

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.12 + phase: 0 /pseudo
(307 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
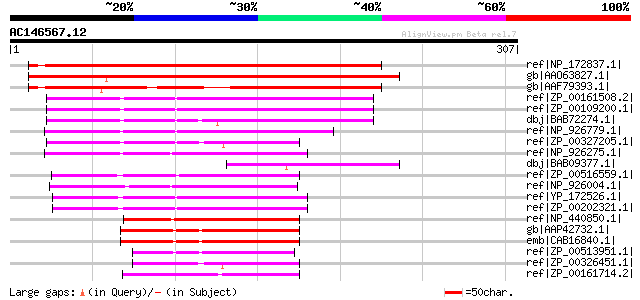
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_172837.1| hydrolase, alpha/beta fold family protein [Arab... 335 1e-90
gb|AAO63827.1| unknown protein [Arabidopsis thaliana] gi|2839357... 285 1e-75
gb|AAF79393.1| F16A14.4 [Arabidopsis thaliana] gi|25402766|pir||... 266 7e-70
ref|ZP_00161508.2| COG0596: Predicted hydrolases or acyltransfer... 131 3e-29
ref|ZP_00109200.1| COG0596: Predicted hydrolases or acyltransfer... 129 1e-28
dbj|BAB72274.1| 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Nos... 127 3e-28
ref|NP_926779.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydr... 118 2e-25
ref|ZP_00327205.1| COG0596: Predicted hydrolases or acyltransfer... 116 9e-25
ref|NP_926275.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydr... 114 4e-24
dbj|BAB09377.1| unnamed protein product [Arabidopsis thaliana] 113 8e-24
ref|ZP_00516559.1| Alpha/beta hydrolase fold [Crocosphaera watso... 112 2e-23
ref|NP_926004.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydr... 102 1e-20
ref|YP_172526.1| 2-hydroxy-6-oxohepta-24-dienoate hydrolase [Syn... 100 5e-20
ref|ZP_00202321.1| COG0596: Predicted hydrolases or acyltransfer... 100 7e-20
ref|NP_440850.1| 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Sy... 91 6e-17
gb|AAP42732.1| At4g36530 [Arabidopsis thaliana] gi|30690680|ref|... 75 3e-12
emb|CAB16840.1| putative protein [Arabidopsis thaliana] gi|72706... 75 3e-12
ref|ZP_00513951.1| Alpha/beta hydrolase fold [Crocosphaera watso... 73 9e-12
ref|ZP_00326451.1| COG0596: Predicted hydrolases or acyltransfer... 72 2e-11
ref|ZP_00161714.2| COG0596: Predicted hydrolases or acyltransfer... 69 2e-10
>ref|NP_172837.1| hydrolase, alpha/beta fold family protein [Arabidopsis thaliana]
gi|48310456|gb|AAT41824.1| At1g13820 [Arabidopsis
thaliana] gi|46518397|gb|AAS99680.1| At1g13820
[Arabidopsis thaliana]
Length = 339
Score = 335 bits (859), Expect = 1e-90
Identities = 159/214 (74%), Positives = 183/214 (85%), Gaps = 4/214 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQN 71
FKV AE FP+FLP DV +IKD FA LA RI+RLPVSV F E+ IMSSCV PL++N
Sbjct: 25 FKVTAE----KFPTFLPTDVEKIKDTFALKLAARIERLPVSVSFREDSIMSSCVTPLMRN 80
Query: 72 KENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHF 131
+ P+VLLHGFDSSCLEWRYTYPLLEEAG+ETWA DILGWGFSDL+KLP CDV SKREHF
Sbjct: 81 ETTPVVLLHGFDSSCLEWRYTYPLLEEAGLETWAFDILGWGFSDLDKLPPCDVASKREHF 140
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAA 191
Y+FWKS+IK+P++LVGPSLG+AVAID A+N+PEAVE LVL+DASVY EGTGNLATLP+AA
Sbjct: 141 YKFWKSHIKRPVVLVGPSLGAAVAIDIAVNHPEAVESLVLMDASVYAEGTGNLATLPKAA 200
Query: 192 AYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWT 225
AYAGVYLLKS+PLR+Y N++ F IS TS DWT
Sbjct: 201 AYAGVYLLKSIPLRLYVNFICFNGISLETSWDWT 234
>gb|AAO63827.1| unknown protein [Arabidopsis thaliana] gi|28393576|gb|AAO42208.1|
unknown protein [Arabidopsis thaliana]
gi|30693366|ref|NP_198738.2| hydrolase, alpha/beta fold
family protein [Arabidopsis thaliana]
Length = 330
Score = 285 bits (729), Expect = 1e-75
Identities = 137/228 (60%), Positives = 172/228 (75%), Gaps = 3/228 (1%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSE--NPIMSSCVKPLV 69
F+ + D FP+FLPK++ IKDPFAR LA RI R+PV ++ +MSSC+KPLV
Sbjct: 17 FRQRVTANGDGFPTFLPKEIQNIKDPFARALAQRIVRIPVPLQMGNFRGSVMSSCIKPLV 76
Query: 70 Q-NKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKR 128
Q + ++P+VLLH FDSSCLEWR TYPLLE+A +ETWAID+LGWGFSDLEKLP CD SKR
Sbjct: 77 QLHDKSPVVLLHCFDSSCLEWRRTYPLLEQACLETWAIDVLGWGFSDLEKLPPCDAASKR 136
Query: 129 EHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLP 188
H ++ WK+YIK+PMILVGPSLG+ VA+DF YPEAV+KLVLI+A+ Y+EGTG L LP
Sbjct: 137 HHLFELWKTYIKRPMILVGPSLGATVAVDFTATYPEAVDKLVLINANAYSEGTGRLKELP 196
Query: 189 RAAAYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWTNATMLLRR*KW 236
++ AYAGV LLKS PLR+ AN L+F + S ++DWTN L + W
Sbjct: 197 KSIAYAGVKLLKSFPLRLLANVLAFCSSPLSENIDWTNIGRLHCQMPW 244
>gb|AAF79393.1| F16A14.4 [Arabidopsis thaliana] gi|25402766|pir||E86271 protein
F16A14.4 [imported] - Arabidopsis thaliana
Length = 633
Score = 266 bits (679), Expect = 7e-70
Identities = 140/221 (63%), Positives = 162/221 (72%), Gaps = 32/221 (14%)
Query: 12 FKVVAETDADSFPSFLPKDVNRIKDPFARTLATRIQRLPVSVK-------FSENPIMSSC 64
FKV AE FP+FLP DV +IKD FA LA RI+RLPVSV F E+ IMSSC
Sbjct: 333 FKVTAE----KFPTFLPTDVEKIKDTFALKLAARIERLPVSVSGLRFHVSFREDSIMSSC 388
Query: 65 VKPLVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDV 124
V PL++N+ P+VLLHGFD RYTYPLLEEAG+ETWA DILGWGFSDL
Sbjct: 389 VTPLMRNETTPVVLLHGFD------RYTYPLLEEAGLETWAFDILGWGFSDLGF------ 436
Query: 125 VSKREHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNL 184
FWKS+IK+P++LVGPSLG+AVAID A+N+PEAVE LVL+DASVY EGTGNL
Sbjct: 437 ---------FWKSHIKRPVVLVGPSLGAAVAIDIAVNHPEAVESLVLMDASVYAEGTGNL 487
Query: 185 ATLPRAAAYAGVYLLKSVPLRVYANYLSFTNISFSTSLDWT 225
ATLP+AAAYAGVYLLKS+PLR+Y N++ F IS TS DWT
Sbjct: 488 ATLPKAAAYAGVYLLKSIPLRLYVNFICFNGISLETSWDWT 528
>ref|ZP_00161508.2| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Anabaena variabilis ATCC 29413]
Length = 295
Score = 131 bits (329), Expect = 3e-29
Identities = 77/198 (38%), Positives = 113/198 (56%), Gaps = 3/198 (1%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGF 82
FPSFLP V ++ + + LA IQ ++ S PI ++ V+ + PI+L+HGF
Sbjct: 2 FPSFLPAAVGQLTESESIALAKTIQTQAIATPLSNQPITTAYVRQ--GSGGTPILLIHGF 59
Query: 83 DSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKP 142
DSS LE+R PLL + ETWA+D+LG+GF+ V+ R H Y FWK+ I +P
Sbjct: 60 DSSVLEFRRLLPLLGKEN-ETWAVDLLGFGFTQRLAGIKFSPVAIRTHLYSFWKTLINQP 118
Query: 143 MILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLKSV 202
+ILVG S+G A AIDF + YPEAV+KLVLID++ G+ + Y L+S
Sbjct: 119 VILVGASMGGAAAIDFTLTYPEAVQKLVLIDSAGLRGGSPLSKFMFPPLDYLAAQFLRSP 178
Query: 203 PLRVYANYLSFTNISFST 220
+R + ++ N + +T
Sbjct: 179 KVRDRVSRAAYKNPNLAT 196
>ref|ZP_00109200.1| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Nostoc punctiforme PCC 73102]
Length = 295
Score = 129 bits (324), Expect = 1e-28
Identities = 73/198 (36%), Positives = 112/198 (55%), Gaps = 3/198 (1%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGF 82
FP+FLP+ V ++ + + LA IQ ++ P+ ++ VK + PI+L+HGF
Sbjct: 2 FPNFLPQAVGQLTESASIALAQNIQTAAIATPLINQPVTTTYVKQ--GSGGTPILLIHGF 59
Query: 83 DSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKP 142
DSS LE+R PLL ETWA+D+LG+GF+D + + + H Y FWKS I +P
Sbjct: 60 DSSVLEFRRLLPLLSRDN-ETWAVDLLGFGFTDRLSGIAYSPTAIKTHLYYFWKSLINQP 118
Query: 143 MILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLKSV 202
+ILVG S+G A AIDF + YPE V+KLVLID++ G+ + Y L ++
Sbjct: 119 VILVGASMGGATAIDFTLTYPEVVKKLVLIDSAGLAGGSPLSKFMFPPLDYFATQFLSNL 178
Query: 203 PLRVYANYLSFTNISFST 220
+R + + + N S ++
Sbjct: 179 KVRDRVSRIGYKNQSLAS 196
>dbj|BAB72274.1| 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Nostoc sp. PCC 7120]
gi|17227812|ref|NP_484360.1|
2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Nostoc sp.
PCC 7120] gi|25288883|pir||AD1846
2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [imported] -
Nostoc sp. (strain PCC 7120)
Length = 295
Score = 127 bits (320), Expect = 3e-28
Identities = 76/200 (38%), Positives = 115/200 (57%), Gaps = 7/200 (3%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGF 82
FPSFLP V ++ + + LA I ++ S PI ++ V+ PI+L+HGF
Sbjct: 2 FPSFLPAAVGQLTESESIALAKSIHTQAIATPLSNQPITTAYVRQ--GGGGTPILLIHGF 59
Query: 83 DSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDV--VSKREHFYQFWKSYIK 140
DSS LE+R PLL + ETWA+D+LG+GF+ ++LP ++ R H + FWK+ I
Sbjct: 60 DSSVLEFRRLLPLLGKEN-ETWAVDLLGFGFT--QRLPGIKFSPIAIRTHLHSFWKTLIN 116
Query: 141 KPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLK 200
+P+ILVG S+G A AIDF + YPEAV+KLVLID++ G+ + Y L+
Sbjct: 117 QPVILVGASMGGAAAIDFTLTYPEAVQKLVLIDSAGLRGGSPLSKFMFPPLDYLAAQFLR 176
Query: 201 SVPLRVYANYLSFTNISFST 220
S +R + ++ N + +T
Sbjct: 177 SPKVRDRVSRAAYKNQNLAT 196
>ref|NP_926779.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Gloeobacter
violaceus PCC 7421] gi|35214406|dbj|BAC91774.1| glr3833
[Gloeobacter violaceus PCC 7421]
Length = 296
Score = 118 bits (296), Expect = 2e-25
Identities = 65/175 (37%), Positives = 105/175 (59%), Gaps = 2/175 (1%)
Query: 22 SFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHG 81
+ P+FLP ++DP + L +QR PVSV F ++ + + PI+LLHG
Sbjct: 3 TLPAFLPPAAALLRDPASIGLLQTLQRDPVSVPFLPQTAETAWTFA-AGSDDPPILLLHG 61
Query: 82 FDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKK 141
FDSS LE+R+ P L + TWA+D+ G+GF++ + + + H + FW+++I++
Sbjct: 62 FDSSALEFRFVLPRLGQRH-PTWAVDLWGFGFTERQSGAAYGRAEIQTHLHAFWQAHIRR 120
Query: 142 PMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEGTGNLATLPRAAAYAGV 196
P+++VG S+G A A+DFA+N+P+AV+KLVLI ++ +T LA L A V
Sbjct: 121 PVLVVGASMGGAAAVDFAVNHPQAVQKLVLIASTGFTGPPAWLAGLAEPLREAAV 175
>ref|ZP_00327205.1| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Trichodesmium erythraeum IMS101]
Length = 294
Score = 116 bits (290), Expect = 9e-25
Identities = 69/155 (44%), Positives = 93/155 (59%), Gaps = 7/155 (4%)
Query: 23 FPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGF 82
FP FLPK + + + A IQ + PI ++ + PI+LLHGF
Sbjct: 4 FPDFLPKLTASLTESISIDFAQNIQSESIFTPLISQPINTTFLSK--GQGGTPILLLHGF 61
Query: 83 DSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKR--EHFYQFWKSYIK 140
DSS LE+R PLL +T A+D+LG+GF+D +LP+ V + H Y FWKS I
Sbjct: 62 DSSILEFRRILPLLAIQN-KTLAVDLLGFGFTD--RLPNLKVNPRAIGTHLYYFWKSLIN 118
Query: 141 KPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+P+ILVG S+G AVAIDF +NYPE V+KLVLID++
Sbjct: 119 QPIILVGASMGGAVAIDFTLNYPEVVQKLVLIDSA 153
>ref|NP_926275.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Gloeobacter
violaceus PCC 7421] gi|35213900|dbj|BAC91270.1| glr3329
[Gloeobacter violaceus PCC 7421]
Length = 300
Score = 114 bits (285), Expect = 4e-24
Identities = 67/159 (42%), Positives = 93/159 (58%), Gaps = 2/159 (1%)
Query: 22 SFPSFLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHG 81
S P+ D I P AR L RI+R+PV + +S V+ + P++LLHG
Sbjct: 6 SVPAPFAPDAADIDHPQARWLLERIERVPVLCPSGSQSVATSFVRA-GRADGPPVLLLHG 64
Query: 82 FDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKK 141
FDSS LE+ PLL A TWAID+LG+GF++ +C + + H + FW I +
Sbjct: 65 FDSSLLEFFRLVPLLA-AHRSTWAIDLLGFGFTERRPDLACSAAALKAHLWSFWLERIGE 123
Query: 142 PMILVGPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEG 180
P++LVG S+G A AIDFA+ +PEAV LVLID+ +T G
Sbjct: 124 PVVLVGASMGGAAAIDFALTHPEAVAGLVLIDSVGFTCG 162
>dbj|BAB09377.1| unnamed protein product [Arabidopsis thaliana]
Length = 239
Score = 113 bits (282), Expect = 8e-24
Identities = 62/137 (45%), Positives = 79/137 (57%), Gaps = 32/137 (23%)
Query: 132 YQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAV------------------------- 166
+Q WK+YIK+PMILVGPSLG+ VA+DF YPEAV
Sbjct: 17 FQLWKTYIKRPMILVGPSLGATVAVDFTATYPEAVNFQFTIIKLFWIKPILIYTSLTKFD 76
Query: 167 -------EKLVLIDASVYTEGTGNLATLPRAAAYAGVYLLKSVPLRVYANYLSFTNISFS 219
+KLVLI+A+ Y+EGTG L LP++ AYAGV LLKS PLR+ AN L+F + S
Sbjct: 77 YFRGFQVDKLVLINANAYSEGTGRLKELPKSIAYAGVKLLKSFPLRLLANVLAFCSSPLS 136
Query: 220 TSLDWTNATMLLRR*KW 236
++DWTN L + W
Sbjct: 137 ENIDWTNIGRLHCQMPW 153
>ref|ZP_00516559.1| Alpha/beta hydrolase fold [Crocosphaera watsonii WH 8501]
gi|67855080|gb|EAM50347.1| Alpha/beta hydrolase fold
[Crocosphaera watsonii WH 8501]
Length = 289
Score = 112 bits (279), Expect = 2e-23
Identities = 63/151 (41%), Positives = 91/151 (59%), Gaps = 5/151 (3%)
Query: 26 FLPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENP-IVLLHGFDS 84
FLP + + + + L +I+ + S PI +S V Q +E P ++LLHGFDS
Sbjct: 3 FLPFEAKNLTEETSLDLVDKIKFENIITPLSPQPIPTSYV---FQGEEKPPVLLLHGFDS 59
Query: 85 SCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPMI 144
S LE+R +P+L ++ TWAID+LG+GF++ + + H Y WK+ IK+P I
Sbjct: 60 SLLEYRRLFPILSQSHA-TWAIDLLGFGFTERSSDLLFSPETIKTHLYYTWKTLIKEPCI 118
Query: 145 LVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
LVG S+G A AIDF + YPE V KLVLID++
Sbjct: 119 LVGASMGGATAIDFTLTYPETVSKLVLIDSA 149
>ref|NP_926004.1| probable 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Gloeobacter
violaceus PCC 7421] gi|35213628|dbj|BAC90999.1| glr3058
[Gloeobacter violaceus PCC 7421]
Length = 297
Score = 102 bits (254), Expect = 1e-20
Identities = 61/151 (40%), Positives = 89/151 (58%), Gaps = 4/151 (2%)
Query: 25 SFLPKDVNRIKDPFARTLATRIQRLPVSVKFSEN-PIMSSCVKPLVQNKENPIVLLHGFD 83
+FLP + +P + +A R++ + V ++ N + + CV E P +LLHGFD
Sbjct: 4 AFLPPGARILSEPGSLEMARRLELVSVRLEGRVNRTVATGCVHSGAG--EPPALLLHGFD 61
Query: 84 SSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPM 143
SS E+R PLL A E WA+D+LG+GF++ + D + +H FW+ YI +P
Sbjct: 62 SSVFEFRRLLPLLA-ARREVWAMDLLGFGFTERPAGIAYDPRAIGDHLASFWEQYIGRPA 120
Query: 144 ILVGPSLGSAVAIDFAINYPEAVEKLVLIDA 174
+LVG S+G A AID A+ PEAV KLVLID+
Sbjct: 121 LLVGASMGGAAAIDLALARPEAVAKLVLIDS 151
>ref|YP_172526.1| 2-hydroxy-6-oxohepta-24-dienoate hydrolase [Synechococcus elongatus
PCC 6301] gi|56686784|dbj|BAD80006.1|
2-hydroxy-6-oxohepta-24-dienoate hydrolase
[Synechococcus elongatus PCC 6301]
Length = 303
Score = 100 bits (249), Expect = 5e-20
Identities = 58/154 (37%), Positives = 90/154 (57%), Gaps = 3/154 (1%)
Query: 27 LPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGFDSSC 86
L V + + ++ LA IQ V+ E PI +S V+ + + P++ LHGFDSS
Sbjct: 10 LGSTVANLTEDTSKNLAAAIQWQAVATDLREAPIETSFVQ--LGQGDRPLLCLHGFDSSV 67
Query: 87 LEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPMILV 146
E+R P L E A+D LG+GF++ + + R+H + W+++ +KP++LV
Sbjct: 68 FEFRRLLPQLAEQH-SVRALDCLGFGFTERPEGIEISPDTIRQHLWGCWQAWYQKPIVLV 126
Query: 147 GPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEG 180
G S+G A+AIDFA+NYPEAV L+LID++ G
Sbjct: 127 GASMGGAIAIDFALNYPEAVAGLILIDSAGIAPG 160
>ref|ZP_00202321.1| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Synechococcus elongatus PCC
7942]
Length = 303
Score = 100 bits (248), Expect = 7e-20
Identities = 57/154 (37%), Positives = 90/154 (58%), Gaps = 3/154 (1%)
Query: 27 LPKDVNRIKDPFARTLATRIQRLPVSVKFSENPIMSSCVKPLVQNKENPIVLLHGFDSSC 86
L V + + ++ LA IQ ++ E PI +S V+ + + P++ LHGFDSS
Sbjct: 10 LGSTVANLTEDTSKNLAAAIQWQAIATDLREAPIETSFVQ--LGQGDRPLLCLHGFDSSV 67
Query: 87 LEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQFWKSYIKKPMILV 146
E+R P L E A+D LG+GF++ + + R+H + W+++ +KP++LV
Sbjct: 68 FEFRRLLPQLAEQH-SVRALDCLGFGFTERPEGIEISPDTIRQHLWGCWQAWYQKPIVLV 126
Query: 147 GPSLGSAVAIDFAINYPEAVEKLVLIDASVYTEG 180
G S+G A+AIDFA+NYPEAV L+LID++ G
Sbjct: 127 GASMGGAIAIDFALNYPEAVAGLILIDSAGIAPG 160
>ref|NP_440850.1| 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase [Synechocystis sp. PCC
6803] gi|1652609|dbj|BAA17530.1|
2-hydroxy-6-oxohepta-2,4-dienoate hydrolase
[Synechocystis sp. PCC 6803] gi|7469251|pir||S77427
2-hydroxy-6-oxohepta-2,4-dienoate hydrolase (EC 3.7.-.-)
- Synechocystis sp. (strain PCC 6803)
Length = 296
Score = 90.5 bits (223), Expect = 6e-17
Identities = 44/106 (41%), Positives = 72/106 (67%), Gaps = 1/106 (0%)
Query: 70 QNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKRE 129
+ + P++ +HGFDSS LE+R PL+++ AID+LG+GF+ K+ + +
Sbjct: 49 EGQGEPMLFIHGFDSSVLEFRRLLPLIKK-NFRAIAIDLLGFGFTTRSKILLPTPANIKI 107
Query: 130 HFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
H FW++ I++P+ LVG S+G AVA+DF +++PE V+KLVLID++
Sbjct: 108 HLDHFWQTIIQEPITLVGVSMGGAVALDFCLSFPERVKKLVLIDSA 153
>gb|AAP42732.1| At4g36530 [Arabidopsis thaliana] gi|30690680|ref|NP_849507.1|
hydrolase, alpha/beta fold family protein [Arabidopsis
thaliana] gi|13877561|gb|AAK43858.1| Unknown protein
[Arabidopsis thaliana]
Length = 321
Score = 74.7 bits (182), Expect = 3e-12
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+VQ + +P+VL+HGF +S WRY P L + + +A+D+LG+G+SD + L D +
Sbjct: 37 VVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSD-KALIEYDAMVW 94
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+ F K +K+P ++VG SLG A+ A+ PE V + L++++
Sbjct: 95 TDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLNSA 142
>emb|CAB16840.1| putative protein [Arabidopsis thaliana] gi|7270601|emb|CAB80319.1|
putative protein [Arabidopsis thaliana]
gi|21593181|gb|AAM65130.1| unknown [Arabidopsis
thaliana] gi|15234433|ref|NP_195371.1| hydrolase,
alpha/beta fold family protein [Arabidopsis thaliana]
gi|25407735|pir||C85431 hypothetical protein AT4g36530
[imported] - Arabidopsis thaliana
Length = 378
Score = 74.7 bits (182), Expect = 3e-12
Identities = 38/108 (35%), Positives = 65/108 (60%), Gaps = 2/108 (1%)
Query: 68 LVQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK 127
+VQ + +P+VL+HGF +S WRY P L + + +A+D+LG+G+SD + L D +
Sbjct: 94 VVQGEGSPLVLIHGFGASVFHWRYNIPELAKK-YKVYALDLLGFGWSD-KALIEYDAMVW 151
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
+ F K +K+P ++VG SLG A+ A+ PE V + L++++
Sbjct: 152 TDQVIDFMKEVVKEPAVVVGNSLGGFTALSVAVGLPEQVTGVALLNSA 199
>ref|ZP_00513951.1| Alpha/beta hydrolase fold [Crocosphaera watsonii WH 8501]
gi|67857915|gb|EAM53154.1| Alpha/beta hydrolase fold
[Crocosphaera watsonii WH 8501]
Length = 305
Score = 73.2 bits (178), Expect = 9e-12
Identities = 37/98 (37%), Positives = 56/98 (56%), Gaps = 2/98 (2%)
Query: 75 PIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSKREHFYQF 134
P++L+HGF + WR+ P L + +A+D+LG+G S + E Y F
Sbjct: 42 PLILIHGFGAGVEHWRHNIPTLRQY-YRVYALDLLGFGRSH-KAATDYTAYLWAEQIYYF 99
Query: 135 WKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLI 172
W+S+I KP++LVG S+GS V + A YPE V LV++
Sbjct: 100 WRSFIGKPVVLVGNSIGSLVCLTAAFKYPEMVSGLVML 137
>ref|ZP_00326451.1| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Trichodesmium erythraeum IMS101]
Length = 271
Score = 72.4 bits (176), Expect = 2e-11
Identities = 36/103 (34%), Positives = 61/103 (58%), Gaps = 5/103 (4%)
Query: 75 PIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLPSCDVVSK--REHFY 132
P++L HGF +S WR PLL ++G + +A+D+LG+G S PS D + E Y
Sbjct: 7 PLLLTHGFGASINHWRNNIPLLAKSGYQVFALDLLGFGAS---SKPSIDYSMELWEELIY 63
Query: 133 QFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
FW ++I++P + VG S+G+ +++ +YPE +LI+ +
Sbjct: 64 DFWSAHIRQPTVFVGNSIGALLSLMILASYPEIATGGILINCA 106
>ref|ZP_00161714.2| COG0596: Predicted hydrolases or acyltransferases (alpha/beta
hydrolase superfamily) [Anabaena variabilis ATCC 29413]
Length = 300
Score = 68.9 bits (167), Expect = 2e-10
Identities = 39/108 (36%), Positives = 62/108 (57%), Gaps = 3/108 (2%)
Query: 69 VQNKENPIVLLHGFDSSCLEWRYTYPLLEEAGIETWAIDILGWGFSDLEKLP-SCDVVSK 127
V P+VL+HGF +S WR P+L AG + +AID+LG+G SD + S DV
Sbjct: 32 VMGTGQPLVLVHGFGASIGHWRKNIPVLANAGYQVFAIDLLGFGGSDKAVIDYSVDVWV- 90
Query: 128 REHFYQFWKSYIKKPMILVGPSLGSAVAIDFAINYPEAVEKLVLIDAS 175
E FW ++I++P + VG S+G+ +++ +PE VLI+++
Sbjct: 91 -ELLKDFWTAHIQQPAVFVGNSIGALLSLIVLAKHPEITSGGVLINSA 137
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.335 0.145 0.460
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 483,648,346
Number of Sequences: 2540612
Number of extensions: 19671105
Number of successful extensions: 68362
Number of sequences better than 10.0: 1012
Number of HSP's better than 10.0 without gapping: 292
Number of HSP's successfully gapped in prelim test: 720
Number of HSP's that attempted gapping in prelim test: 67520
Number of HSP's gapped (non-prelim): 1035
length of query: 307
length of database: 863,360,394
effective HSP length: 127
effective length of query: 180
effective length of database: 540,702,670
effective search space: 97326480600
effective search space used: 97326480600
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146567.12


