

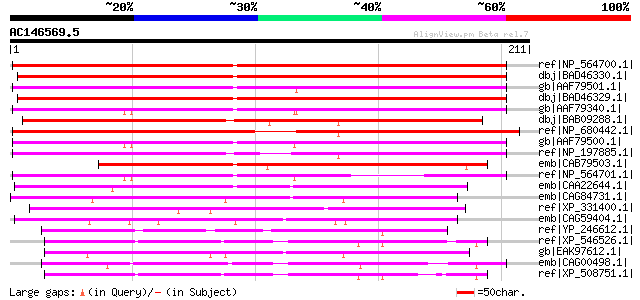
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146569.5 + phase: 0
(211 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_564700.1| hypothetical protein [Arabidopsis thaliana] 201 1e-50
dbj|BAD46330.1| unknown protein [Oryza sativa (japonica cultivar... 190 3e-47
gb|AAF79501.1| F20N2.17 [Arabidopsis thaliana] 187 1e-46
dbj|BAD46329.1| unknown protein [Oryza sativa (japonica cultivar... 187 1e-46
gb|AAF79340.1| F14J16.3 [Arabidopsis thaliana] 187 2e-46
dbj|BAB09288.1| unnamed protein product [Arabidopsis thaliana] g... 179 4e-44
ref|NP_680442.1| hypothetical protein [Arabidopsis thaliana] 179 5e-44
gb|AAF79500.1| F20N2.18 [Arabidopsis thaliana] 171 1e-41
ref|NP_197885.1| hypothetical protein [Arabidopsis thaliana] 155 9e-37
emb|CAB79503.1| putative protein [Arabidopsis thaliana] gi|29824... 142 5e-33
ref|NP_564701.1| hypothetical protein [Arabidopsis thaliana] 132 5e-30
emb|CAA22644.1| SPCC1919.13c [Schizosaccharomyces pombe] gi|1907... 101 2e-20
emb|CAG84731.1| unnamed protein product [Debaryomyces hansenii C... 77 4e-13
ref|XP_331400.1| hypothetical protein [Neurospora crassa] gi|289... 75 9e-13
emb|CAG59404.1| unnamed protein product [Candida glabrata CBS138... 64 2e-09
ref|YP_246612.1| hypothetical protein RF_0596 [Rickettsia felis ... 64 2e-09
ref|XP_546526.1| PREDICTED: similar to disrupted in bipolar diso... 64 4e-09
gb|EAK97612.1| hypothetical protein CaO19.7366 [Candida albicans... 62 1e-08
emb|CAG00498.1| unnamed protein product [Tetraodon nigroviridis] 62 1e-08
ref|XP_508751.1| PREDICTED: similar to disrupted in bipolar diso... 60 4e-08
>ref|NP_564700.1| hypothetical protein [Arabidopsis thaliana]
Length = 414
Score = 201 bits (510), Expect = 1e-50
Identities = 100/201 (49%), Positives = 134/201 (65%), Gaps = 1/201 (0%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD + KY+ A NL
Sbjct: 17 EEVWVKHYSSNHQILLVGEGDFSFSHSLATLFGSASNICASSLDSYDVVVRKYKKARSNL 76
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
L+ LG ++H VD ++ H L++ F DR+IFNFPH+GF ESD+ +I +H++LV
Sbjct: 77 KTLKRLGALLLHGVDATTLHFHPDLRYRRF-DRVIFNFPHAGFHGRESDSSLIRKHRELV 135
Query: 122 SGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNK 181
GF A +L GE+H++HK PFS WN++ LA L+ I+ V F ++ YPGY NK
Sbjct: 136 FGFFNGASRLLRANGEVHVSHKNKAPFSEWNLEELASRCFLVLIQRVAFEKNNYPGYENK 195
Query: 182 KGAGFKCDKSFPIGKCSTFKF 202
+G G +CD+ F +G+CSTFKF
Sbjct: 196 RGDGRRCDQPFLLGECSTFKF 216
>dbj|BAD46330.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 575
Score = 190 bits (482), Expect = 3e-47
Identities = 96/199 (48%), Positives = 126/199 (63%), Gaps = 1/199 (0%)
Query: 4 KIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIE 63
K +KHYSS +IL VG+GDFSF+L LA AFGS N+VATSLD L KY A N++E
Sbjct: 187 KWLKHYSSLQSILTVGDGDFSFSLALATAFGSGDNLVATSLDTIEDLRGKYSKAESNIME 246
Query: 64 LEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSG 123
L+ +G T++H +D M H LK F DRIIFNFPH+GF E D +I H++LV G
Sbjct: 247 LKRMGATVLHGIDAKRMKDHTNLKLRRF-DRIIFNFPHAGFKGKEDDLHMINLHRELVWG 305
Query: 124 FLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
F +A+++L GEIH++HK P+ W I++LA L I +V F + YPGY K+G
Sbjct: 306 FFQNARHLLRPYGEIHVSHKIGLPYDRWCIEHLAYESSLTMIAKVDFRKEDYPGYNQKRG 365
Query: 184 AGFKCDKSFPIGKCSTFKF 202
KCD+ F +G C TF F
Sbjct: 366 DSAKCDQPFELGACCTFMF 384
>gb|AAF79501.1| F20N2.17 [Arabidopsis thaliana]
Length = 435
Score = 187 bits (476), Expect = 1e-46
Identities = 100/224 (44%), Positives = 134/224 (59%), Gaps = 24/224 (10%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD + KY+ A NL
Sbjct: 17 EEVWVKHYSSNHQILLVGEGDFSFSHSLATLFGSASNICASSLDSYDVVVRKYKKARSNL 76
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIG------ 115
L+ LG ++H VD ++ H L++ F DR+IFNFPH+GF ESD+ +I
Sbjct: 77 KTLKRLGALLLHGVDATTLHFHPDLRYRRF-DRVIFNFPHAGFHGRESDSSLIRFVLLAA 135
Query: 116 -----------------EHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAE 158
+H++LV GF A +L GE+H++HK PFS WN++ LA
Sbjct: 136 ECVDQLLQLRGICVKSLKHRELVFGFFNGASRLLRANGEVHVSHKNKAPFSEWNLEELAS 195
Query: 159 NEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
L+ I+ V F ++ YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 196 RCFLVLIQRVAFEKNNYPGYENKRGDGRRCDQPFLLGECSTFKF 239
>dbj|BAD46329.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 470
Score = 187 bits (476), Expect = 1e-46
Identities = 93/199 (46%), Positives = 125/199 (62%), Gaps = 1/199 (0%)
Query: 4 KIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIE 63
K +KHYSS +IL+VG+GDFSF+ LA AF S N+V+TSLD +L KY A N++
Sbjct: 74 KWLKHYSSMQSILVVGDGDFSFSRALAVAFCSGENLVSTSLDSYEALRGKYANAESNIMV 133
Query: 64 LEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSG 123
L+ +G T +H VD M H LK F DRI+FN PH+GF E D +I HK LV G
Sbjct: 134 LKLMGATTLHGVDAKTMKHHTDLKMRRF-DRIVFNLPHAGFKAKEGDMRMINLHKDLVRG 192
Query: 124 FLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
F +A+ +L GEIH++HK + NW I+ LA L+ +E+V F+ YPGY +K+G
Sbjct: 193 FFRNARCLLRPSGEIHVSHKRGKVYENWEIEKLASESSLIMVEKVDFHIEDYPGYNHKRG 252
Query: 184 AGFKCDKSFPIGKCSTFKF 202
G +CD+ FP+G C FKF
Sbjct: 253 DGPRCDEPFPLGPCCIFKF 271
>gb|AAF79340.1| F14J16.3 [Arabidopsis thaliana]
Length = 1033
Score = 187 bits (475), Expect = 2e-46
Identities = 100/225 (44%), Positives = 134/225 (59%), Gaps = 25/225 (11%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD + KY+ A NL
Sbjct: 539 EEVWVKHYSSNHQILLVGEGDFSFSHSLATLFGSASNICASSLDSYDVVVRKYKKARSNL 598
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIG------ 115
L+ LG ++H VD ++ H L++ F DR+IFNFPH+GF ESD+ +I
Sbjct: 599 KTLKRLGALLLHGVDATTLHFHPDLRYRRF-DRVIFNFPHAGFHGRESDSSLIRPAAATS 657
Query: 116 ------------------EHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLA 157
+H++LV GF A +L GE+H++HK PFS WN++ LA
Sbjct: 658 RNMCEEFVHVLYLFVCCRKHRELVFGFFNGASRLLRANGEVHVSHKNKAPFSEWNLEELA 717
Query: 158 ENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
L+ I+ V F ++ YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 718 SRCFLVLIQRVAFEKNNYPGYENKRGDGRRCDQPFLLGECSTFKF 762
Score = 171 bits (433), Expect = 1e-41
Identities = 98/232 (42%), Positives = 130/232 (55%), Gaps = 32/232 (13%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLD-------DRG------ 48
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD ++G
Sbjct: 156 EEVWVKHYSSKHQILLVGEGDFSFSCSLATCFGSASNIYASSLDSYDYKPVNKGCSFKLD 215
Query: 49 --------------SLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDR 94
+ KY+ A NL L+ LG ++H VD ++ H L++ F DR
Sbjct: 216 FLSCCMSFMVIKPDDVVRKYKNARSNLETLKRLGAFLLHGVDATTLHFHPDLRYRRF-DR 274
Query: 95 IIFNFPHSGFFQNESDAWVI----GEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSN 150
+IFNFPH+GF + ESD I + L FL A +ML GE+H++HK PF
Sbjct: 275 VIFNFPHTGFHRKESDPCQIQPAAATLRNLFKDFLHGASHMLRADGEVHVSHKNKAPFCY 334
Query: 151 WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
WN++ LA L+ I+ F + YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 335 WNLEELASRCFLVLIQLEAFEKRNYPGYENKRGDGSRCDQPFLLGECSTFKF 386
>dbj|BAB09288.1| unnamed protein product [Arabidopsis thaliana]
gi|15241118|ref|NP_200417.1| hypothetical protein
[Arabidopsis thaliana]
Length = 220
Score = 179 bits (455), Expect = 4e-44
Identities = 95/189 (50%), Positives = 126/189 (66%), Gaps = 5/189 (2%)
Query: 6 VKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELE 65
++ YS+ ILLVGEGDFSF+L LA+ FGSA N+ ATSLD R L +KY N+ LE
Sbjct: 9 LQQYSNKQKILLVGEGDFSFSLSLARVFGSATNITATSLDTREELGIKYTDGKANVEGLE 68
Query: 66 GLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGF-FQNESDAWVIGEHKKLVSGF 124
GCT++H V+VH+M+ ++L +DRIIFNFPHSG F +E D + I H+ LV GF
Sbjct: 69 LFGCTVVHGVNVHSMSSDYRLGR---YDRIIFNFPHSGLGFGSEHDIFFIMLHQGLVRGF 125
Query: 125 LGSAKYML-NVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
L SA+ ML + GEIH+THKT PF+ W I+ LA + L I E+ F++ +PGY NKKG
Sbjct: 126 LESARKMLKDEDGEIHVTHKTTDPFNRWGIETLAGEKGLRLIGEIEFHKWAFPGYSNKKG 185
Query: 184 AGFKCDKSF 192
G C+ +F
Sbjct: 186 GGSNCNSTF 194
>ref|NP_680442.1| hypothetical protein [Arabidopsis thaliana]
Length = 256
Score = 179 bits (454), Expect = 5e-44
Identities = 95/209 (45%), Positives = 129/209 (61%), Gaps = 19/209 (9%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
E K ++HY++ ILLVGEGDFSF+L LA+AFGSA N+ ATSLD +G L K++ N+
Sbjct: 63 ETKRLRHYTNKQKILLVGEGDFSFSLSLARAFGSASNLTATSLDTQGELEQKFKNGKANV 122
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
ELE LGC++++ V+VH+M + +DR+IFNFP +LV
Sbjct: 123 EELERLGCSVVYGVNVHSMITKPSVGGSAIYDRVIFNFP----------------THELV 166
Query: 122 SGFLGSAKYML---NVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGY 178
GF+ SA+ ++ + GGEIH+ HKT +PFS W +K L E E L I EV F YPGY
Sbjct: 167 RGFMKSARVLVKDEDKGGEIHVIHKTEYPFSEWKLKTLGEKEGLDLIREVEFCLSHYPGY 226
Query: 179 GNKKGAGFKCDKSFPIGKCSTFKFGSLYY 207
NK+G+G D SFP+GK STF F ++
Sbjct: 227 FNKRGSGGYSDSSFPVGKSSTFMFTKQFF 255
>gb|AAF79500.1| F20N2.18 [Arabidopsis thaliana]
Length = 512
Score = 171 bits (433), Expect = 1e-41
Identities = 98/232 (42%), Positives = 130/232 (55%), Gaps = 32/232 (13%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLD-------DRG------ 48
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD ++G
Sbjct: 135 EEVWVKHYSSKHQILLVGEGDFSFSCSLATCFGSASNIYASSLDSYDYKPVNKGCSFKLD 194
Query: 49 --------------SLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDR 94
+ KY+ A NL L+ LG ++H VD ++ H L++ F DR
Sbjct: 195 FLSCCMSFMVIKPDDVVRKYKNARSNLETLKRLGAFLLHGVDATTLHFHPDLRYRRF-DR 253
Query: 95 IIFNFPHSGFFQNESDAWVI----GEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSN 150
+IFNFPH+GF + ESD I + L FL A +ML GE+H++HK PF
Sbjct: 254 VIFNFPHTGFHRKESDPCQIQPAAATLRNLFKDFLHGASHMLRADGEVHVSHKNKAPFCY 313
Query: 151 WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
WN++ LA L+ I+ F + YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 314 WNLEELASRCFLVLIQLEAFEKRNYPGYENKRGDGSRCDQPFLLGECSTFKF 365
>ref|NP_197885.1| hypothetical protein [Arabidopsis thaliana]
Length = 193
Score = 155 bits (391), Expect = 9e-37
Identities = 86/202 (42%), Positives = 118/202 (57%), Gaps = 16/202 (7%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNL 61
E K + YS+ IL+VGEG+FSF+L LAKA GSA N+ A SLD R L Y N+
Sbjct: 5 ESKRLSRYSNEQKILVVGEGEFSFSLSLAKALGSATNITAISLDIREDLGRNYNNGKGNV 64
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
ELE LGCT++ V+VH+M +L H +D IIFNFPH+ G+ K+
Sbjct: 65 EELERLGCTVVRGVNVHSMKSDDRLAH---YDIIIFNFPHA------------GKRNKVF 109
Query: 122 SGFLGSAKYML-NVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGN 180
GF+ SA+ M+ + GEIHIT T +PF+ W++K LAE L I+ + F + +P N
Sbjct: 110 GGFMESAREMMKDEDGEIHITLNTLNPFNKWDLKALAEESGLRLIQRMQFIKWAFPSSSN 169
Query: 181 KKGAGFKCDKSFPIGKCSTFKF 202
K+ +G CD +PIG T+ F
Sbjct: 170 KRESGSNCDFIYPIGSAITYMF 191
>emb|CAB79503.1| putative protein [Arabidopsis thaliana] gi|2982458|emb|CAA18222.1|
putative protein [Arabidopsis thaliana]
gi|15236738|ref|NP_194378.1| KH domain-containing
protein [Arabidopsis thaliana] gi|7486790|pir||T05056
hypothetical protein M3E9.90 - Arabidopsis thaliana
Length = 555
Score = 142 bits (359), Expect = 5e-33
Identities = 76/161 (47%), Positives = 101/161 (62%), Gaps = 4/161 (2%)
Query: 37 VNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRII 96
+N+ ATSLD L++KY A+ N+ L+ GC I HEVDVH M+ + L + DRI+
Sbjct: 1 MNITATSLDSEDELSIKYMDAVDNINILKRYGCDIQHEVDVHTMSFDNSLSLQRY-DRIV 59
Query: 97 FNFPHSG--FFQNESDAWVIGEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIK 154
FNFPH+G FF E + I HK+LV GFL +AK ML GEIHITHKT +PFS+W IK
Sbjct: 60 FNFPHAGSRFFGRELSSRAIESHKELVRGFLENAKEMLEEDGEIHITHKTTYPFSDWGIK 119
Query: 155 NLAENEKLLFIEEVTFYQHFYPGYGNKKGA-GFKCDKSFPI 194
L + E L +++ F YPGY K+G+ G + D FP+
Sbjct: 120 KLGKGEGLKLLKKSKFELSHYPGYITKRGSGGRRSDDYFPV 160
>ref|NP_564701.1| hypothetical protein [Arabidopsis thaliana]
Length = 314
Score = 132 bits (333), Expect = 5e-30
Identities = 87/232 (37%), Positives = 113/232 (48%), Gaps = 61/232 (26%)
Query: 2 EEKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLD-------DRG------ 48
EE VKHYSS H ILLVGEGDFSF+ LA FGSA N+ A+SLD ++G
Sbjct: 15 EEVWVKHYSSKHQILLVGEGDFSFSCSLATCFGSASNIYASSLDSYDYKPVNKGCSFKLD 74
Query: 49 --------------SLAMKYRGAIRNLIELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDR 94
+ KY+ A NL L+ LG ++H VD ++ H L++ F DR
Sbjct: 75 FLSCCMSFMVIKPDDVVRKYKNARSNLETLKRLGAFLLHGVDATTLHFHPDLRYRRF-DR 133
Query: 95 IIFNFPHSGFFQNESDAWVI----GEHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSN 150
+IFNFPH+GF + ESD I + L FL A +ML GE+
Sbjct: 134 VIFNFPHTGFHRKESDPCQIQPAAATLRNLFKDFLHGASHMLRADGELE----------- 182
Query: 151 WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFKCDKSFPIGKCSTFKF 202
F + YPGY NK+G G +CD+ F +G+CSTFKF
Sbjct: 183 ------------------AFEKRNYPGYENKRGDGSRCDQPFLLGECSTFKF 216
>emb|CAA22644.1| SPCC1919.13c [Schizosaccharomyces pombe]
gi|19075995|ref|NP_588495.1| hypothetical protein
SPCC1919.13c [Schizosaccharomyces pombe 972h-]
gi|7491878|pir||T41238 hypothetical protein SPCC1919.13c
- fission yeast (Schizosaccharomyces pombe)
Length = 282
Score = 101 bits (251), Expect = 2e-20
Identities = 61/185 (32%), Positives = 100/185 (53%), Gaps = 3/185 (1%)
Query: 3 EKIVKHYSSFHNILLVGEGDFSFALCLAKAFGSAVNMV-ATSLDDRGSLAMKYRGAIRNL 61
E+ V + + LL+GEG+FSFA L S+ V ATS D + L KY A +
Sbjct: 48 ERYVLPFEKNNRFLLLGEGNFSFAFSLLLHHVSSEGFVLATSYDSKEDLKQKYPDAAEYI 107
Query: 62 IELEGLGCTIMHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLV 121
++E G +MHE+D ++ H +LK F D I +NFPHSG + D ++ +++K++
Sbjct: 108 SKIEINGGKVMHEIDATKLHLHKKLKTQKF-DTIFWNFPHSGKGIKDQDRNIL-DNQKML 165
Query: 122 SGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNK 181
F ++K++L+ G I IT P++ WN+K LA++ + F FYP Y ++
Sbjct: 166 LAFFKASKFLLSEKGVIVITLAETKPYTLWNLKGLAKDAGYTSLMTEKFDSSFYPEYSHR 225
Query: 182 KGAGF 186
+ G+
Sbjct: 226 RTIGW 230
>emb|CAG84731.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50408283|ref|XP_456768.1| unnamed protein product
[Debaryomyces hansenii]
Length = 322
Score = 76.6 bits (187), Expect = 4e-13
Identities = 57/218 (26%), Positives = 99/218 (45%), Gaps = 37/218 (16%)
Query: 1 MEEKIVKHYSSFHNILLVGEGDFSFALCLAKA-FGSAVNMVATSLDDRGSLAMKYRGAIR 59
+++K ++ ++ +LLVGEGDFSFA+ + K F N++ATS D + + KY +
Sbjct: 65 IQQKGLQPFNIDDKLLLVGEGDFSFAVSIIKENFIKPENLIATSFDSKDEIIQKYPTVEQ 124
Query: 60 NLIELEGLGCTIMHEVDVHNMNQHHQL---------------KHHNFFDRIIFNFPHSGF 104
NL L G ++H +D ++ +L + D I+FNFPH+G
Sbjct: 125 NLNFLIDEGVQLLHSIDATDLVSSLKLNTTAKNKKKAKARLFSDNRNLDYIMFNFPHTGK 184
Query: 105 FQNESDAWVIGEHKKLVSGFLGSAKYMLNV--------------------GGEIHITHKT 144
+ D +I +H+KLV + S + + N+ G+I +T
Sbjct: 185 GIKDVDRNII-DHQKLVLNYFKSCRAVFNLVNNDMENDFAGYASSAVNENKGKILLTLFE 243
Query: 145 AHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKK 182
P+++WNIK L +E F +P Y +++
Sbjct: 244 GEPYTSWNIKILGRSEDYKVERSGKFSWQMFPDYHHRR 281
>ref|XP_331400.1| hypothetical protein [Neurospora crassa] gi|28919310|gb|EAA28770.1|
hypothetical protein [Neurospora crassa]
Length = 388
Score = 75.5 bits (184), Expect = 9e-13
Identities = 56/190 (29%), Positives = 84/190 (43%), Gaps = 14/190 (7%)
Query: 9 YSSFHNILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELEGL- 67
+S NILLVG+GD SFA L + + D L+ KY N+ ++ +
Sbjct: 105 FSPDENILLVGDGDLSFAASLVEHHHCTHLTASVYEKDLDELSAKYPHVRANVDKILAVP 164
Query: 68 GCTIMHEVDVHNM------------NQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIG 115
GC +++ VD M Q +++ DRIIFNFPH G + + V
Sbjct: 165 GCKVLYNVDARRMAPFAHKAKTKDNKQAGRVEQVGTMDRIIFNFPHVGGKSTDVNRQVRY 224
Query: 116 EHKKLVSGFLGSAKYMLNVGGEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFY 175
+ LV F S L GG + +T P++ WNI++LA + L + F Y
Sbjct: 225 NQELLVDFFKRSL-LSLAPGGSVIVTLFEGEPYTLWNIRDLARHSDLAVEKSFKFQARAY 283
Query: 176 PGYGNKKGAG 185
PGY + + G
Sbjct: 284 PGYHHARTLG 293
>emb|CAG59404.1| unnamed protein product [Candida glabrata CBS138]
gi|50288097|ref|XP_446477.1| unnamed protein product
[Candida glabrata]
Length = 323
Score = 64.3 bits (155), Expect = 2e-09
Identities = 60/228 (26%), Positives = 96/228 (41%), Gaps = 49/228 (21%)
Query: 3 EKIVKHYSSFHNILLVGEGDFSFALCLAK-AFGSAVNMVATSLDDR-GSLAMKYRGAIR- 59
EK + ++LVGEGDFSFA + + ++ N++ TS D+ L +KY +
Sbjct: 58 EKSFIPFEKNETLMLVGEGDFSFARSIIEESYILPENLIVTSYDNSYNELKLKYPHSFEE 117
Query: 60 NLIELEGLGCTIMHEVDVHNMNQHHQLKHHN-------------FFDRIIFNFPHSGFFQ 106
N L I+++VD + + +L H + I+FNFPH+G
Sbjct: 118 NFKFLVDNNVKILYQVDATKLIKSLKLSKHTPWSKLMGPSWKFKYLQNIMFNFPHTGKGV 177
Query: 107 NESDAWVIGEHKKLVSGFLGSAKYM----------LNVG--------------------- 135
+ D I +H++L+ GF SAK + L VG
Sbjct: 178 KDQDR-NIRDHQELLFGFFDSAKQLYSLVNSNKKNLEVGQTMGYNLSNNHDAKSNITEEG 236
Query: 136 -GEIHITHKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKK 182
G I ++ T P+ +W+IK LA+N L F +P Y +K+
Sbjct: 237 YGRIILSLFTGEPYDSWSIKILAKNNGLQLERSNKFQWENFPQYSHKR 284
>ref|YP_246612.1| hypothetical protein RF_0596 [Rickettsia felis URRWXCal2]
gi|67004521|gb|AAY61447.1| unknown [Rickettsia felis
URRWXCal2]
Length = 247
Score = 64.3 bits (155), Expect = 2e-09
Identities = 49/166 (29%), Positives = 80/166 (47%), Gaps = 10/166 (6%)
Query: 14 NILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMH 73
N LLVGEG+ SF++ L K + ++ +D L+ A N +L G ++H
Sbjct: 66 NSLLVGEGNLSFSVSLMKKLQQLPRCITSTYEDYDDLS---ETAQLNTYKLRKFGINVLH 122
Query: 74 EVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSGFLGSAKYMLN 133
+D + H+ +HN FD IIF FPHSG + + + + LV F+ SA Y+L
Sbjct: 123 NIDATKL---HKNFNHNSFDTIIFQFPHSG---SREEINGLNPNYILVRDFIVSASYVLK 176
Query: 134 VGGEIHITHKTAHPFSN-WNIKNLAENEKLLFIEEVTFYQHFYPGY 178
G I IT + +++ + + L++ K+ + F YP Y
Sbjct: 177 KHGLILITIVDSDFYNSIFQFEKLSQELKISTPIKYKFDPKDYPEY 222
>ref|XP_546526.1| PREDICTED: similar to disrupted in bipolar disorder 1 [Canis
familiaris]
Length = 1165
Score = 63.5 bits (153), Expect = 4e-09
Identities = 58/192 (30%), Positives = 86/192 (44%), Gaps = 23/192 (11%)
Query: 15 ILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMHE 74
+LLVGEG+FSFA L++ + + AT L +A + A NL L G I+
Sbjct: 46 LLLVGEGNFSFAAALSETLDGSTRVTATCLQRAADVA-RDPVARENLRRLRERGTEILFC 104
Query: 75 VDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSGFLGSAKYMLNV 134
VD + +L H FDRI FNFPH G + ++++L++ F S K +L
Sbjct: 105 VDCTRLADALEL-HPREFDRIYFNFPHCGRKAG------VAKNRELLAKFFQSCKDVLAE 157
Query: 135 GGEIHI----------THKTAHPFSN-WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
GE+H+ K + N W + +A + E F PGY K
Sbjct: 158 EGEVHVALCRGQGGTSADKPRREWHNSWQVVAMAALGGFILSEVHPFSCESVPGY---KC 214
Query: 184 AGFKC-DKSFPI 194
G++ DKSF +
Sbjct: 215 TGYRSQDKSFHV 226
>gb|EAK97612.1| hypothetical protein CaO19.7366 [Candida albicans SC5314]
Length = 353
Score = 62.0 bits (149), Expect = 1e-08
Identities = 56/224 (25%), Positives = 92/224 (41%), Gaps = 52/224 (23%)
Query: 15 ILLVGEGDFSFALCLA-KAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMH 73
+LL+GEGDFSFA L + F N++ATS D L KY + EL+ +G IMH
Sbjct: 85 VLLIGEGDFSFAKSLILQNFIQPENLIATSFDSFEQLINKYENVNEIIEELKNMGVIIMH 144
Query: 74 EVDVHNM--------------NQHHQL---------KHHNFFDRIIFNFPHSGFFQNESD 110
E+D N+ NQ+ + K + + I+FNFP +G + D
Sbjct: 145 EIDGTNLLKSLKLNPNKLKRNNQNSDVGKVKKLKLFKDYGNVNYIMFNFPRNGKGIKDVD 204
Query: 111 AWVIGEHKKLVSGFLGSAKYMLNV---------------------------GGEIHITHK 143
I +H++L+ F + + + +V G+I I+
Sbjct: 205 R-NIRDHQRLMLSFFENCQQLFDVINTDTISGYNTFNSNNNNNNNASGSSSTGKIIISMF 263
Query: 144 TAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKGAGFK 187
P+ +W IK L +++ F +P Y +++ K
Sbjct: 264 EGEPYHSWGIKILGKSQGWKVERSGKFDWSMFPEYHHRRTTSMK 307
>emb|CAG00498.1| unnamed protein product [Tetraodon nigroviridis]
Length = 612
Score = 62.0 bits (149), Expect = 1e-08
Identities = 53/208 (25%), Positives = 91/208 (43%), Gaps = 35/208 (16%)
Query: 14 NILLVGEGDFSFALCLAKAFGSAVN--MVATSLDDRGSLAMKYRGAIRNLIELEGLGCTI 71
+ILLVGEG+FSF+ L++ A + ++AT L A++ GA N+ + G +
Sbjct: 6 SILLVGEGNFSFSASLSQQHNEAASGSVIATCLQSEEE-ALRQEGAAENIQIITDSGGAV 64
Query: 72 MHEVDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSGFLGSAKYM 131
+ VD + + L+ FDR++FNFPH G + +++ L+ F S +
Sbjct: 65 LFGVDCTRLGECASLQGC-LFDRVVFNFPHCGRKSG------VKKNRDLLKTFFLSCVQV 117
Query: 132 LNVGGEIHIT-----------HKTAHPFSNWNIKNLAENEKLLFIEEVTFYQHFYPGYGN 180
L GGE+H++ ++W + +A +L+ E F +
Sbjct: 118 LAEGGEVHVSLCNGQGGTPADQPKREWHNSWQVTAMAAEAQLILTEVRPF--------ES 169
Query: 181 KKGAGFKC------DKSFPIGKCSTFKF 202
+K +KC DK F + K F
Sbjct: 170 EKNQSYKCTGYRSQDKGFHVEKALVHVF 197
>ref|XP_508751.1| PREDICTED: similar to disrupted in bipolar disorder 1; disrupted in
bipolar affective disorder 1 [Pan troglodytes]
Length = 1517
Score = 60.1 bits (144), Expect = 4e-08
Identities = 57/197 (28%), Positives = 87/197 (43%), Gaps = 33/197 (16%)
Query: 15 ILLVGEGDFSFALCLAKAFGSAVNMVATSLDDRGSLAMKYRGAIRNLIELEGLGCTIMHE 74
+LLVGEG+FSFA L++ + + AT L LA + A NL L G +
Sbjct: 191 LLLVGEGNFSFAAALSETLDQSTQLTATCLQRPAELA-RDPLAWENLQRLRERGIDVRFG 249
Query: 75 VDVHNMNQHHQLKHHNFFDRIIFNFPHSGFFQNESDAWVIGEHKKLVSGFLGSAKYMLNV 134
VD ++ +L H FD+I FNFPH G + ++++L++ F S +L
Sbjct: 250 VDCTHLADVFEL-HEREFDQIYFNFPHCGRKAG------VAKNRELLAKFFQSCADVLAE 302
Query: 135 GGEIHI----------THKTAHPFSN-WNIKNLAENEKLLFIEEVTFYQHFYPGYGNKKG 183
GE+H+ K + N W + +A L+ + YP + K
Sbjct: 303 EGEVHVALCRGQGGTPADKPQREWHNSWQVVAMAALGGLILSD-------VYP-FSCKAV 354
Query: 184 AGFKC------DKSFPI 194
AG+KC DKSF +
Sbjct: 355 AGYKCTGYRSQDKSFHV 371
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.141 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 389,155,169
Number of Sequences: 2540612
Number of extensions: 16323671
Number of successful extensions: 41692
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 41588
Number of HSP's gapped (non-prelim): 74
length of query: 211
length of database: 863,360,394
effective HSP length: 122
effective length of query: 89
effective length of database: 553,405,730
effective search space: 49253109970
effective search space used: 49253109970
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146569.5


