

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146568.7 - phase: 0
(703 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
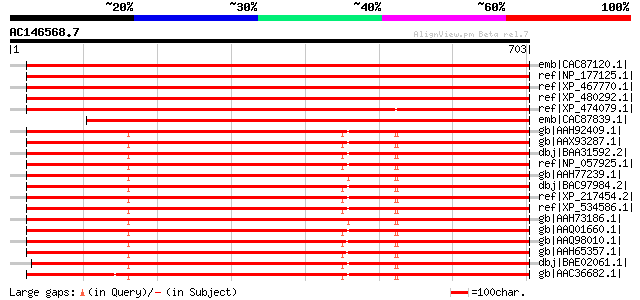
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC87120.1| cullin 3a [Arabidopsis thaliana] gi|15223361|ref... 1141 0.0
ref|NP_177125.1| cullin, putative [Arabidopsis thaliana] gi|1232... 1132 0.0
ref|XP_467770.1| putative cullin 3 [Oryza sativa (japonica culti... 1091 0.0
ref|XP_480292.1| putative cullin 3B [Oryza sativa (japonica cult... 1007 0.0
ref|XP_474079.1| OSJNBa0063C18.16 [Oryza sativa (japonica cultiv... 1006 0.0
emb|CAC87839.1| cullin 3B [Arabidopsis thaliana] 999 0.0
gb|AAH92409.1| Cullin 3 [Homo sapiens] gi|24660078|gb|AAH39598.1... 694 0.0
gb|AAX93287.1| unknown [Homo sapiens] 694 0.0
dbj|BAA31592.2| KIAA0617 protein [Homo sapiens] 694 0.0
ref|NP_057925.1| cullin 3 [Mus musculus] gi|20071136|gb|AAH27304... 693 0.0
gb|AAH77239.1| Cul3-prov protein [Xenopus laevis] 693 0.0
dbj|BAC97984.2| mKIAA0617 protein [Mus musculus] 693 0.0
ref|XP_217454.2| PREDICTED: similar to mKIAA0617 protein [Rattus... 693 0.0
ref|XP_534586.1| PREDICTED: similar to Cullin homolog 3 (CUL-3) ... 693 0.0
gb|AAH73186.1| MGC80402 protein [Xenopus laevis] 692 0.0
gb|AAQ01660.1| cullin 3 isoform [Homo sapiens] 691 0.0
gb|AAQ98010.1| cullin 3 [Danio rerio] gi|41055488|ref|NP_955985.... 687 0.0
gb|AAH65357.1| Cullin 3 [Danio rerio] 687 0.0
dbj|BAE02061.1| unnamed protein product [Macaca fascicularis] 684 0.0
gb|AAC36682.1| cullin 3 [Homo sapiens] 666 0.0
>emb|CAC87120.1| cullin 3a [Arabidopsis thaliana] gi|15223361|ref|NP_174005.1|
cullin, putative [Arabidopsis thaliana]
gi|4262186|gb|AAD14503.1| Highly similar to cullin 3
[Arabidopsis thaliana] gi|25354602|pir||A86395
hypothetical protein T2P11.2 [imported] - Arabidopsis
thaliana gi|9295728|gb|AAF87034.1| T24P13.25
[Arabidopsis thaliana]
Length = 732
Score = 1141 bits (2952), Expect = 0.0
Identities = 553/680 (81%), Positives = 621/680 (91%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+RNAYNMVLHKFG++LY+G +ATMT+HLKE +K IEAAQGGSFLEELN+KWN+HNKAL+M
Sbjct: 53 YRNAYNMVLHKFGEKLYTGFIATMTSHLKEKSKLIEAAQGGSFLEELNKKWNEHNKALEM 112
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRT+I S KKT VH +GLNLWR++V++ +I TRLLNTLL+LVQ ER GEVI
Sbjct: 113 IRDILMYMDRTYIESTKKTHVHPMGLNLWRDNVVHFTKIHTRLLNTLLDLVQKERIGEVI 172
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
DRG+MRN+ KM MDLG +VY +DFE FL S+EFY+VESQ FIE CDCGDYLKK+E+RL
Sbjct: 173 DRGLMRNVIKMFMDLGESVYQEDFEKPFLDASSEFYKVESQEFIESCDCGDYLKKSEKRL 232
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
EE++RV HY+D ++E+KI VVE +MI NHM RL+HMENSGLVNML +DKYEDLGRMYN
Sbjct: 233 TEEIERVAHYLDAKSEEKITSVVEKEMIANHMQRLVHMENSGLVNMLLNDKYEDLGRMYN 292
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LFRRV +GL+ +R+VMT H+RE GKQLVTDPE+ KDPVEFVQRLLDE+DKYDKIIN AF
Sbjct: 293 LFRRVTNGLVTVRDVMTSHLREMGKQLVTDPEKSKDPVEFVQRLLDERDKYDKIINTAFG 352
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFEYFINLN RSPEFISLFVDDKLRKGLKG+ + DVEV LDKVMMLFRY
Sbjct: 353 NDKTFQNALNSSFEYFINLNARSPEFISLFVDDKLRKGLKGITDVDVEVILDKVMMLFRY 412
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTS+
Sbjct: 413 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSE 472
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DTM+GFY SHP+L +GPTL VQVLTTGSWPTQ ++ CNLP E+S LCEKFRSYYLGTHTG
Sbjct: 473 DTMRGFYGSHPELSEGPTLIVQVLTTGSWPTQPAVPCNLPAEVSVLCEKFRSYYLGTHTG 532
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRLSWQTNMG AD+KA FGKGQKHELNVST+QMCVLMLFNN+D+LSYKEIEQATEIPA D
Sbjct: 533 RRLSWQTNMGTADIKAIFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIEQATEIPAAD 592
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQSLA VKG+NV++KEPMSKD+GE+D F VNDKF+SK YKVKIGTVVAQKE+EPEK
Sbjct: 593 LKRCLQSLACVKGKNVIKKEPMSKDIGEEDLFVVNDKFTSKFYKVKIGTVVAQKETEPEK 652
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
QETRQRVEEDRKPQIEAAIVRIMKSR++LDHNN+IAEVTKQLQ RFLANPTE+KKRIESL
Sbjct: 653 QETRQRVEEDRKPQIEAAIVRIMKSRKILDHNNIIAEVTKQLQPRFLANPTEIKKRIESL 712
Query: 684 IERDFLERDDNDRKMYRYLA 703
IERDFLERD DRK+YRYLA
Sbjct: 713 IERDFLERDSTDRKLYRYLA 732
>ref|NP_177125.1| cullin, putative [Arabidopsis thaliana] gi|12325193|gb|AAG52544.1|
putative cullin; 66460-68733 [Arabidopsis thaliana]
gi|25354600|pir||E96718 probable cullin T6C23.13
[imported] - Arabidopsis thaliana
Length = 732
Score = 1132 bits (2928), Expect = 0.0
Identities = 550/680 (80%), Positives = 621/680 (90%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+RNAYNMVLHK+GD+LY+GLV TMT HLKEI KSIE AQGG+FLE LNRKWNDHNKALQM
Sbjct: 53 YRNAYNMVLHKYGDKLYTGLVTTMTFHLKEICKSIEEAQGGAFLELLNRKWNDHNKALQM 112
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRT++ + KKT VHELGL+LWR++V+YS++I+TRLLNTLL+LV ERTGEVI
Sbjct: 113 IRDILMYMDRTYVSTTKKTHVHELGLHLWRDNVVYSSKIQTRLLNTLLDLVHKERTGEVI 172
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
DR +MRN+ KM MDLG +VY DFE FL+ SAEFY+VES FIE CDCG+YLKKAE+ L
Sbjct: 173 DRVLMRNVIKMFMDLGESVYQDDFEKPFLEASAEFYKVESMEFIESCDCGEYLKKAEKPL 232
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
EE++RV +Y+D ++E KI VVE +MI NH+ RL+HMENSGLVNML +DKYED+GRMY+
Sbjct: 233 VEEVERVVNYLDAKSEAKITSVVEREMIANHVQRLVHMENSGLVNMLLNDKYEDMGRMYS 292
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LFRRVA+GL+ +R+VMTLH+RE GKQLVTDPE+ KDPVEFVQRLLDE+DKYD+IIN AFN
Sbjct: 293 LFRRVANGLVTVRDVMTLHLREMGKQLVTDPEKSKDPVEFVQRLLDERDKYDRIINMAFN 352
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFEYF+NLN RSPEFISLFVDDKLRKGLKGV E+DV++ LDKVMMLFRY
Sbjct: 353 NDKTFQNALNSSFEYFVNLNTRSPEFISLFVDDKLRKGLKGVGEEDVDLILDKVMMLFRY 412
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKTVSDDAER+LIVKLKTECGYQFTSKLEGMFTDMKTS
Sbjct: 413 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERNLIVKLKTECGYQFTSKLEGMFTDMKTSH 472
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DT+ GFY SHP+L +GPTL VQVLTTGSWPTQ +I CNLP E+S LCEKFRSYYLGTHTG
Sbjct: 473 DTLLGFYNSHPELSEGPTLVVQVLTTGSWPTQPTIQCNLPAEVSVLCEKFRSYYLGTHTG 532
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRLSWQTNMG AD+KA FGKGQKHELNVST+QMCVLMLFNN+D+LSYKEIEQATEIP PD
Sbjct: 533 RRLSWQTNMGTADIKAVFGKGQKHELNVSTFQMCVLMLFNNSDRLSYKEIEQATEIPTPD 592
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQS+A VKG+NVLRKEPMSK++ E+D F VND+F+SK YKVKIGTVVAQKE+EPEK
Sbjct: 593 LKRCLQSMACVKGKNVLRKEPMSKEIAEEDWFVVNDRFASKFYKVKIGTVVAQKETEPEK 652
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
QETRQRVEEDRKPQIEAAIVRIMKSRR+LDHNN+IAEVTKQLQ RFLANPTE+KKRIESL
Sbjct: 653 QETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNNIIAEVTKQLQTRFLANPTEIKKRIESL 712
Query: 684 IERDFLERDDNDRKMYRYLA 703
IERDFLERD+ DRK+YRYLA
Sbjct: 713 IERDFLERDNTDRKLYRYLA 732
>ref|XP_467770.1| putative cullin 3 [Oryza sativa (japonica cultivar-group)]
gi|46390815|dbj|BAD16320.1| putative cullin 3 [Oryza
sativa (japonica cultivar-group)]
gi|46390116|dbj|BAD15552.1| putative cullin 3 [Oryza
sativa (japonica cultivar-group)]
Length = 736
Score = 1091 bits (2822), Expect = 0.0
Identities = 540/681 (79%), Positives = 602/681 (88%), Gaps = 1/681 (0%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+R+AYNMVLHK+G++LY GL TMT LKEI+KSIEAAQGG FLEELN KW DHNKALQM
Sbjct: 56 YRSAYNMVLHKYGEKLYDGLERTMTWRLKEISKSIEAAQGGLFLEELNAKWMDHNKALQM 115
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRT++P +++TPVHELGLNLWR+ +I+S I +RLL+TLL+L+ ER GE+I
Sbjct: 116 IRDILMYMDRTYVPQSRRTPVHELGLNLWRDHIIHSPMIHSRLLDTLLDLIHRERMGEMI 175
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
+RG+MR+ITKMLMDLG AVY DFE FL V+A FY ESQ FIECCDCG+YLKK+ERRL
Sbjct: 176 NRGLMRSITKMLMDLGAAVYQDDFEKPFLDVTASFYSGESQEFIECCDCGNYLKKSERRL 235
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
NEEM+RV HY+D TE KI VVE +MI NHM RL+HMENSGLVNML DDKY+DL RMYN
Sbjct: 236 NEEMERVSHYLDSGTEAKITSVVEKEMIANHMHRLVHMENSGLVNMLVDDKYDDLARMYN 295
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LFRRV DGL IR+VMT ++RE+GKQLVTDPERLKDPVEFVQRLL+EKDK+DKIIN AF
Sbjct: 296 LFRRVFDGLSTIRDVMTSYLRETGKQLVTDPERLKDPVEFVQRLLNEKDKHDKIINVAFG 355
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFEYFINLN RSPEFISL+VDDKLRKGLKG E+DVEV LDKVMMLFRY
Sbjct: 356 NDKTFQNALNSSFEYFINLNNRSPEFISLYVDDKLRKGLKGATEEDVEVILDKVMMLFRY 415
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERS+IVKLKTECGYQFTSKLEGMFTDMKTSQ
Sbjct: 416 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSMIVKLKTECGYQFTSKLEGMFTDMKTSQ 475
Query: 444 DTMQGFYA-SHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHT 502
DTM FYA +LGDGPTL V +LTTGSWPTQ CNLP EI A+C+KFR+YYLGTH+
Sbjct: 476 DTMIDFYAKKSEELGDGPTLDVHILTTGSWPTQPCPPCNLPTEILAICDKFRTYYLGTHS 535
Query: 503 GRRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAP 562
GRRL+WQTNMG AD+KATFGKGQKHELNVSTYQMCVLMLFN+ D L+YK+IEQ T IPA
Sbjct: 536 GRRLTWQTNMGTADIKATFGKGQKHELNVSTYQMCVLMLFNSTDGLTYKDIEQDTAIPAS 595
Query: 563 DLKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPE 622
DLKRCLQSLA VKG+NVLRKEPMSKD+ EDD F NDKF+SKL KVKIGTVVAQKESEPE
Sbjct: 596 DLKRCLQSLACVKGKNVLRKEPMSKDISEDDTFYFNDKFTSKLVKVKIGTVVAQKESEPE 655
Query: 623 KQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIES 682
KQETRQRVEEDRKPQIEAAIVRIMKSRR+LDHN+++AEVTKQLQ RF+ NP +KKRIES
Sbjct: 656 KQETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNSIVAEVTKQLQARFMPNPVVIKKRIES 715
Query: 683 LIERDFLERDDNDRKMYRYLA 703
LIER+FLERD DRK+YRYLA
Sbjct: 716 LIEREFLERDKADRKLYRYLA 736
>ref|XP_480292.1| putative cullin 3B [Oryza sativa (japonica cultivar-group)]
gi|40253773|dbj|BAD05712.1| putative cullin 3B [Oryza
sativa (japonica cultivar-group)]
gi|40253859|dbj|BAD05794.1| putative cullin 3B [Oryza
sativa (japonica cultivar-group)]
Length = 731
Score = 1007 bits (2604), Expect = 0.0
Identities = 492/680 (72%), Positives = 573/680 (83%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+R AYN+VLHK G +LY L M HL+E+ SIEAAQGG FL EL RKW+DHNKALQM
Sbjct: 52 YRTAYNLVLHKHGPKLYDKLTENMEDHLQEMRVSIEAAQGGLFLVELQRKWDDHNKALQM 111
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDR FIP+ KKTPV +LGL+LWR++++ S +I RLL+TLL+L+ ERTGEVI
Sbjct: 112 IRDILMYMDRVFIPTNKKTPVFDLGLDLWRDTIVRSPKIHGRLLDTLLDLIHRERTGEVI 171
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
+R +MR+ TKMLMDLG +VY DFE FL+VSA FY ESQ+FIECC CG+YLKKA++RL
Sbjct: 172 NRSLMRSTTKMLMDLGSSVYQDDFERPFLEVSASFYSGESQKFIECCSCGEYLKKAQQRL 231
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
+EE +RV YMD +T++KI VV +M+ NHM RLI MENSGLVNML +DKYEDL MY+
Sbjct: 232 DEEAERVSQYMDAKTDEKITAVVVKEMLANHMQRLILMENSGLVNMLVEDKYEDLTMMYS 291
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LF+RV DG I+ VM H++E+GK +V DPERLKDPV+FVQRLL+EKDKYD I+ +F+
Sbjct: 292 LFQRVPDGHSTIKSVMNSHVKETGKDMVMDPERLKDPVDFVQRLLNEKDKYDSIVTTSFS 351
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDKSFQNALNSSFE+FINLN R PEFISL+VDDKLRKG+K NE+DVE LDKVMMLFRY
Sbjct: 352 NDKSFQNALNSSFEHFINLNNRCPEFISLYVDDKLRKGMKEANEEDVETVLDKVMMLFRY 411
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKD+FEKYYKQHLAKRLLSGK SDD+ERS++VKLKTECGYQFTSKLEGMF D+KTS
Sbjct: 412 LQEKDLFEKYYKQHLAKRLLSGKAASDDSERSMLVKLKTECGYQFTSKLEGMFNDLKTSH 471
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DT Q FYA PDLGD PT++VQ+LTTGSWPTQ TCNLP EI + E FR +YLGTH G
Sbjct: 472 DTTQRFYAGTPDLGDAPTISVQILTTGSWPTQPCNTCNLPPEILGVSEMFRGFYLGTHNG 531
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRL+WQTNMG AD+KA FG G KHELNVSTYQMCVLMLFN+AD LSY++IEQ T IP+ D
Sbjct: 532 RRLTWQTNMGTADIKAVFGNGSKHELNVSTYQMCVLMLFNSADCLSYRDIEQTTAIPSAD 591
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQSLALVKG+NVLRKEPMS+D+ +DD F VNDKF+SKL+KVKIGTV QKESEPEK
Sbjct: 592 LKRCLQSLALVKGKNVLRKEPMSRDISDDDNFYVNDKFTSKLFKVKIGTVATQKESEPEK 651
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
ETRQRVEEDRKPQIEAAIVRIMKSRR+LDHN+++ EVTKQLQ RF+ NP +KKR+ESL
Sbjct: 652 METRQRVEEDRKPQIEAAIVRIMKSRRVLDHNSIVTEVTKQLQPRFMPNPVVIKKRVESL 711
Query: 684 IERDFLERDDNDRKMYRYLA 703
IER+FLERD DRK+YRYLA
Sbjct: 712 IEREFLERDKTDRKLYRYLA 731
>ref|XP_474079.1| OSJNBa0063C18.16 [Oryza sativa (japonica cultivar-group)]
gi|38347325|emb|CAE05975.2| OSJNBa0063C18.16 [Oryza
sativa (japonica cultivar-group)]
gi|38344878|emb|CAD41901.2| OSJNBa0033G05.2 [Oryza
sativa (japonica cultivar-group)]
Length = 731
Score = 1006 bits (2602), Expect = 0.0
Identities = 504/680 (74%), Positives = 575/680 (84%), Gaps = 1/680 (0%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHLKEIAKSIEAAQGGSFLEELNRKWNDHNKALQM 83
+R AYN+VLHK G +LY L + HLKE+ +SIE AQG FLEEL R+W DHNKALQM
Sbjct: 53 YRTAYNLVLHKHGLKLYDKLTENLKGHLKEMCRSIEDAQGSLFLEELQRRWADHNKALQM 112
Query: 84 IRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVI 143
IRDILMYMDRTFI + KKTPV +LGL LWR+ V+ + +I RLL+TLLEL+ ER GE+I
Sbjct: 113 IRDILMYMDRTFIATNKKTPVFDLGLELWRDIVVRTPKIHGRLLDTLLELIHRERMGEMI 172
Query: 144 DRGIMRNITKMLMDLGPAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRL 203
+RG+MR+ TKMLMDLG +VY DFE FL+VSA FY ESQ+FIECCDCG+YLKKAERRL
Sbjct: 173 NRGLMRSTTKMLMDLGSSVYHDDFEKPFLEVSASFYSGESQQFIECCDCGEYLKKAERRL 232
Query: 204 NEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYN 263
EE++RV YMD +T KI VV+T+M+ NHM RLI MENSGLVNML DDK+EDL RMYN
Sbjct: 233 AEELERVSQYMDAKTADKITSVVDTEMLANHMQRLILMENSGLVNMLVDDKHEDLSRMYN 292
Query: 264 LFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFN 323
LF+RV DG IR VM H++ESGK LV+DPE++KDPVEFVQRLL+EKDKYD+II+ +F+
Sbjct: 293 LFKRVPDGHSTIRSVMASHVKESGKALVSDPEKIKDPVEFVQRLLNEKDKYDEIISISFS 352
Query: 324 NDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRY 383
NDK+FQNALNSSFE FINLN RSPEFISLFVDDKLRKG+KG NE+DVE LDKVMMLFRY
Sbjct: 353 NDKAFQNALNSSFENFINLNNRSPEFISLFVDDKLRKGVKGANEEDVETVLDKVMMLFRY 412
Query: 384 LQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQ 443
LQEKDVFEKYYKQHLAKRLLSGKT SD+AERS++VKLKTECGYQFTSKLEGMF D+KTS
Sbjct: 413 LQEKDVFEKYYKQHLAKRLLSGKTTSDEAERSMLVKLKTECGYQFTSKLEGMFNDLKTSH 472
Query: 444 DTMQGFYASHPDLGDGPTLTVQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTG 503
DTMQ FYA+ D PT++VQ+LTTGSWPTQ C LP EI + EKFR++YLGTH G
Sbjct: 473 DTMQSFYANLSGDTDSPTISVQILTTGSWPTQPCTPCKLPPEIVDISEKFRAFYLGTHNG 532
Query: 504 RRLSWQTNMGFADLKATFGKGQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPD 563
RRL+WQTNMG AD+KATFG G++HELNVSTYQMCVLMLFN+AD L+Y +IEQAT IP D
Sbjct: 533 RRLTWQTNMGNADIKATFG-GRRHELNVSTYQMCVLMLFNSADGLTYGDIEQATGIPHAD 591
Query: 564 LKRCLQSLALVKGRNVLRKEPMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEK 623
LKRCLQSLA VKG+NVLRKEPMSKD+ EDD F NDKF+SKL KVKIGTVVAQKE+EPEK
Sbjct: 592 LKRCLQSLACVKGKNVLRKEPMSKDISEDDTFYYNDKFTSKLVKVKIGTVVAQKETEPEK 651
Query: 624 QETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESL 683
ETRQRVEEDRKPQIEAAIVRIMKSRR+LDHN++I EVTKQLQ RFL NP +KKRIESL
Sbjct: 652 LETRQRVEEDRKPQIEAAIVRIMKSRRVLDHNSIITEVTKQLQSRFLPNPVVIKKRIESL 711
Query: 684 IERDFLERDDNDRKMYRYLA 703
IER+FLERD DRKMYRYLA
Sbjct: 712 IEREFLERDKVDRKMYRYLA 731
>emb|CAC87839.1| cullin 3B [Arabidopsis thaliana]
Length = 601
Score = 999 bits (2583), Expect = 0.0
Identities = 486/600 (81%), Positives = 549/600 (91%)
Query: 104 VHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIMRNITKMLMDLGPAVY 163
VHELGL+LWR++V+YS++I+TRLLNTLL+LV ERTGEVIDR +MRN+ KM MDLG +VY
Sbjct: 2 VHELGLHLWRDNVVYSSKIQTRLLNTLLDLVHKERTGEVIDRVLMRNVIKMFMDLGESVY 61
Query: 164 GQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNEEMDRVGHYMDPETEKKIN 223
DFE FL+ SAEFY+VES FIE CDCG+YLKKAE+ L EE++RV +Y+D ++E KI
Sbjct: 62 QDDFEKPFLEASAEFYKVESMEFIESCDCGEYLKKAEKPLVEEVERVVNYLDAKSEAKIT 121
Query: 224 KVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLFRRVADGLLKIREVMTLHI 283
VVE +MI NH+ RL+HMENSGLVNML +DKYED+GRMY+LFRRVA+GL+ +R+VMTLH+
Sbjct: 122 SVVEREMIANHVQRLVHMENSGLVNMLLNDKYEDMGRMYSLFRRVANGLVTVRDVMTLHL 181
Query: 284 RESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNNDKSFQNALNSSFEYFINLN 343
RE GKQLVTDPE+ KDPVEFVQRLLDE+DKYD+IIN AFNNDK+FQNALNSSFEYF+NLN
Sbjct: 182 REMGKQLVTDPEKSKDPVEFVQRLLDERDKYDRIINMAFNNDKTFQNALNSSFEYFVNLN 241
Query: 344 PRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQEKDVFEKYYKQHLAKRLL 403
RSPEFISLFVDDKLRKGLKGV E+DV++ LDKVMMLFRYLQEKDVFEKYYKQHLAKRLL
Sbjct: 242 TRSPEFISLFVDDKLRKGLKGVGEEDVDLILDKVMMLFRYLQEKDVFEKYYKQHLAKRLL 301
Query: 404 SGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDTMQGFYASHPDLGDGPTLT 463
SGKTVSDDAER+LIVKLKTECGYQFTSKLEGMFTDMKTS DT+ GFY SHP+L +GPTL
Sbjct: 302 SGKTVSDDAERNLIVKLKTECGYQFTSKLEGMFTDMKTSHDTLLGFYNSHPELSEGPTLV 361
Query: 464 VQVLTTGSWPTQSSITCNLPVEISALCEKFRSYYLGTHTGRRLSWQTNMGFADLKATFGK 523
VQVLTTGSWPTQ +I CNLP E+S LCEKFRSYYLGTHTGRRLSWQTNMG AD+KA FGK
Sbjct: 362 VQVLTTGSWPTQPTIQCNLPAEVSVLCEKFRSYYLGTHTGRRLSWQTNMGTADIKAVFGK 421
Query: 524 GQKHELNVSTYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVKGRNVLRKE 583
GQKHELNVST+QMCVLMLFNN+D+LSYKEIEQATEIP PDLKRCLQS+A VKG+NVLRKE
Sbjct: 422 GQKHELNVSTFQMCVLMLFNNSDRLSYKEIEQATEIPTPDLKRCLQSMACVKGKNVLRKE 481
Query: 584 PMSKDVGEDDAFSVNDKFSSKLYKVKIGTVVAQKESEPEKQETRQRVEEDRKPQIEAAIV 643
PMSK++ E+D F VND+F+SK YKVKIGTVVAQKE+EPEKQETRQRVEEDRKPQIEAAIV
Sbjct: 482 PMSKEIAEEDWFVVNDRFASKFYKVKIGTVVAQKETEPEKQETRQRVEEDRKPQIEAAIV 541
Query: 644 RIMKSRRLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
RIMKSRR+LDHNN+IAEVTKQLQ RFLANPTE+KKRIESLIERDFLERD+ DRK+YRYLA
Sbjct: 542 RIMKSRRVLDHNNIIAEVTKQLQTRFLANPTEIKKRIESLIERDFLERDNTDRKLYRYLA 601
>gb|AAH92409.1| Cullin 3 [Homo sapiens] gi|24660078|gb|AAH39598.1| Cullin 3 [Homo
sapiens] gi|12643396|sp|Q13618|CUL3_HUMAN Cullin-3
(CUL-3) gi|3639052|gb|AAC36304.1| cullin 3 [Homo
sapiens] gi|4503165|ref|NP_003581.1| cullin 3 [Homo
sapiens]
Length = 768
Score = 694 bits (1791), Expect = 0.0
Identities = 371/712 (52%), Positives = 502/712 (70%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLG 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLL 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 536
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 537 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 596
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 597 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 656
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 657 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 716
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 717 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>gb|AAX93287.1| unknown [Homo sapiens]
Length = 746
Score = 694 bits (1791), Expect = 0.0
Identities = 371/712 (52%), Positives = 502/712 (70%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 36 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 95
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 96 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 155
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 156 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 215
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG
Sbjct: 216 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLG 275
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 276 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLL 335
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 336 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 395
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 396 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 455
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 456 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 514
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 515 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 574
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 575 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 634
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 635 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 694
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 695 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 746
>dbj|BAA31592.2| KIAA0617 protein [Homo sapiens]
Length = 786
Score = 694 bits (1791), Expect = 0.0
Identities = 371/712 (52%), Positives = 502/712 (70%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 76 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 135
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 136 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 195
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 196 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 255
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG
Sbjct: 256 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLG 315
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 316 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLL 375
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 376 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 435
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 436 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 495
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 496 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 554
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 555 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 614
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 615 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 674
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 675 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 734
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 735 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 786
>ref|NP_057925.1| cullin 3 [Mus musculus] gi|20071136|gb|AAH27304.1| Cullin 3 [Mus
musculus] gi|7108617|gb|AAF36500.1| cullin 3 [Mus
musculus] gi|13124074|sp|Q9JLV5|CUL3_MOUSE Cullin-3
(CUL-3)
Length = 768
Score = 693 bits (1789), Expect = 0.0
Identities = 370/712 (51%), Positives = 501/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSCYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 536
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 537 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 596
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 597 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 656
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 657 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 716
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 717 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>gb|AAH77239.1| Cul3-prov protein [Xenopus laevis]
Length = 768
Score = 693 bits (1789), Expect = 0.0
Identities = 369/711 (51%), Positives = 499/711 (69%), Gaps = 31/711 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+L++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSLYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++F+ND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFSNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVESILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGFYASHPDLG---DGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFRS 495
S TM F G G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQTTGVSLGGVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFRR 537
Query: 496 YYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVSTY 534
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST+
Sbjct: 538 FYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVSTF 597
Query: 535 QMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGEDD 593
QM +LMLFNN DK +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 598 QMTILMLFNNRDKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESGH 657
Query: 594 AFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLL 652
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+ +
Sbjct: 658 MFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKKM 717
Query: 653 DHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 718 QHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>dbj|BAC97984.2| mKIAA0617 protein [Mus musculus]
Length = 792
Score = 693 bits (1789), Expect = 0.0
Identities = 370/712 (51%), Positives = 501/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 82 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 141
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 142 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 201
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 202 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 261
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 262 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 321
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 322 CMYKLFSRVPNGLKTMCECMSCYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 381
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 382 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 441
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 442 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 501
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 502 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 560
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 561 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 620
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 621 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 680
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 681 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 740
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 741 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 792
>ref|XP_217454.2| PREDICTED: similar to mKIAA0617 protein [Rattus norvegicus]
Length = 768
Score = 693 bits (1788), Expect = 0.0
Identities = 370/712 (51%), Positives = 501/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 536
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 537 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 596
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 597 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 656
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 657 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 716
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 717 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>ref|XP_534586.1| PREDICTED: similar to Cullin homolog 3 (CUL-3) [Canis familiaris]
Length = 999
Score = 693 bits (1788), Expect = 0.0
Identities = 370/712 (51%), Positives = 501/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 289 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 348
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 349 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 408
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 409 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 468
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 469 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 528
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 529 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 588
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 589 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMV 648
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 649 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 708
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 709 SISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 767
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 768 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 827
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 828 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 887
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 888 HIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 947
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 948 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 999
>gb|AAH73186.1| MGC80402 protein [Xenopus laevis]
Length = 768
Score = 692 bits (1786), Expect = 0.0
Identities = 368/711 (51%), Positives = 499/711 (69%), Gaps = 31/711 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 177
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 178 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 237
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 238 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 297
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+L++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 298 CMYKLFSRVPNGLKTMCECMSLYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 357
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++F+ND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 358 ESFSNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVESILDKAMV 417
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 418 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 477
Query: 440 KTSQDTMQGFYASHPDLG---DGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFRS 495
S TM F G G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 478 SISNTTMDEFRQHLQTTGVSLGGVDLTVRVLTTGYWPTQSATPKCNIPPSPRHAFEIFRR 537
Query: 496 YYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVSTY 534
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST+
Sbjct: 538 FYLAKHSGRQLTLQHHMGSADLNATFYGAVKKEDGSEVGVGGAQVTGSNTRKHILQVSTF 597
Query: 535 QMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGEDD 593
QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 598 QMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESGH 657
Query: 594 AFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLL 652
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+ +
Sbjct: 658 MFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKKM 717
Query: 653 DHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 718 QHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
>gb|AAQ01660.1| cullin 3 isoform [Homo sapiens]
Length = 744
Score = 691 bits (1784), Expect = 0.0
Identities = 371/712 (52%), Positives = 500/712 (70%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 34 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 93
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 94 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 153
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK
Sbjct: 154 VDRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKV 213
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG
Sbjct: 214 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLG 273
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 274 CMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLL 333
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL KG+KG+ E +VE LDK M+
Sbjct: 334 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLTKGVKGLTEQEVETILDKAMV 393
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 394 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 453
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 454 SISTTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFR 512
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 513 RFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVST 572
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 573 FQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENG 632
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 633 HIFTVNDQFISKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 692
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 693 MQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 744
>gb|AAQ98010.1| cullin 3 [Danio rerio] gi|41055488|ref|NP_955985.1| cullin 3 [Danio
rerio]
Length = 766
Score = 687 bits (1774), Expect = 0.0
Identities = 368/712 (51%), Positives = 497/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 56 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLHSLNNNFLQTLNQAWNDHQTAMV 115
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 116 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEV 175
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFE FL +SAEF+Q+ESQ+F+ Y+KK
Sbjct: 176 VDRGAIRNACQMLMVLGLKGRSVYEEDFEIPFLDMSAEFFQMESQKFLAENSASVYIKKV 235
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 236 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 295
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 296 CMYKLFGRVPNGLKTMCECMSWYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 355
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 356 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVESILDKAMV 415
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 416 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 475
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F S L G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 476 SISNTTMDEFRHHLQTSQVSL-CGVDLTVRVLTTGYWPTQSATPKCNIPPSPRHAFEVFR 534
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 535 RFYLAKHSGRQLTLQHHMGSADLNATFYGPIKKEDGSDVGVGGALLTGSNTRKHILQVST 594
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 595 FQMTILMLFNNREKFTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 654
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 655 HVFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 714
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 715 MQHNVLVAEVTQQLRARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 766
>gb|AAH65357.1| Cullin 3 [Danio rerio]
Length = 766
Score = 687 bits (1774), Expect = 0.0
Identities = 368/712 (51%), Positives = 497/712 (69%), Gaps = 33/712 (4%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 56 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLHSLNNNFLQTLNQAWNDHQTAMV 115
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEV 142
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV
Sbjct: 116 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERRGEV 175
Query: 143 IDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKA 199
+DRG +RN +MLM LG +VY +DFE FL +SAEF+Q+ESQ+F+ Y+KK
Sbjct: 176 VDRGAIRNACQMLMVLGLEGRSVYEEDFEIPFLDMSAEFFQMESQKFLAENSASVYIKKV 235
Query: 200 ERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLG 259
E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDL
Sbjct: 236 EARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLA 295
Query: 260 RMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIIN 319
MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ +
Sbjct: 296 CMYKLFGRVPNGLKTMCECMSWYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLQ 355
Query: 320 QAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMM 379
++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+
Sbjct: 356 ESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVESILDKAMV 415
Query: 380 LFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDM 439
LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM
Sbjct: 416 LFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDM 475
Query: 440 KTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFR 494
S TM F S L G LTV+VLTTG WPTQS+ CN+P E FR
Sbjct: 476 SISNTTMDEFRHHLQTSQVSL-CGVDLTVRVLTTGYWPTQSATPKCNIPPSPRHAFEVFR 534
Query: 495 SYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVST 533
+YL H+GR+L+ Q +MG ADL ATF G G +KH L VST
Sbjct: 535 RFYLAKHSGRQLTLQHHMGSADLNATFYGPIKKEDGSDVGVGGALLTGSNTRKHILQVST 594
Query: 534 YQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGED 592
+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 595 FQMTILMLFNNREKFTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIESG 654
Query: 593 DAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRL 651
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+
Sbjct: 655 HVFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKK 714
Query: 652 LDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 715 MQHNVLVAEVTQQLRARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 766
>dbj|BAE02061.1| unnamed protein product [Macaca fascicularis]
Length = 705
Score = 684 bits (1766), Expect = 0.0
Identities = 367/706 (51%), Positives = 497/706 (69%), Gaps = 33/706 (4%)
Query: 30 MVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQMIRDIL 88
MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+ MIRDIL
Sbjct: 1 MVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMVMIRDIL 60
Query: 89 MYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIRTRLLNTLLELVQSERTGEVIDRGIM 148
MYMDR ++ V+ LGL ++R+ V+ IR L TLL+++ ER GEV+DRG +
Sbjct: 61 MYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRDHLRQTLLDMIARERKGEVVDRGAI 120
Query: 149 RNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLKKAERRLNE 205
RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+KK E R+NE
Sbjct: 121 RNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIKKVEARINE 180
Query: 206 EMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYEDLGRMYNLF 265
E++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K EDLG MY LF
Sbjct: 181 EIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTEDLGCMYKLF 240
Query: 266 RRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKIINQAFNND 325
RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+ + ++FNND
Sbjct: 241 SRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRFLLESFNND 300
Query: 326 KSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKVMMLFRYLQ 385
+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK M+LFR++Q
Sbjct: 301 RLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKAMVLFRFMQ 360
Query: 386 EKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFTDMKTSQDT 445
EKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF DM S T
Sbjct: 361 EKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFRDMSISNTT 420
Query: 446 MQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEKFRSYYLGT 500
M F A+ LG G LTV+VLTTG WPTQS+ CN+P E FR +YL
Sbjct: 421 MDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEIFRRFYLAK 479
Query: 501 HTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNVSTYQMCVL 539
H+GR+L+ Q +MG ADL ATF G G +KH L VST+QM +L
Sbjct: 480 HSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQVSTFQMTIL 539
Query: 540 MLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVGEDDAFSVN 598
MLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++ F+VN
Sbjct: 540 MLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIENGHIFTVN 599
Query: 599 DKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSRRLLDHNNL 657
D+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR+ + HN L
Sbjct: 600 DQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSRKKMQHNVL 659
Query: 658 IAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 660 VAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 705
>gb|AAC36682.1| cullin 3 [Homo sapiens]
Length = 768
Score = 666 bits (1718), Expect = 0.0
Identities = 364/714 (50%), Positives = 497/714 (68%), Gaps = 37/714 (5%)
Query: 24 FRNAYNMVLHKFGDRLYSGLVATMTAHL-KEIAKSIEAAQGGSFLEELNRKWNDHNKALQ 82
+RNAY MVLHK G++LY+GL +T HL ++ + + + +FL+ LN+ WNDH A+
Sbjct: 58 YRNAYTMVLHKHGEKLYTGLREVVTEHLINKVREDVLNSLNNNFLQTLNQAWNDHQTAMV 117
Query: 83 MIRDILMYMDRTFIPSAKKTPVHELGLNLWRESVIYSNQIR-TRLLNTL-LELVQSERTG 140
MIRDILMYMDR ++ V+ LGL ++R+ V+ IR + N++ + ++ER
Sbjct: 118 MIRDILMYMDRVYVQQNNVENVYNLGLIIFRDQVVRYGCIRGSSTANSIGYDCKRAERRS 177
Query: 141 EVIDRGIMRNITKMLMDLG---PAVYGQDFEAHFLQVSAEFYQVESQRFIECCDCGDYLK 197
RG +RN +MLM LG +VY +DFEA FL++SAEF+Q+ESQ+F+ Y+K
Sbjct: 178 R--SRGAIRNACQMLMILGLEGRSVYEEDFEAPFLEMSAEFFQMESQKFLAENSASVYIK 235
Query: 198 KAERRLNEEMDRVGHYMDPETEKKINKVVETQMIENHMLRLIHMENSGLVNMLCDDKYED 257
K E R+NEE++RV H +D TE+ I KVVE ++I HM ++ MENSGLV+ML + K ED
Sbjct: 236 KVEARINEEIERVMHCLDKSTEEPIVKVVERELISKHMKTIVEMENSGLVHMLKNGKTED 295
Query: 258 LGRMYNLFRRVADGLLKIREVMTLHIRESGKQLVTDPERLKDPVEFVQRLLDEKDKYDKI 317
LG MY LF RV +GL + E M+ ++RE GK LV++ K+PV+++Q LLD K ++D+
Sbjct: 296 LGCMYKLFSRVPNGLKTMCECMSSYLREQGKALVSEEGEGKNPVDYIQGLLDLKSRFDRF 355
Query: 318 INQAFNNDKSFQNALNSSFEYFINLNPRSPEFISLFVDDKLRKGLKGVNEDDVEVTLDKV 377
+ ++FNND+ F+ + FEYF+NLN RSPE++SLF+DDKL+KG+KG+ E +VE LDK
Sbjct: 356 LLESFNNDRLFKQTIAGDFEYFLNLNSRSPEYLSLFIDDKLKKGVKGLTEQEVETILDKA 415
Query: 378 MMLFRYLQEKDVFEKYYKQHLAKRLLSGKTVSDDAERSLIVKLKTECGYQFTSKLEGMFT 437
M+LFR++QEKDVFE+YYKQHLA+RLL+ K+VSDD+E+++I KLKTECG QFTSKLEGMF
Sbjct: 416 MVLFRFMQEKDVFERYYKQHLARRLLTNKSVSDDSEKNMISKLKTECGCQFTSKLEGMFR 475
Query: 438 DMKTSQDTMQGF----YASHPDLGDGPTLTVQVLTTGSWPTQSSI-TCNLPVEISALCEK 492
DM S TM F A+ LG G LTV+VLTTG WPTQS+ CN+P E
Sbjct: 476 DMSISNTTMDEFRQHLQATGVSLG-GVDLTVRVLTTGYWPTQSATPKCNIPPAPRHAFEI 534
Query: 493 FRSYYLGTHTGRRLSWQTNMGFADLKATF------------GKG---------QKHELNV 531
FR +YL H+GR+L+ Q +MG ADL ATF G G +KH L V
Sbjct: 535 FRRFYLAKHSGRQLTLQHHMGSADLNATFYGPVKKEDGSEVGVGGAQVTGSNTRKHILQV 594
Query: 532 STYQMCVLMLFNNADKLSYKEIEQATEIPAPDLKRCLQSLALVK-GRNVLRKEPMSKDVG 590
ST+QM +LMLFNN +K +++EI+Q T+IP +L R LQSLA K + VL KEP SK++
Sbjct: 595 STFQMTILMLFNNREKYTFEEIQQETDIPERELVRALQSLACGKPTQRVLTKEPKSKEIE 654
Query: 591 EDDAFSVNDKFSSKLYKVKIGTVVA-QKESEPEKQETRQRVEEDRKPQIEAAIVRIMKSR 649
F+VND+F+SKL++VKI TV A Q ES+PE++ETRQ+V++DRK +IEAAIVRIMKSR
Sbjct: 655 NGHIFTVNDQFTSKLHRVKIQTVAAKQGESDPERKETRQKVDDDRKHEIEAAIVRIMKSR 714
Query: 650 RLLDHNNLIAEVTKQLQLRFLANPTEVKKRIESLIERDFLERDDNDRKMYRYLA 703
+ + HN L+AEVT+QL+ RFL +P +KKRIE LIER++L R DRK+Y Y+A
Sbjct: 715 KKMQHNVLVAEVTQQLKARFLPSPVVIKKRIEGLIEREYLARTPEDRKVYTYVA 768
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,131,807,672
Number of Sequences: 2540612
Number of extensions: 46563140
Number of successful extensions: 149388
Number of sequences better than 10.0: 507
Number of HSP's better than 10.0 without gapping: 333
Number of HSP's successfully gapped in prelim test: 174
Number of HSP's that attempted gapping in prelim test: 147055
Number of HSP's gapped (non-prelim): 679
length of query: 703
length of database: 863,360,394
effective HSP length: 135
effective length of query: 568
effective length of database: 520,377,774
effective search space: 295574575632
effective search space used: 295574575632
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC146568.7


