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCY 135
I SCC+ C C + P S CP C + C+
Sbjct: 117 CCISSCCHPSCCVSSC--RCPFS-----CPTTCCRTTCF 148
>dbj|BAD01541.1| keratin associated protein [Homo sapiens]
gi|38490571|ref|NP_941962.1| keratin associated protein
10-7 [Homo sapiens] gi|42558953|sp|P60409|K107_HUMAN
Keratin-associated protein KAP10-7 (Keratin-associated
protein 10.7) (High sulfur keratin-associated protein
10.7) (Keratin-associated protein KAP18-7)
(Keratin-associated protein 18.7)
Length = 370
Score = 52.4 bits (124), Expect = 5e-06
Identities = 30/115 (26%), Positives = 39/115 (33%), Gaps = 18/115 (15%)
Query: 52 CNSCCKTNC--NGKCEKCTCGSAKCDGIQCVTICFK------------CENPKSCCECKP 97
C S C +C C+ C S+ C CV +C K C SCC+
Sbjct: 81 CTSSCTPSCCQQSSCQLACCASSPCQQACCVPVCCKTVCCKPVYCVPVCSGDSSCCQQSS 140
Query: 98 IIKSCCNNKKCDDGCTPK---KPPSPKV-QQCPQWCTCSRCYGYAPYQPYCNPCP 148
+CC + C C KP + C Q +C C Q C P P
Sbjct: 141 CQSACCTSSPCQQACCVPICCKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSP 195
Score = 45.8 bits (107), Expect = 4e-04
Identities = 23/72 (31%), Positives = 32/72 (43%), Gaps = 7/72 (9%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKPIIK 100
C+ V G +SCC+ + C C A C+ C + C P SCC+ CKP
Sbjct: 161 CKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSPCQSGCISSCTP-SCCQQSSCKP--- 216
Query: 101 SCCNNKKCDDGC 112
+CC + C C
Sbjct: 217 ACCTSSPCQQAC 228
Score = 44.7 bits (104), Expect = 0.001
Identities = 32/115 (27%), Positives = 42/115 (35%), Gaps = 13/115 (11%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--CENPKSCCECKPIIKS 101
C V G + C +++C C C S+ C CV IC K C S C + S
Sbjct: 125 CVPVCSGDSSCCQQSSCQSAC----CTSSPCQQACCVPICCKPVCSGISSSCCQQSSCVS 180
Query: 102 CCNNKKCDDGCTPKKPPSPKVQQC-PQWCTCSRCYGYAPYQPYCNPCPPPNYMVC 155
C ++ C C P S + C P C S C +P C P C
Sbjct: 181 CVSSPCCQAVCEPSPCQSGCISSCTPSCCQQSSC------KPACCTSSPCQQACC 229
Score = 43.1 bits (100), Expect = 0.003
Identities = 37/145 (25%), Positives = 52/145 (35%), Gaps = 34/145 (23%)
Query: 44 CRTVLFGSCN-SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCE 94
C++ SC SCC+ + C+ C S+ C CV +C K ++ SCC+
Sbjct: 196 CQSGCISSCTPSCCQQS---SCKPACCTSSPCQQACCVPVCCKPVCCVPTCSDDSGSCCQ 252
Query: 95 -------------CKPIIKSCCNNKKCDDGCTPKKPPSPKVQQC-PQWCTCSRCYGYAPY 140
C P+ CC C C+ + C P CT S C +
Sbjct: 253 PACCTSSQSQQGCCVPV---CCKPVCCVPVCSGASTSCCQQSSCQPACCTTSCCRPSSSV 309
Query: 141 QPYCNP-CPPPNYMVCYDPNPEPCS 164
C P C P C P P C+
Sbjct: 310 SLLCRPVCRP----ACCVPVPSCCA 330
Score = 40.8 bits (94), Expect = 0.014
Identities = 33/114 (28%), Positives = 41/114 (35%), Gaps = 28/114 (24%)
Query: 45 RTVLFGSCNSC--------CKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECK 96
R L GSC+SC C +C CE C A C + C + + CC
Sbjct: 18 RVCLPGSCDSCSDSWQVDDCPESC---CEPPCCAPAPCLSLVCTPVSYV---SSPCC--- 68
Query: 97 PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCS-RCYGYAPYQPYCNPCPP 149
+ C C GCT PS C Q +C C +P Q C C P
Sbjct: 69 ---RVTCEPSPCQSGCTSSCTPS-----CCQQSSCQLACCASSPCQQAC--CVP 112
>dbj|BAD01546.1| keratin associated protein [Homo sapiens]
gi|42558956|sp|P60412|K10B_HUMAN Keratin-associated
protein KAP10-11 (Keratin-associated protein 10.11)
(High sulfur keratin-associated protein 10.11)
(Keratin-associated protein KAP18-11)
gi|38490579|ref|NP_941965.1| keratin associated protein
10-11 [Homo sapiens]
Length = 298
Score = 51.6 bits (122), Expect = 8e-06
Identities = 32/114 (28%), Positives = 43/114 (37%), Gaps = 16/114 (14%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCEC 95
C V G+ +SCC+ + C+ C S+ C CV +C K E+ SCC+
Sbjct: 115 CVPVCCGAASSCCRQS---SCQPACCASSSCQPACCVPVCCKPVCCVSTCSEDSSSCCQQ 171
Query: 96 KPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWC---TCSRCYGYAPYQPYCNP 146
+CC + C P K C C CSR QP C P
Sbjct: 172 SSCQPACCTSSSYQQACC--VPVCCKTVYCKPICCVPVCSRASYSRCQQPSCQP 223
Score = 47.4 bits (111), Expect = 1e-04
Identities = 41/161 (25%), Positives = 54/161 (33%), Gaps = 44/161 (27%)
Query: 43 ACRTVLFGSCN-SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-------------CEN 88
AC++ SC SCC+ + C+ C S+ C CV +C K C
Sbjct: 66 ACQSGCTSSCTPSCCQQS---SCQPACCTSSPCQQACCVPVCCKTVCCKPVCCVPVCCGA 122
Query: 89 PKSCCECKPIIKSCCNNKKCDDGC-------------TPKKPPSPKVQQ--CPQWCTCSR 133
SCC +CC + C C T + S QQ C C S
Sbjct: 123 ASSCCRQSSCQPACCASSSCQPACCVPVCCKPVCCVSTCSEDSSSCCQQSSCQPACCTSS 182
Query: 134 CYGYAPYQP------YCNP------CPPPNYMVCYDPNPEP 162
Y A P YC P C +Y C P+ +P
Sbjct: 183 SYQQACCVPVCCKTVYCKPICCVPVCSRASYSRCQQPSCQP 223
Score = 33.9 bits (76), Expect = 1.7
Identities = 25/105 (23%), Positives = 34/105 (31%), Gaps = 32/105 (30%)
Query: 51 SCNSCCKTNCNGK---CEKCTCGSAKCDGIQCVTICFK-----CENPK--------SCCE 94
SC C T+ + + C C + C I CV +C + C+ P SCC
Sbjct: 173 SCQPACCTSSSYQQACCVPVCCKTVYCKPICCVPVCSRASYSRCQQPSCQPACCTTSCCR 232
Query: 95 --------CKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTC 131
C P+ +S C C P C C C
Sbjct: 233 PSSSVSLLCHPVCRSTC--------CVPVSSCCAPTSSCQSSCCC 269
>gb|AAL35411.1| PFTAIRE-interacting factor 2 [Drosophila melanogaster]
gi|28381164|gb|AAF54113.3| CG31483-PA [Drosophila
melanogaster] gi|28571535|ref|NP_731093.2| CG31483-PA
[Drosophila melanogaster]
Length = 118
Score = 51.2 bits (121), Expect = 1e-05
Identities = 41/125 (32%), Positives = 46/125 (36%), Gaps = 31/125 (24%)
Query: 52 CNSCCKTNCNGKCEKC-------TCGS--AKCDGIQCVTICFKCENPKSCCE--CKPIIK 100
C SCC C+ C C CGS + C G C C C +P CC C P
Sbjct: 6 CGSCCGPCCSPCCSPCCPPCCNDCCGSCCSPCCGPCCSPCCGPCCSP--CCSPCCTPCCT 63
Query: 101 SCCNNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYCNPCPPPNYMVCYDP 158
CC C CTP P C CT C+ C P C+PC P C P
Sbjct: 64 PCCT--PCCKCCTPCCVPC-----CTPCCTPCCTPC-----CTPCCSPCCGP----CCSP 107
Query: 159 NPEPC 163
PC
Sbjct: 108 CCSPC 112
Score = 43.1 bits (100), Expect = 0.003
Identities = 32/110 (29%), Positives = 36/110 (32%), Gaps = 27/110 (24%)
Query: 42 EACRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE-CKPIIK 100
+ C + C CC C C C C C C C P CC+ C P
Sbjct: 28 DCCGSCCSPCCGPCCSPCCGPCCSPC-CSPC------CTPCCTPCCTP--CCKCCTPCCV 78
Query: 101 SCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPP 150
CC C CTP P CS C G P C+PC P
Sbjct: 79 PCC-TPCCTPCCTPCCTP-----------CCSPCCG-----PCCSPCCSP 111
Score = 39.7 bits (91), Expect = 0.031
Identities = 28/87 (32%), Positives = 31/87 (35%), Gaps = 21/87 (24%)
Query: 79 CVTICFKCENPKSCCE--CKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYG 136
C C C P CC C P CCN+ C C+P P CS C G
Sbjct: 2 CSPCCGSCCGP--CCSPCCSPCCPPCCND-CCGSCCSPCCGP-----------CCSPCCG 47
Query: 137 YAPYQPYCNPCPPPNYMVCYDPNPEPC 163
P C+PC P C P PC
Sbjct: 48 -----PCCSPCCSPCCTPCCTPCCTPC 69
Score = 39.3 bits (90), Expect = 0.041
Identities = 32/109 (29%), Positives = 37/109 (33%), Gaps = 24/109 (22%)
Query: 74 CDGIQCVTICFKCENP----------KSCCE--CKPIIKSCCN---NKKCDDGCTPKKPP 118
C G C C C +P SCC C P CC + C CTP P
Sbjct: 5 CCGSCCGPCCSPCCSPCCPPCCNDCCGSCCSPCCGPCCSPCCGPCCSPCCSPCCTPCCTP 64
Query: 119 --SPKVQQCPQWCT--CSRCYGYAPYQPYCNPCPPPNYMVCYDPNPEPC 163
+P + C C C+ C P C PC P C P PC
Sbjct: 65 CCTPCCKCCTPCCVPCCTPC-----CTPCCTPCCTPCCSPCCGPCCSPC 108
>dbj|BAD38070.1| putative keratin associated protein [Oryza sativa (japonica
cultivar-group)]
Length = 426
Score = 51.2 bits (121), Expect = 1e-05
Identities = 41/131 (31%), Positives = 47/131 (35%), Gaps = 32/131 (24%)
Query: 51 SCNSCC-----KTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSC---CECK-PIIKS 101
SC+SCC K NC+ C C C C C C+ P C C C P S
Sbjct: 122 SCSSCCDEPCCKPNCSACCAGSCCSPDCCS--CCKPNCSCCKTPSCCKPNCSCSCPSCSS 179
Query: 102 CCNNKKCDDGCT-------------PKKPPSPKVQQC----PQWCTCS----RCYGYAPY 140
CC+ C CT K PS QC P CTC+ C G A
Sbjct: 180 CCDTSCCKPSCTCFNIFSCFKSLYSCFKIPSCFKSQCNCSSPNCCTCTLPSCSCKGCACP 239
Query: 141 QPYCNPCPPPN 151
CN C P+
Sbjct: 240 SCGCNGCGCPS 250
Score = 50.1 bits (118), Expect = 2e-05
Identities = 34/118 (28%), Positives = 47/118 (39%), Gaps = 37/118 (31%)
Query: 51 SCNSCCKTNC-------NGKCEKCT-------CGSAKCDG----IQCVTICFKCEN---- 88
SCN CC C + C +C+ C A C C T CF C++
Sbjct: 301 SCNGCCGEQCCRCADCFSCSCPRCSSCFNIFKCSCAGCCSSLCKCPCTTQCFSCQSSCCK 360
Query: 89 -PKSCCECKPIIKSCCNNK-KCDDG--CTPKKPPSPK--------VQQCPQWCTCSRC 134
SCC+C+ SCC + C +G C+ KP P+ + C + C C RC
Sbjct: 361 RQPSCCKCQ---SSCCEGQPSCCEGHCCSLPKPSCPECSCGCVWSCKNCTEGCRCPRC 415
Score = 45.4 bits (106), Expect = 6e-04
Identities = 33/111 (29%), Positives = 44/111 (38%), Gaps = 29/111 (26%)
Query: 51 SCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTI-CFKCENPK---------SCCECKPIIK 100
SC C +C C C C S C+G C + C C P SC +CKP
Sbjct: 232 SCKGCACPSCG--CNGCGCPSCGCNGCGCPSCGCNGCGLPSCGCNGCGSCSCAQCKPDCG 289
Query: 101 S----CCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPC 147
S CC+ K +GC +QC C C+ C+ + P C+ C
Sbjct: 290 SCSTNCCSCKPSCNGCCG--------EQC---CRCADCFSCS--CPRCSSC 327
Score = 38.5 bits (88), Expect = 0.069
Identities = 31/99 (31%), Positives = 38/99 (38%), Gaps = 23/99 (23%)
Query: 55 CCKTNCNGKCEK-----CTCGSAKCDGIQCVTICFKCENPK----SCCECKPIIKSCCNN 105
CCK C+ KC++ C+C S CD C C C CC C SCC
Sbjct: 106 CCK--CSPKCKRPRCLNCSCSSC-CDEPCCKPNCSACCAGSCCSPDCCSCCKPNCSCCKT 162
Query: 106 KKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC 144
C C P S CP +CS C + +P C
Sbjct: 163 PSC---CKPNCSCS-----CP---SCSSCCDTSCCKPSC 190
Score = 37.0 bits (84), Expect = 0.20
Identities = 29/91 (31%), Positives = 36/91 (38%), Gaps = 20/91 (21%)
Query: 70 GSAKCDGIQCVTICFKCENPKSCCECK-PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQW 128
GS C I C+ C KC +PK CK P +C + CD+ C KP
Sbjct: 93 GSKLCICISCLCYCCKC-SPK----CKRPRCLNCSCSSCCDEPCC--KP----------- 134
Query: 129 CTCSRCYGYAPYQPYCNPCPPPNYMVCYDPN 159
CS C + P C C PN C P+
Sbjct: 135 -NCSACCAGSCCSPDCCSCCKPNCSCCKTPS 164
>ref|XP_487191.1| PREDICTED: similar to keratin associated protein 9.3 [Mus musculus]
Length = 427
Score = 51.2 bits (121), Expect = 1e-05
Identities = 44/146 (30%), Positives = 52/146 (35%), Gaps = 43/146 (29%)
Query: 44 CRTVLFGSC-------NSCCKTNC-NGKCEKCTCGSAKCDGIQCVTICFK------CENP 89
C+ SC SCC+T C C K C ++ C C T C+K C P
Sbjct: 166 CKPACVASCCSPPCCQPSCCETTCCRTTCYKPPCVTSCCHSSNCETTCYKPTCITSCCQP 225
Query: 90 KSCCE--------CKP-IIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPY 140
SCCE CKP + SCC C+ C + C C S C
Sbjct: 226 -SCCETSCCRTTCCKPTCVISCCQPSCCETTCC-------RTTCCKPTCVISCC------ 271
Query: 141 QPYC---NPCPPPNYMVCYDPNPEPC 163
QP C C P CY P PC
Sbjct: 272 QPSCCETTCCKPVCVTSCYSP---PC 294
Score = 50.4 bits (119), Expect = 2e-05
Identities = 35/106 (33%), Positives = 41/106 (38%), Gaps = 16/106 (15%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--CENPKSCCECKPIIKS 101
CRT +CCK C C + +C C CVT C+ C P SCCE +
Sbjct: 257 CRT-------TCCKPTCVISCCQPSCCETTCCKPVCVTSCYSPPCCQP-SCCET-----T 303
Query: 102 CCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCY-GYAPYQPYCNP 146
CC C C P S C C + Y Y YQP C P
Sbjct: 304 CCRTTCCKPTCRPTCVTSCCHPNCCGPSCCGQIYTNYGFYQPCCQP 349
Score = 47.8 bits (112), Expect = 1e-04
Identities = 38/136 (27%), Positives = 48/136 (34%), Gaps = 26/136 (19%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--CENPKSC--------C 93
C T + + C K C C + +C C CVT C C P SC C
Sbjct: 86 CETTCYRTTCCCYKPTCATNCCQPSCYETTCCKPACVTSCCSPPCCQPSSCETTCYRTTC 145
Query: 94 ECKPI-IKSCCNNKKCDDGCTPK-------KPPSPKVQQCPQWCTCSRCYGYAPYQPYC- 144
CKP + SCC C+ C PP + C C + C Y+P C
Sbjct: 146 CCKPTCVTSCCQPSCCETTCCKPACVASCCSPPCCQPSCCETTCCRTTC-----YKPPCV 200
Query: 145 -NPCPPPN-YMVCYDP 158
+ C N CY P
Sbjct: 201 TSCCHSSNCETTCYKP 216
Score = 46.2 bits (108), Expect = 3e-04
Identities = 37/119 (31%), Positives = 45/119 (37%), Gaps = 21/119 (17%)
Query: 44 CRTVLFGSCN------SCCKTNC-NGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE-- 94
CR SC+ SCC+T C C K T ++ C C T C+K SCC
Sbjct: 20 CRPTYVTSCSPSRYQPSCCETTCCRTTCCKPTSINSCCQPSMCETTCYKPICITSCCSPP 79
Query: 95 -CKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC--NPCPPP 150
C+P SCC C KP C C CY +P C + C PP
Sbjct: 80 CCQP---SCCETTCYRTTCCCYKP------TCATNCCQPSCYETTCCKPACVTSCCSPP 129
Score = 45.1 bits (105), Expect = 7e-04
Identities = 42/146 (28%), Positives = 49/146 (32%), Gaps = 39/146 (26%)
Query: 44 CRTVLFGS--CNSCC-KTNCNGKCEKCTCGSAKCDGIQCVTICFK--CENPK-------- 90
CRT + SCC +NC C K TC ++ C C T C + C P
Sbjct: 190 CRTTCYKPPCVTSCCHSSNCETTCYKPTCITSCCQPSCCETSCCRTTCCKPTCVISCCQP 249
Query: 91 SCCE--------CKPII------KSCCNNKKCDDGCTPK--KPPSPKVQQCPQWCTCSRC 134
SCCE CKP SCC C C PP + C C + C
Sbjct: 250 SCCETTCCRTTCCKPTCVISCCQPSCCETTCCKPVCVTSCYSPPCCQPSCCETTCCRTTC 309
Query: 135 YGYAPYQPYCNP-CPPPNYMVCYDPN 159
C P C P C PN
Sbjct: 310 ---------CKPTCRPTCVTSCCHPN 326
Score = 44.3 bits (103), Expect = 0.001
Identities = 31/114 (27%), Positives = 44/114 (38%), Gaps = 18/114 (15%)
Query: 43 ACRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENP---KSCCE---CK 96
+C T + + CCK C C + +C C CV C C P SCCE C+
Sbjct: 135 SCETTCYRT-TCCCKPTCVTSCCQPSCCETTCCKPACVASC--CSPPCCQPSCCETTCCR 191
Query: 97 ------PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC 144
P + SCC++ C+ C P+ C C + C +P C
Sbjct: 192 TTCYKPPCVTSCCHSSNCETTC---YKPTCITSCCQPSCCETSCCRTTCCKPTC 242
Score = 43.1 bits (100), Expect = 0.003
Identities = 39/136 (28%), Positives = 43/136 (30%), Gaps = 33/136 (24%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE--------CKPI-IKSCC 103
NSCC C + C C VT C SCCE CKP I SCC
Sbjct: 3 NSCCSP-----CSQLGCCRTTCCRPTYVTSCSPSRYQPSCCETTCCRTTCCKPTSINSCC 57
Query: 104 NNKKCDDGCTPK-------KPPSPKVQQCPQWCTCSRCYGYAP------YQPYC---NPC 147
C+ C PP + C C + C Y P QP C C
Sbjct: 58 QPSMCETTCYKPICITSCCSPPCCQPSCCETTCYRTTCCCYKPTCATNCCQPSCYETTCC 117
Query: 148 PPPNYMVCYDPNPEPC 163
P C P PC
Sbjct: 118 KPACVTSCCSP---PC 130
Score = 37.7 bits (86), Expect = 0.12
Identities = 35/124 (28%), Positives = 42/124 (33%), Gaps = 22/124 (17%)
Query: 54 SCCKTNCNGK---CEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKCDD 110
SCC+T C C K TC + C T C K SCC CC C+
Sbjct: 84 SCCETTCYRTTCCCYKPTCATNCCQPSCYETTCCKPACVTSCCS-----PPCCQPSSCET 138
Query: 111 GC---TPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCN-PCPPPN-------YMVCYDPN 159
C T P+ C C + C A C+ PC P+ CY P
Sbjct: 139 TCYRTTCCCKPTCVTSCCQPSCCETTCCKPACVASCCSPPCCQPSCCETTCCRTTCYKP- 197
Query: 160 PEPC 163
PC
Sbjct: 198 --PC 199
>dbj|BAD01540.1| keratin associated protein [Homo sapiens]
gi|42558951|sp|P60371|K106_HUMAN Keratin-associated
protein 10-6 (Keratin-associated protein 10.6) (High
sulfur keratin-associated protein 10.6)
(Keratin-associated protein 18-6) (Keratin-associated
protein 18.6) gi|38490575|ref|NP_941961.1| keratin
associated protein 10-6 [Homo sapiens]
Length = 365
Score = 50.8 bits (120), Expect = 1e-05
Identities = 30/115 (26%), Positives = 39/115 (33%), Gaps = 18/115 (15%)
Query: 52 CNSCCKTNC--NGKCEKCTCGSAKCDGIQCVTICFK------------CENPKSCCECKP 97
C S C +C C+ C S+ C CV +C K C SCC+
Sbjct: 81 CTSSCTPSCCQQSSCQLACCASSPCQQACCVPVCCKTVCCKPVCCVSVCCGDSSCCQQSS 140
Query: 98 IIKSCCNNKKCDDGCTPK---KPPSPKV-QQCPQWCTCSRCYGYAPYQPYCNPCP 148
+CC + C C KP + C Q +C C Q C P P
Sbjct: 141 CQSACCTSSPCQQACCVPVCCKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSP 195
Score = 47.4 bits (111), Expect = 1e-04
Identities = 30/114 (26%), Positives = 42/114 (36%), Gaps = 5/114 (4%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCC 103
C+ V G +SCC+ + C C A C+ C + C P SCC+ +CC
Sbjct: 161 CKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSPCQSGCTSSCTP-SCCQQSSCQPTCC 219
Query: 104 NNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYCNPCPPPNYMVC 155
+ C C P C C+ S C + QP C P + C
Sbjct: 220 TSSPCQQACCVPVCCVPVC--CVPTCSEDSSSCCQQSSCQPACCTSSPCQHACC 271
Score = 40.0 bits (92), Expect = 0.024
Identities = 35/114 (30%), Positives = 44/114 (37%), Gaps = 28/114 (24%)
Query: 45 RTVLFGSCNSC--------CKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECK 96
R L GSC+SC C +C CE C A C + C + + +P CC
Sbjct: 18 RVCLPGSCDSCSDSWQVDDCPESC---CEPPCCAPAPCLSLVCTPVS-RVSSP--CC--- 68
Query: 97 PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCS-RCYGYAPYQPYCNPCPP 149
P+ C C GCT PS C Q +C C +P Q C C P
Sbjct: 69 PV---TCEPSPCQSGCTSSCTPS-----CCQQSSCQLACCASSPCQQAC--CVP 112
Score = 35.4 bits (80), Expect = 0.59
Identities = 27/119 (22%), Positives = 39/119 (32%), Gaps = 24/119 (20%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKCDDG- 111
+SCC+ + C+ C S+ C CV +C SCC+ +CC C
Sbjct: 248 SSCCQQS---SCQPACCTSSPCQHACCVPVCSGAST--SCCQQSSCQPACCTASCCRSSS 302
Query: 112 --------------CTPKKPPSPKVQQC-PQWCTCSRCYGYAPYQPYCNPCPPPNYMVC 155
C P C P C + C +P C+ P Y +C
Sbjct: 303 SVSLLCHPVCKSTCCVPVPSCGASASSCQPSCCRTASCVSLL-CRPMCS--RPACYSLC 358
>dbj|BAD01538.1| keratin associated protein [Homo sapiens]
gi|38490568|ref|NP_941960.1| keratin associated protein
10-4 [Homo sapiens] gi|42558952|sp|P60372|K104_HUMAN
Keratin-associated protein 10-4 (Keratin-associated
protein 10.4) (High sulfur keratin-associated protein
10.4) (Keratin-associated protein 18-4)
(Keratin-associated protein 18.4)
Length = 401
Score = 50.8 bits (120), Expect = 1e-05
Identities = 30/115 (26%), Positives = 39/115 (33%), Gaps = 18/115 (15%)
Query: 52 CNSCCKTNC--NGKCEKCTCGSAKCDGIQCVTICFK------------CENPKSCCECKP 97
C S C +C C+ C S+ C CV +C K C SCC+
Sbjct: 81 CTSSCTPSCCQQSSCQLACCASSPCQQACCVPVCCKTVCCKPVCCVPVCCGDSSCCQQSS 140
Query: 98 IIKSCCNNKKCDDGCTPK---KPPSPKV-QQCPQWCTCSRCYGYAPYQPYCNPCP 148
+CC + C C KP + C Q +C C Q C P P
Sbjct: 141 CQSACCTSSPCQQACCVPICCKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSP 195
Score = 50.4 bits (119), Expect = 2e-05
Identities = 31/114 (27%), Positives = 42/114 (36%), Gaps = 5/114 (4%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCC 103
C+ V G +SCC+ + C C A C+ C + C P SCC+ +CC
Sbjct: 161 CKPVCSGISSSCCQQSSCVSCVSSPCCQAVCEPSPCQSGCISSCTP-SCCQQSSCQPACC 219
Query: 104 NNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYCNPCPPPNYMVC 155
+ C C P K C C+ S C + QP C P C
Sbjct: 220 TSSSCQQACC--VPVCCKTVCCKPVCSEDSSSCCQQSSCQPACCTSSPCQQACC 271
Score = 43.1 bits (100), Expect = 0.003
Identities = 30/116 (25%), Positives = 43/116 (36%), Gaps = 20/116 (17%)
Query: 44 CRTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK------------CENPKS 91
C+ V +SCC+ + C+ C S+ C CV +C K C S
Sbjct: 239 CKPVCSEDSSSCCQQS---SCQPACCTSSPCQQACCVPVCCKPVCCKPVGSVPICSGASS 295
Query: 92 -CCECKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYC 144
CC+ +CC + + GC P K C C+ S C + QP C
Sbjct: 296 LCCQQSSCQPACCTSSQSQQGCC--VPVCCKPVSCVPVCSGASSSCCQQSSCQPAC 349
Score = 38.5 bits (88), Expect = 0.069
Identities = 34/115 (29%), Positives = 43/115 (36%), Gaps = 25/115 (21%)
Query: 45 RTVLFGSCNSC--------CKTNCNGKCEKCTCGSAKCDGIQCVT-ICFKCENPKSCCEC 95
R L GSC+SC C +C CE C + C C++ +C S C
Sbjct: 13 RVCLPGSCDSCSDSWQVDDCPESC---CEPPCCAPSCCAPAPCLSLVCTPVSRVSS--PC 67
Query: 96 KPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCS-RCYGYAPYQPYCNPCPP 149
P+ C C GCT PS C Q +C C +P Q C C P
Sbjct: 68 CPV---TCEPSPCQSGCTSSCTPS-----CCQQSSCQLACCASSPCQQAC--CVP 112
>ref|XP_484079.1| PREDICTED: similar to keratin associated protein 9-1 [Mus musculus]
Length = 278
Score = 50.8 bits (120), Expect = 1e-05
Identities = 34/126 (26%), Positives = 43/126 (33%), Gaps = 8/126 (6%)
Query: 44 CRTVLFGS--CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKS 101
C+ GS C CC+T C C + C S+ C C C + CC+ S
Sbjct: 105 CQPSCCGSSCCQPCCQTTCCRTCFQPCCVSSCCRTPCCQPCCCVSSCCQPCCQPSCCQSS 164
Query: 102 CCNNKKCDDGCTPKKPPSPKVQQC----PQWCTCSRCYGYAPYQPYCNPCPPPNYMVCYD 157
CC C C P Q P+ C S C C PC P+ C
Sbjct: 165 CCQPSCCQPSCCQSSCCQPSCCQSSCCQPRCCISSCCQPRCCISSCCQPCCRPS--CCQS 222
Query: 158 PNPEPC 163
+PC
Sbjct: 223 SCCKPC 228
Score = 48.5 bits (114), Expect = 7e-05
Identities = 35/109 (32%), Positives = 42/109 (38%), Gaps = 17/109 (15%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-CENP----KSCCE-------CKPIIK 100
NSCC C C + TC C CVT C + C P SCC+ C+ +
Sbjct: 70 NSCCSPCCQPTCCRTTCCRTTCWRPSCVTSCCQPCCQPSCCGSSCCQPCCQTTCCRTCFQ 129
Query: 101 SCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPP 149
CC + C TP P V C Q C C + QP C C P
Sbjct: 130 PCCVSSCCR---TPCCQPCCCVSSCCQPCCQPSCCQSSCCQPSC--CQP 173
Score = 43.5 bits (101), Expect = 0.002
Identities = 31/100 (31%), Positives = 43/100 (43%), Gaps = 13/100 (13%)
Query: 54 SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-CENPKSCCE---CKPIIKSCCNNKKCD 109
SCC+++C C+ C S+ C C++ C + C P SCC+ CKP + C N C
Sbjct: 184 SCCQSSC---CQPRCCISSCCQPRCCISSCCQPCCRP-SCCQSSCCKPCCQPFCLNLCCQ 239
Query: 110 DGCTPKKPPSPKVQQCPQ-WCTC-SRCYGYAPYQPYCNPC 147
C+ P + C Q C C C C PC
Sbjct: 240 PACS---GPVTCTRTCYQPTCVCVPGCLSQGCGSSCCEPC 276
>ref|NP_056623.1| keratin associated protein 5-1 [Mus musculus] gi|111230|pir||A38346
ultra-high-sulfur keratin 1 - mouse
gi|200962|gb|AAA40106.1| serine 1 ultra high sulfur
protein
Length = 230
Score = 50.4 bits (119), Expect = 2e-05
Identities = 33/110 (30%), Positives = 40/110 (36%), Gaps = 8/110 (7%)
Query: 44 CRTVLFGSCNSCC-KTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCC----ECKPI 98
C G C CC +++C C CGS+ C C C + K CC CKP
Sbjct: 99 CSCSSCGGCKPCCCQSSCCKPCCSSGCGSSCCQSSCCKPCCCQSSCCKPCCCQSSCCKPC 158
Query: 99 IKSCCNNKKCDDGCTPKKPPSPKVQQC-PQWCTCSRCYGYAPYQPYCNPC 147
S C + C C KP + C P C S C C PC
Sbjct: 159 CSSGCGSSCCQSSCC--KPCCCQSSCCKPCCCQSSCCKPCCCQSSCCKPC 206
Score = 45.4 bits (106), Expect = 6e-04
Identities = 35/113 (30%), Positives = 43/113 (37%), Gaps = 16/113 (14%)
Query: 44 CRTVLFGSCNSC--CKTNCNGKCEKCT-----CGSAKCDGIQCVTICFKCENPKSCCECK 96
C V SC+SC C ++C G C C CGS+ C + C C SC CK
Sbjct: 53 CCCVPVCSCSSCGGCGSSCGG-CGSCGSSCGGCGSSCCKPVCCCVPVCSCS---SCGGCK 108
Query: 97 PIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTC--SRCYGYAPYQPYCNPC 147
P CC + C C+ S C + C C S C C PC
Sbjct: 109 PC---CCQSSCCKPCCSSGCGSSCCQSSCCKPCCCQSSCCKPCCCQSSCCKPC 158
Score = 43.5 bits (101), Expect = 0.002
Identities = 25/79 (31%), Positives = 32/79 (39%), Gaps = 19/79 (24%)
Query: 53 NSCCKTNCNGK----------CEKCTCGSAKCDGIQCVTICFK-CENPKSCCE------- 94
+SCCK C+ C+ C C S+ C C + C K C SCC+
Sbjct: 152 SSCCKPCCSSGCGSSCCQSSCCKPCCCQSSCCKPCCCQSSCCKPCCCQSSCCKPCCCQSS 211
Query: 95 -CKPIIKSCCNNKKCDDGC 112
CKP S C + C D C
Sbjct: 212 CCKPCCSSGCGSSCCQDSC 230
Score = 41.6 bits (96), Expect = 0.008
Identities = 32/107 (29%), Positives = 36/107 (32%), Gaps = 16/107 (14%)
Query: 51 SCNSC------CKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCN 104
SC C C +NC G CGS+ C + C C SC C SC
Sbjct: 20 SCGGCGSGCGGCGSNCGG------CGSSCCKPVCCCKPVCCCVPVCSCSSCGGCGSSCGG 73
Query: 105 NKKCDDGCTPKKPPSPK-VQQCPQWCTCSRCYGYAP---YQPYCNPC 147
C C K V C C+CS C G P C PC
Sbjct: 74 CGSCGSSCGGCGSSCCKPVCCCVPVCSCSSCGGCKPCCCQSSCCKPC 120
>dbj|BAD01539.1| keratin associated protein [Homo sapiens]
gi|42558950|sp|P60370|KR105_HUMAN Keratin-associated
protein 10-5 (Keratin-associated protein 10.5) (High
sulfur keratin-associated protein 10.5)
(Keratin-associated protein 18-5) (Keratin-associated
protein 18.5)
Length = 271
Score = 50.4 bits (119), Expect = 2e-05
Identities = 34/121 (28%), Positives = 48/121 (39%), Gaps = 19/121 (15%)
Query: 52 CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCECKPIIKSCC 103
C S C +C C+ C S+ C CV +C K ++ SCC+ +CC
Sbjct: 65 CTSSCTPSC---CQPACCASSPCQQACCVPVCCKPVCCLPTCSKDSSSCCQQSSCQPTCC 121
Query: 104 NNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYC---NPCPPPNYM-VCYD 157
+ C C P K C C+ S C ++ QP C +PC Y+ VC
Sbjct: 122 ASSSCQQSCC--VPVCCKPVCCVPTCSEDSSSCCQHSSCQPTCCTSSPCQQSCYVPVCCK 179
Query: 158 P 158
P
Sbjct: 180 P 180
Score = 43.5 bits (101), Expect = 0.002
Identities = 28/105 (26%), Positives = 41/105 (38%), Gaps = 16/105 (15%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCECKPIIKSCCN 104
+SCC+ + C+ C S+ C CV +C K E+ SCC+ +CC
Sbjct: 108 SSCCQQS---SCQPTCCASSSCQQSCCVPVCCKPVCCVPTCSEDSSSCCQHSSCQPTCCT 164
Query: 105 NKKCDDGCTPK---KPPSPKVQQCPQWCT--CSRCYGYAPYQPYC 144
+ C C KP K C C+ + C + QP C
Sbjct: 165 SSPCQQSCYVPVCCKPVCFKPICCVPVCSGASTSCCQQSSCQPAC 209
Score = 38.5 bits (88), Expect = 0.069
Identities = 29/112 (25%), Positives = 38/112 (33%), Gaps = 29/112 (25%)
Query: 52 CNSCCKTN-----CNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNK 106
C+S C + C C + CG+A C + C + C P ++ C
Sbjct: 9 CSSACSDSWRVDDCPESCCEPPCGTAPCLTLVCTPV---------SCVSSPCCQAACEPS 59
Query: 107 KCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPPNYMVCYDP 158
C GCT PS C C +P Q C C P VC P
Sbjct: 60 PCQSGCTSSCTPS---------CCQPACCASSPCQQAC--CVP----VCCKP 96
>ref|NP_941967.2| keratin associated protein 10-5 [Homo sapiens]
Length = 271
Score = 50.4 bits (119), Expect = 2e-05
Identities = 34/121 (28%), Positives = 48/121 (39%), Gaps = 19/121 (15%)
Query: 52 CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCECKPIIKSCC 103
C S C +C C+ C S+ C CV +C K ++ SCC+ +CC
Sbjct: 65 CTSSCTPSC---CQPACCASSPCQQACCVPVCCKPVCCLPTCSKDSSSCCQQSSCQPTCC 121
Query: 104 NNKKCDDGCTPKKPPSPKVQQCPQWCT--CSRCYGYAPYQPYC---NPCPPPNYM-VCYD 157
+ C C P K C C+ S C ++ QP C +PC Y+ VC
Sbjct: 122 ASSSCQQSCC--VPVCCKPVCCVPTCSEDSSSCCQHSSCQPTCCTSSPCQQSCYVPVCCK 179
Query: 158 P 158
P
Sbjct: 180 P 180
Score = 43.5 bits (101), Expect = 0.002
Identities = 28/105 (26%), Positives = 41/105 (38%), Gaps = 16/105 (15%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK--------CENPKSCCECKPIIKSCCN 104
+SCC+ + C+ C S+ C CV +C K E+ SCC+ +CC
Sbjct: 108 SSCCQQS---SCQPTCCASSSCQQSCCVPVCCKPVCCVPTCSEDSSSCCQHSSCQPTCCT 164
Query: 105 NKKCDDGCTPK---KPPSPKVQQCPQWCT--CSRCYGYAPYQPYC 144
+ C C KP K C C+ + C + QP C
Sbjct: 165 SSPCQQSCYVPVCCKPVCFKPICCVPVCSGASTSCCQQSSCQPAC 209
Score = 38.5 bits (88), Expect = 0.069
Identities = 29/112 (25%), Positives = 38/112 (33%), Gaps = 29/112 (25%)
Query: 52 CNSCCKTN-----CNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNK 106
C+S C + C C + CG+A C + C + C P ++ C
Sbjct: 9 CSSACSDSWRVDDCPESCCEPPCGTAPCLTLVCTPV---------SCVSSPCCQAACEPS 59
Query: 107 KCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPPNYMVCYDP 158
C GCT PS C C +P Q C C P VC P
Sbjct: 60 PCQSGCTSSCTPS---------CCQPACCASSPCQQAC--CVP----VCCKP 96
>dbj|BAD20200.1| keratin associated protein [Homo sapiens]
gi|60593038|ref|NP_001012727.1| keratin associated
protein 5-4 [Homo sapiens]
gi|56749058|sp|Q6L8H1|KR54_HUMAN Keratin-associated
protein 5-4 (Keratin-associated protein 5.4) (Ultrahigh
sulfur keratin-associated protein 5.4)
Length = 288
Score = 49.7 bits (117), Expect = 3e-05
Identities = 31/104 (29%), Positives = 36/104 (33%), Gaps = 16/104 (15%)
Query: 50 GSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKPIIKSCCNNK 106
G C SC G C C C C C C SCC+ CKP CC++
Sbjct: 179 GGCGSC--GGSKGGCGSCGCSQCSC----CKPCCCSSGCGSSCCQSSCCKP----CCSSS 228
Query: 107 KCDDGCTPK---KPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPC 147
C C KP + C C+ S C CNPC
Sbjct: 229 GCGSSCCQSSCCKPYCCQSSCCKPCCSSSGCGSSCCQSSCCNPC 272
Score = 46.6 bits (109), Expect = 3e-04
Identities = 23/77 (29%), Positives = 32/77 (40%), Gaps = 1/77 (1%)
Query: 50 GSCNSCCKTNC-NGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKC 108
G +SCC+++C C CGS+ C C C + K CC SCC + C
Sbjct: 210 GCGSSCCQSSCCKPCCSSSGCGSSCCQSSCCKPYCCQSSCCKPCCSSSGCGSSCCQSSCC 269
Query: 109 DDGCTPKKPPSPKVQQC 125
+ C+ P QC
Sbjct: 270 NPCCSQSSCCVPVCCQC 286
Score = 43.5 bits (101), Expect = 0.002
Identities = 29/99 (29%), Positives = 37/99 (37%), Gaps = 18/99 (18%)
Query: 50 GSCNSCCKTNCNGKCEKCTC----GSAKCDGIQCVTICFKCENPKSCCE---CKPII--- 99
G C SC + C+ C+ C C GS+ C C C SCC+ CKP
Sbjct: 189 GGCGSCGCSQCSC-CKPCCCSSGCGSSCCQSSCCKPCCSSSGCGSSCCQSSCCKPYCCQS 247
Query: 100 ---KSCCNNKKCDDGCTPKKPPSPKVQQ----CPQWCTC 131
K CC++ C C +P Q P C C
Sbjct: 248 SCCKPCCSSSGCGSSCCQSSCCNPCCSQSSCCVPVCCQC 286
Score = 34.3 bits (77), Expect = 1.3
Identities = 32/105 (30%), Positives = 33/105 (30%), Gaps = 27/105 (25%)
Query: 50 GSCNSC--CKTNCNGKC--EKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNN 105
G C SC K C G C K CGS C QC SCC K CC +
Sbjct: 169 GGCGSCGGSKGGC-GSCGGSKGGCGSCGCS--QC-----------SCC------KPCCCS 208
Query: 106 KKCDDGCTPK---KPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPC 147
C C KP C S C Y C PC
Sbjct: 209 SGCGSSCCQSSCCKPCCSSSGCGSSCCQSSCCKPYCCQSSCCKPC 253
>emb|CAI74087.1| hypothetical protein [Theileria annulata]
Length = 1506
Score = 49.7 bits (117), Expect = 3e-05
Identities = 25/75 (33%), Positives = 33/75 (43%), Gaps = 4/75 (5%)
Query: 45 RTVLFGSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKC--ENPKSC-CECKPIIKS 101
+ V C CCK N C+ CTC ++ +C C C +N K C C+C K
Sbjct: 784 QVVATSECTCCCKKQGNNGCQ-CTCSTSPPANCKCCLCCLCCPADNSKGCSCDCAGTPKK 842
Query: 102 CCNNKKCDDGCTPKK 116
CC C + C KK
Sbjct: 843 CCICCICCEACQTKK 857
Score = 42.0 bits (97), Expect = 0.006
Identities = 30/78 (38%), Positives = 37/78 (46%), Gaps = 18/78 (23%)
Query: 42 EACRTVLFGSCNSCCKTNCNGKC------------EKCTCGSAKCDGIQCVTICFKCENP 89
EAC+T G C+ CC N GKC EKC C S C G C C C+
Sbjct: 851 EACQTKK-GGCD-CCNGN-TGKCCVCCEKCPKTCSEKCACCSKDCSGC-CCMCCLICKKK 906
Query: 90 KSCCECKPIIKSCCNNKK 107
+S E P++K C N+KK
Sbjct: 907 ES--ERYPLLKLCENSKK 922
Score = 32.0 bits (71), Expect = 6.5
Identities = 19/66 (28%), Positives = 24/66 (35%), Gaps = 3/66 (4%)
Query: 52 CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCEC--KPIIKSCCNNKKCD 109
C CC + N K C C C C C+ K C+C K C +KC
Sbjct: 820 CCLCCPAD-NSKGCSCDCAGTPKKCCICCICCEACQTKKGGCDCCNGNTGKCCVCCEKCP 878
Query: 110 DGCTPK 115
C+ K
Sbjct: 879 KTCSEK 884
>emb|CAC27576.1| keratin associated protein 4.5 [Homo sapiens]
gi|37999766|sp|Q9BYR2|KR45_HUMAN Keratin-associated
protein 4-5 (Keratin-associated protein 4.5) (Ultrahigh
sulfur keratin-associated protein 4.5)
Length = 186
Score = 49.7 bits (117), Expect = 3e-05
Identities = 33/127 (25%), Positives = 44/127 (33%), Gaps = 22/127 (17%)
Query: 52 CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNKKCDDG 111
C SCC+T C C C + C C ++C++ +CC I SCC C+
Sbjct: 22 CPSCCQTTC---CRTTCCRPSCCKPQCCQSVCYQ----PTCCHPSCCISSCCRPYCCESS 74
Query: 112 CTPKKPPSPKVQQ--------------CPQWCTCSRCYGYAPYQPYCNP-CPPPNYMVCY 156
C P Q CP C S C C P C P+Y +
Sbjct: 75 CCRPCCCRPSCCQTTCCRTTCCRTTCCCPSCCVSSCCRPQCCQSVCCQPTCCRPSYCISS 134
Query: 157 DPNPEPC 163
+P C
Sbjct: 135 CCHPSCC 141
Score = 41.2 bits (95), Expect = 0.011
Identities = 31/105 (29%), Positives = 46/105 (43%), Gaps = 18/105 (17%)
Query: 54 SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKS-CCE---CKP--IIKSCCNNKK 107
SCC+T C C C + C CV+ C + + +S CC+ C+P I SCC+
Sbjct: 84 SCCQTTC---CRTTCCRTTCCCPSCCVSSCCRPQCCQSVCCQPTCCRPSYCISSCCHPSC 140
Query: 108 CDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC--NPCPPP 150
C+ C +P C + + C Y+P C + CP P
Sbjct: 141 CESSCC--RPCCCVRPVCGRVSCHTTC-----YRPTCVISTCPRP 178
Score = 35.0 bits (79), Expect = 0.77
Identities = 17/57 (29%), Positives = 23/57 (39%), Gaps = 16/57 (28%)
Query: 54 SCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-------CENPKSCCECKPIIKSCC 103
SCC+++C C C C C + C T C++ C P C SCC
Sbjct: 139 SCCESSC---CRPCCCVRPVCGRVSCHTTCYRPTCVISTCPRPLCCA------SSCC 186
>ref|NP_056556.1| keratin associated protein 9-1 [Mus musculus] gi|321286|pir||A45910
ultra-high-sulfur keratin - mouse
gi|1066818|gb|AAA81560.1| ultra-high sulphur keratin
Length = 186
Score = 49.3 bits (116), Expect = 4e-05
Identities = 29/105 (27%), Positives = 40/105 (37%), Gaps = 5/105 (4%)
Query: 44 CRTVLFGS--CNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKS 101
C+ GS C CC+T C C + C S+ C C C + CC+ S
Sbjct: 38 CQPSCCGSSCCQPCCQTTCCRTCFQPCCVSSCCRTPCCQPCCCVSSCCQPCCQPSCCQSS 97
Query: 102 CCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNP 146
CC + C+ C P + C Q C C + +P C P
Sbjct: 98 CCQPRCCESSCC---QPRCCISSCCQPCCRPSCCQSSCCRPCCQP 139
Score = 49.3 bits (116), Expect = 4e-05
Identities = 38/125 (30%), Positives = 47/125 (37%), Gaps = 19/125 (15%)
Query: 53 NSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFK-CENP----KSCCE-------CKPIIK 100
NSCC C C + TC C CVT C + C P SCC+ C+ +
Sbjct: 3 NSCCSPCCQPTCCRTTCCRTTCWRPSCVTSCCQPCCQPSCCGSSCCQPCCQTTCCRTCFQ 62
Query: 101 SCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYC--NPCPPPNYMVCYDP 158
CC + C TP P V C Q C C + QP C + C P C
Sbjct: 63 PCCVSSCCR---TPCCQPCCCVSSCCQPCCQPSCCQSSCCQPRCCESSCCQPR--CCISS 117
Query: 159 NPEPC 163
+PC
Sbjct: 118 CCQPC 122
Score = 48.1 bits (113), Expect = 9e-05
Identities = 36/129 (27%), Positives = 43/129 (32%), Gaps = 21/129 (16%)
Query: 44 CRTVLF------GSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENP---KSCCE 94
CRT + C CC+ +C CGS+ C T C C P SCC
Sbjct: 20 CRTTCWRPSCVTSCCQPCCQPSC--------CGSSCCQPCCQTTCCRTCFQPCCVSSCCR 71
Query: 95 CKPIIKSCCNNKKCDDGCTPKKPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPCPPPNYMV 154
CC + C C P S Q P+ C S C C PC P+
Sbjct: 72 TPCCQPCCCVSSCCQPCCQPSCCQSSCCQ--PRCCESSCCQPRCCISSCCQPCCRPS--C 127
Query: 155 CYDPNPEPC 163
C PC
Sbjct: 128 CQSSCCRPC 136
>ref|NP_001012726.1| keratin associated protein 5-3 [Homo sapiens]
gi|47522529|dbj|BAD20199.1| keratin associated protein
[Homo sapiens] gi|56749059|sp|Q6L8H2|KR53_HUMAN
Keratin-associated protein 5-3 (Keratin-associated
protein 5.3) (Ultrahigh sulfur keratin-associated
protein 5.3) (Keratin-associated protein 5-9)
(Keratin-associated protein 5.9) (UHS KerB-like)
Length = 238
Score = 49.3 bits (116), Expect = 4e-05
Identities = 29/104 (27%), Positives = 35/104 (32%), Gaps = 6/104 (5%)
Query: 50 GSCNSCCKTNCNGK---CEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCCNNK 106
G C SC + C+ C CGS+ C C C + K CC K CC +
Sbjct: 119 GGCGSCGCSQCSCYKPCCCSSGCGSSCCQSSCCKPSCSQSSCCKPCCSQSSCCKPCCCSS 178
Query: 107 KCDDGCTPK---KPPSPKVQQCPQWCTCSRCYGYAPYQPYCNPC 147
C C KP + C C S C C PC
Sbjct: 179 GCGSSCCQSSCCKPCCSQSSCCKPCCCSSGCGSSCCQSSCCKPC 222
Score = 43.1 bits (100), Expect = 0.003
Identities = 29/96 (30%), Positives = 37/96 (38%), Gaps = 12/96 (12%)
Query: 44 CRTVLFGSC--NSCCKTNCN-GKCEKCTCGSAKCDGIQCVTICFKCENPKSCCE---CKP 97
C + SC +SCCK +C+ C K C + C C C SCC+ CKP
Sbjct: 137 CSSGCGSSCCQSSCCKPSCSQSSCCKPCCSQSSC----CKPCCCSSGCGSSCCQSSCCKP 192
Query: 98 IIK--SCCNNKKCDDGCTPKKPPSPKVQQCPQWCTC 131
SCC C GC S + C +C
Sbjct: 193 CCSQSSCCKPCCCSSGCGSSCCQSSCCKPCSSQSSC 228
Score = 40.0 bits (92), Expect = 0.024
Identities = 30/131 (22%), Positives = 35/131 (25%), Gaps = 35/131 (26%)
Query: 50 GSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTI--------------------------- 82
G C SC C G C C C CV +
Sbjct: 65 GVCGSC--GGCKGGCGSCGGSKGGCGSSCCVPVCCSSSCGSCGGSKGVCGFRGGSKGGCG 122
Query: 83 ---CFKCENPKSCCECKPIIKSCCNNKKCDDGCTPK---KPPSPKVQQCPQWCTCSRCYG 136
C +C K CC SCC + C C+ KP + C C S C
Sbjct: 123 SCGCSQCSCYKPCCCSSGCGSSCCQSSCCKPSCSQSSCCKPCCSQSSCCKPCCCSSGCGS 182
Query: 137 YAPYQPYCNPC 147
C PC
Sbjct: 183 SCCQSSCCKPC 193
Score = 33.5 bits (75), Expect = 2.2
Identities = 17/54 (31%), Positives = 25/54 (45%), Gaps = 3/54 (5%)
Query: 50 GSCNSCCKTNCNGKCEKCTCGSAKCDGIQCVTICFKCENPKSCCECKPIIKSCC 103
G +SCC+++C C+ C S+ C C + C SCC+ SCC
Sbjct: 179 GCGSSCCQSSC---CKPCCSQSSCCKPCCCSSGCGSSCCQSSCCKPCSSQSSCC 229
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.135 0.478
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 347,471,040
Number of Sequences: 2540612
Number of extensions: 17000250
Number of successful extensions: 397947
Number of sequences better than 10.0: 3267
Number of HSP's better than 10.0 without gapping: 1559
Number of HSP's successfully gapped in prelim test: 1787
Number of HSP's that attempted gapping in prelim test: 337497
Number of HSP's gapped (non-prelim): 33859
length of query: 166
length of database: 863,360,394
effective HSP length: 118
effective length of query: 48
effective length of database: 563,568,178
effective search space: 27051272544
effective search space used: 27051272544
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146569.7


