

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146567.1 - phase: 0
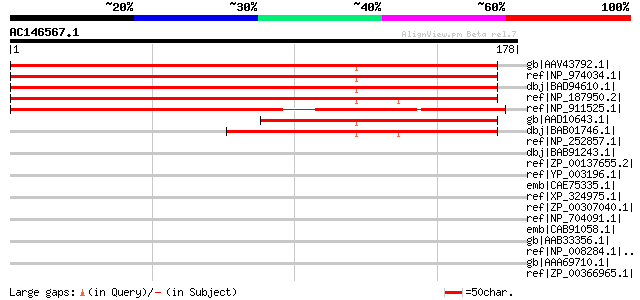
(178 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAV43792.1| At1g55535 [Arabidopsis thaliana] gi|53828513|gb|A... 255 4e-67
ref|NP_974034.1| expressed protein [Arabidopsis thaliana] 255 4e-67
dbj|BAD94610.1| putative protein [Arabidopsis thaliana] 251 5e-66
ref|NP_187950.2| expressed protein [Arabidopsis thaliana] 215 5e-55
ref|NP_911525.1| hypothetical protein [Oryza sativa (japonica cu... 171 8e-42
gb|AAD10643.1| Hypothetical protein [Arabidopsis thaliana] gi|25... 120 2e-26
dbj|BAB01746.1| unnamed protein product [Arabidopsis thaliana] 99 7e-20
ref|NP_252857.1| probable TonB-dependent receptor [Pseudomonas a... 34 2.1
dbj|BAB91243.1| LBP (LPS binding protein)/BPI (bactericidal/perm... 34 2.1
ref|ZP_00137655.2| COG4773: Outer membrane receptor for ferric c... 34 2.1
ref|YP_003196.1| histidine kinase sensor protein [Leptospira int... 33 2.7
emb|CAE75335.1| Hypothetical protein CBG23315 [Caenorhabditis br... 33 2.7
ref|XP_324975.1| hypothetical protein [Neurospora crassa] gi|289... 33 2.7
ref|ZP_00307040.1| COG1524: Uncharacterized proteins of the AP s... 33 4.7
ref|NP_704091.1| hypothetical protein [Plasmodium falciparum 3D7... 33 4.7
emb|CAB91058.1| NADH dehydrogenase subunit 5 [Drosophila melanog... 32 6.1
gb|AAB33356.1| NADH dehydrogenase subunit 5; ND5 [Drosophila mel... 32 6.1
ref|NP_008284.1|ND5_10704 NADH dehydrogenase subunit 5 [Drosophi... 32 6.1
gb|AAA69710.1| NADH dehydrogenase 5 [Drosophila melanogaster] 32 6.1
ref|ZP_00366965.1| type IV prepilin peptidase, probable , putati... 32 6.1
>gb|AAV43792.1| At1g55535 [Arabidopsis thaliana] gi|53828513|gb|AAU94366.1|
At1g55535 [Arabidopsis thaliana]
gi|42570096|ref|NP_683436.3| expressed protein
[Arabidopsis thaliana]
Length = 260
Score = 255 bits (651), Expect = 4e-67
Identities = 129/172 (75%), Positives = 150/172 (87%), Gaps = 1/172 (0%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LFALVAIES+SQSLGRTYA LLFC++LLDIS
Sbjct: 28 LIYAQIGCALIGSLGALYNGVLLINLAIALFALVAIESNSQSLGRTYAVLLFCALLLDIS 87
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILFT EIW+IS++ Y FFIFSVKLT+AM++IGF VRLSSSLLW QIYRLGA+ VDT+
Sbjct: 88 WFILFTEEIWSISAETYGTFFIFSVKLTMAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTS 147
Query: 121 -SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
R D DLR+SFL+P TPA+ARQ S + EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 148 LPRETDSDLRNSFLNPPTPAIARQCSGAEEILGGSIYDPAYYTSLFEESQTN 199
>ref|NP_974034.1| expressed protein [Arabidopsis thaliana]
Length = 250
Score = 255 bits (651), Expect = 4e-67
Identities = 129/172 (75%), Positives = 150/172 (87%), Gaps = 1/172 (0%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LFALVAIES+SQSLGRTYA LLFC++LLDIS
Sbjct: 28 LIYAQIGCALIGSLGALYNGVLLINLAIALFALVAIESNSQSLGRTYAVLLFCALLLDIS 87
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILFT EIW+IS++ Y FFIFSVKLT+AM++IGF VRLSSSLLW QIYRLGA+ VDT+
Sbjct: 88 WFILFTEEIWSISAETYGTFFIFSVKLTMAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTS 147
Query: 121 -SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
R D DLR+SFL+P TPA+ARQ S + EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 148 LPRETDSDLRNSFLNPPTPAIARQCSGAEEILGGSIYDPAYYTSLFEESQTN 199
>dbj|BAD94610.1| putative protein [Arabidopsis thaliana]
Length = 260
Score = 251 bits (642), Expect = 5e-66
Identities = 128/172 (74%), Positives = 149/172 (86%), Gaps = 1/172 (0%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LFALVAIES+SQSLGRTYA LLFC++LLDIS
Sbjct: 28 LIYAQIGCALIGSLGALYNGVLLINLAIALFALVAIESNSQSLGRTYAVLLFCALLLDIS 87
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILFT EIW+IS++ Y FFIFSVKLT+AM++IGF VRLSSSLLW QIYRLGA+ VDT+
Sbjct: 88 WFILFTEEIWSISAETYGTFFIFSVKLTMAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTS 147
Query: 121 -SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
R D DLR+SFL+P TPA+ARQ S + EILGGSI DPAYY+SLFE+SQ N
Sbjct: 148 LPRETDSDLRNSFLNPPTPAIARQCSGAEEILGGSINDPAYYTSLFEESQTN 199
>ref|NP_187950.2| expressed protein [Arabidopsis thaliana]
Length = 242
Score = 215 bits (547), Expect = 5e-55
Identities = 118/190 (62%), Positives = 139/190 (73%), Gaps = 19/190 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I +IGCAL+GSLGA YNGV LINLAI+LF LVAIES+SQSLGRTYA LLFC+ILLD+S
Sbjct: 27 LIYAQIGCALIGSLGALYNGVVLINLAIALFGLVAIESNSQSLGRTYAVLLFCAILLDVS 86
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILF+ EIWNISSD Y F+IFSVKLTLAM+I GF+VRLSSSLLW QIYRLGAS +D+
Sbjct: 87 WFILFSKEIWNISSDMYQVFYIFSVKLTLAMEIAGFVVRLSSSLLWFQIYRLGASIIDSP 146
Query: 121 -SRAADFDLRSSFLSP------------------VTPAVARQISNSNEILGGSIYDPAYY 161
R +D DLR+SFL P PA+A+Q S S+EIL SI +PA Y
Sbjct: 147 FPRQSDSDLRNSFLEPPLLARQRSRDPELRNSFLQAPAIAKQRSRSDEILEDSIDEPASY 206
Query: 162 SSLFEDSQEN 171
+ L + N
Sbjct: 207 TPLLDGGLSN 216
>ref|NP_911525.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|22093623|dbj|BAC06919.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 247
Score = 171 bits (433), Expect = 8e-42
Identities = 94/174 (54%), Positives = 121/174 (69%), Gaps = 12/174 (6%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDIS 60
+I ++IGCAL+GSLGA +NGV +INL I LFA+VAIESSSQ+LGRTYA LLF +I+LD++
Sbjct: 39 LIYLQIGCALIGSLGALFNGVLVINLVIGLFAVVAIESSSQTLGRTYAVLLFFAIVLDVA 98
Query: 61 WFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA 120
WFILF+H I + Y + F+ S+KL L MQIIGF +YRLG S
Sbjct: 99 WFILFSHAINITPEEKYGQLFVLSLKLALWMQIIGF-----------SMYRLGVSSSTPT 147
Query: 121 SRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQENKST 174
++D R+SFLSP + +V R S +++ILGGSIYDPAYYSSLF D + N T
Sbjct: 148 YHEVNYDGRNSFLSPRSSSVRRN-SMADDILGGSIYDPAYYSSLFADVRNNTCT 200
>gb|AAD10643.1| Hypothetical protein [Arabidopsis thaliana] gi|25373300|pir||G96597
hypothetical protein T5A14.6 [imported] - Arabidopsis
thaliana
Length = 185
Score = 120 bits (300), Expect = 2e-26
Identities = 60/84 (71%), Positives = 71/84 (84%), Gaps = 1/84 (1%)
Query: 89 LAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA-SRAADFDLRSSFLSPVTPAVARQISNS 147
+AM++IGF VRLSSSLLW QIYRLGA+ VDT+ R D DLR+SFL+P TPA+ARQ S +
Sbjct: 1 MAMEMIGFFVRLSSSLLWFQIYRLGAAIVDTSLPRETDSDLRNSFLNPPTPAIARQCSGA 60
Query: 148 NEILGGSIYDPAYYSSLFEDSQEN 171
EILGGSIYDPAYY+SLFE+SQ N
Sbjct: 61 EEILGGSIYDPAYYTSLFEESQTN 84
>dbj|BAB01746.1| unnamed protein product [Arabidopsis thaliana]
Length = 163
Score = 98.6 bits (244), Expect = 7e-20
Identities = 58/114 (50%), Positives = 71/114 (61%), Gaps = 19/114 (16%)
Query: 77 YAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGASYVDTA-SRAADFDLRSSFLSP 135
Y F+IFSVKLTLAM+I GF+VRLSSSLLW QIYRLGAS +D+ R +D DLR+SFL P
Sbjct: 2 YQVFYIFSVKLTLAMEIAGFVVRLSSSLLWFQIYRLGASIIDSPFPRQSDSDLRNSFLEP 61
Query: 136 ------------------VTPAVARQISNSNEILGGSIYDPAYYSSLFEDSQEN 171
PA+A+Q S S+EIL SI +PA Y+ L + N
Sbjct: 62 PLLARQRSRDPELRNSFLQAPAIAKQRSRSDEILEDSIDEPASYTPLLDGGLSN 115
>ref|NP_252857.1| probable TonB-dependent receptor [Pseudomonas aeruginosa PAO1]
gi|9950376|gb|AAG07555.1| probable TonB-dependent
receptor [Pseudomonas aeruginosa PAO1]
gi|11351936|pir||A83125 probable TonB-dependent receptor
PA4168 [imported] - Pseudomonas aeruginosa (strain PAO1)
Length = 802
Score = 33.9 bits (76), Expect = 2.1
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query: 108 QIYRLGASYVDTASRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFED 167
Q Y + ASY D ++FD L P+T +I E GG++ +LF+
Sbjct: 570 QTYSVYASYTDIFKPQSNFDAGGGLLDPIT-GKNYEIGLKGEHFGGALNSQI---ALFQI 625
Query: 168 SQENKSTCGVG 178
QEN++T VG
Sbjct: 626 DQENRATEDVG 636
>dbj|BAB91243.1| LBP (LPS binding protein)/BPI (bactericidal/permeability-increasing
protein)-1 [Oncorhynchus mykiss]
Length = 473
Score = 33.9 bits (76), Expect = 2.1
Identities = 37/154 (24%), Positives = 64/154 (41%), Gaps = 26/154 (16%)
Query: 9 ALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF--------LLFCSILLDIS 60
A +G + S G++++NL + AL + + SL T AF + + S + D
Sbjct: 62 APIGKVKYSLTGITIVNLGLPYSALALVPDTGISLSITNAFISLHGNWKIRYLSFIKDSG 121
Query: 61 WF------ILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWIQIYRLGA 114
F + T I I SD+ + + SV + ++ W +Y L +
Sbjct: 122 SFDLEVDGLTVTDSI-TIKSDETGRPTVSSV--NCVANVGSASIKFHGGASW--LYNLFS 176
Query: 115 SYVDTASRAADFDLRSSFLSPVTPAVARQISNSN 148
+Y+D A LRS+ + P VA I++ N
Sbjct: 177 AYIDKA-------LRSALQKQICPLVADTITDMN 203
>ref|ZP_00137655.2| COG4773: Outer membrane receptor for ferric coprogen and
ferric-rhodotorulic acid [Pseudomonas aeruginosa
UCBPP-PA14]
Length = 793
Score = 33.9 bits (76), Expect = 2.1
Identities = 23/71 (32%), Positives = 34/71 (47%), Gaps = 4/71 (5%)
Query: 108 QIYRLGASYVDTASRAADFDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFED 167
Q Y + ASY D ++FD L P+T +I E GG++ +LF+
Sbjct: 561 QTYSVYASYTDIFKPQSNFDAGGGLLDPIT-GKNYEIGLKGEHFGGALNSQI---ALFQI 616
Query: 168 SQENKSTCGVG 178
QEN++T VG
Sbjct: 617 DQENRATEDVG 627
>ref|YP_003196.1| histidine kinase sensor protein [Leptospira interrogans serovar
Copenhageni str. Fiocruz L1-130]
gi|24216826|ref|NP_714307.1| Sensory transduction
histidine kinases [Leptospira interrogans serovar Lai
str. 56601] gi|24198195|gb|AAN51325.1| Sensory
transduction histidine kinases [Leptospira interrogans
serovar lai str. 56601] gi|45602356|gb|AAS71833.1|
histidine kinase sensor protein [Leptospira interrogans
serovar Copenhageni str. Fiocruz L1-130]
Length = 376
Score = 33.5 bits (75), Expect = 2.7
Identities = 22/81 (27%), Positives = 45/81 (55%), Gaps = 3/81 (3%)
Query: 25 NLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDYAKFFIFS 84
N+ +S+F ++ + + + +T +L+C IL+ I F L H++ ++D FF+FS
Sbjct: 109 NIILSIFIMIFLFFTD--VKKTIFIMLYCLILIFIDEFYLKEHDV-VYTADRIGLFFMFS 165
Query: 85 VKLTLAMQIIGFIVRLSSSLL 105
V L L ++ + V+ + L+
Sbjct: 166 VILILTVRALYGSVQEKNELI 186
>emb|CAE75335.1| Hypothetical protein CBG23315 [Caenorhabditis briggsae]
Length = 315
Score = 33.5 bits (75), Expect = 2.7
Identities = 23/100 (23%), Positives = 45/100 (45%), Gaps = 19/100 (19%)
Query: 19 NGVSLINLAISLFALVAIESSSQ-----------SLGRTYAFLLFCSILLDISWFILFTH 67
N V L+ I F + ++ SS S Y + +C +L+ ++ F
Sbjct: 60 NSVILVWGFIGTFVPITLKESSSFSNVYETIVIISCNNIYIAVQYCGLLIALNRFCAMFF 119
Query: 68 EIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLLWI 107
+W YAKFF S+K+T + + F ++L+ ++ ++
Sbjct: 120 PLW------YAKFF--SIKITFVIVVFHFALKLADNIYYL 151
>ref|XP_324975.1| hypothetical protein [Neurospora crassa] gi|28926749|gb|EAA35715.1|
hypothetical protein [Neurospora crassa]
Length = 671
Score = 33.5 bits (75), Expect = 2.7
Identities = 19/32 (59%), Positives = 22/32 (68%), Gaps = 2/32 (6%)
Query: 118 DTASRAADFDLRSSFLSPVTPAVARQISNSNE 149
DTASRAA F LRS L P++ AVA + N NE
Sbjct: 408 DTASRAAPFLLRS--LRPISTAVASMLENPNE 437
>ref|ZP_00307040.1| COG1524: Uncharacterized proteins of the AP superfamily
[Ferroplasma acidarmanus]
Length = 366
Score = 32.7 bits (73), Expect = 4.7
Identities = 43/163 (26%), Positives = 72/163 (43%), Gaps = 19/163 (11%)
Query: 22 SLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLD-ISWFIL------FTHEIWNIS- 73
+L+NL+ +F + + S+ +G Y C ILLD + W I F +EI S
Sbjct: 12 TLVNLSSDIFGHFGLRTESEGIGLNYKNKKLCFILLDGLGWNIYRQTGIEFKNEIKGTSV 71
Query: 74 -----SDDYAKFFIFSVKLTLAMQIIGF--IVRLSSSLLWIQIYRLGASYV-DTASRAAD 125
S+ + FF+ K IIG+ ++ S++ I Y A+Y+ D+ +A
Sbjct: 72 FPSTTSNALSSFFL--NKFPGQHGIIGYQLYIKQLGSIVNILGYTSSAAYMRDSMEKAYP 129
Query: 126 FDLRSSFLSPVTPAVARQISNSNEILGGSIYDPAYYSSLFEDS 168
+F S VT S N I+ G I ++ + L+ D+
Sbjct: 130 MSSIFNFESKVTSLNRAGYSTVN-IIPGFINKTSFSNILYGDN 171
>ref|NP_704091.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23498829|emb|CAD50906.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 434
Score = 32.7 bits (73), Expect = 4.7
Identities = 20/77 (25%), Positives = 39/77 (49%), Gaps = 6/77 (7%)
Query: 19 NGVSLINLAISLF-ALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDDY 77
+ V++I I +F AL++ + + AF +F S + + T EIWN+ + D
Sbjct: 227 HNVAIIQPTIPIFTALISYYLKIEKMNYITAFSIFLSF-----FGLAITAEIWNLKTFDL 281
Query: 78 AKFFIFSVKLTLAMQII 94
+ + +V +T +Q+I
Sbjct: 282 GFYLLLTVPITKGLQVI 298
>emb|CAB91058.1| NADH dehydrogenase subunit 5 [Drosophila melanogaster]
Length = 577
Score = 32.3 bits (72), Expect = 6.1
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF----LLFCSIL 56
+ISM + C LL L+N I + +S S+ T+ F LLF S +
Sbjct: 14 LISMSLSCFLLSLY-------FLLNDMIYFIEWELVSLNSMSIVMTFLFDWMSLLFMSFV 66
Query: 57 LDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLL 105
L IS ++F + + ++ + +F + + L+M ++ L S LL
Sbjct: 67 LMISSLVIFYSKEYMMNDNHINRFIMLVLMFVLSMMLLIISPNLISILL 115
>gb|AAB33356.1| NADH dehydrogenase subunit 5; ND5 [Drosophila melanogaster]
Length = 505
Score = 32.3 bits (72), Expect = 6.1
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF----LLFCSIL 56
+ISM + C LL L+N I + +S S+ T+ F LLF S +
Sbjct: 4 LISMSLSCFLLSLY-------FLLNDMIYFIEWELVSLNSMSIVMTFLFDWMSLLFMSFV 56
Query: 57 LDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLL 105
L IS ++F + + ++ + +F + + L+M ++ L S LL
Sbjct: 57 LMISSLVIFYSKEYMMNDNHINRFIMLVLMFVLSMMLLIISPNLISILL 105
>ref|NP_008284.1|ND5_10704 NADH dehydrogenase subunit 5 [Drosophila melanogaster]
gi|1166537|gb|AAC47818.1| NADH dehydrogenase subunit 5
[Drosophila melanogaster]
Length = 574
Score = 32.3 bits (72), Expect = 6.1
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF----LLFCSIL 56
+ISM + C LL L+N I + +S S+ T+ F LLF S +
Sbjct: 9 LISMSLSCFLLSLY-------FLLNDMIYFIEWELVSLNSMSIVMTFLFDWMSLLFMSFV 61
Query: 57 LDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLL 105
L IS ++F + + ++ + +F + + L+M ++ L S LL
Sbjct: 62 LMISSLVIFYSKEYMMNDNHINRFIMLVLMFVLSMMLLIISPNLISILL 110
>gb|AAA69710.1| NADH dehydrogenase 5 [Drosophila melanogaster]
Length = 580
Score = 32.3 bits (72), Expect = 6.1
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 1 MISMKIGCALLGSLGASYNGVSLINLAISLFALVAIESSSQSLGRTYAF----LLFCSIL 56
+ISM + C LL L+N I + +S S+ T+ F LLF S +
Sbjct: 9 LISMSLSCFLLSLY-------FLLNDMIYFIEWELVSLNSMSIVMTFLFDWMSLLFMSFV 61
Query: 57 LDISWFILFTHEIWNISSDDYAKFFIFSVKLTLAMQIIGFIVRLSSSLL 105
L IS ++F + + ++ + +F + + L+M ++ L S LL
Sbjct: 62 LMISSLVIFYSKEYMMNDNHINRFIMLVLMFVLSMMLLIISPNLISILL 110
>ref|ZP_00366965.1| type IV prepilin peptidase, probable , putative [Campylobacter coli
RM2228] gi|57020947|gb|EAL57611.1| type IV prepilin
peptidase, probable , putative [Campylobacter coli
RM2228]
Length = 237
Score = 32.3 bits (72), Expect = 6.1
Identities = 34/129 (26%), Positives = 57/129 (43%), Gaps = 21/129 (16%)
Query: 17 SYNGVSLINLAISLFALVAIESSSQSLGRTYAFLLFCSILLDISWFILFTHEIWNISSDD 76
S N I+L I LF L A+ L F ++ + W I F + +S++
Sbjct: 74 SQNIGEFIHLLIFLFVLFALSCID---------LKFQAVPQKLLWLIFFIAFSFAFNSNE 124
Query: 77 YAKFFIF----SVKLTLAMQIIGFIVRLSSSLLWIQIYR-------LG-ASYVDTASRAA 124
+A FFIF + + A GF+ L + + +++ +R LG A + AS A
Sbjct: 125 FAYFFIFENFQNGFMVQAFIFAGFLFLLKNFIAFLKNFRTKDIQENLGDADIIILASMAG 184
Query: 125 DFDLRSSFL 133
F +S+F+
Sbjct: 185 VFGFKSAFI 193
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.324 0.137 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 266,019,283
Number of Sequences: 2540612
Number of extensions: 9305345
Number of successful extensions: 32426
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 32406
Number of HSP's gapped (non-prelim): 30
length of query: 178
length of database: 863,360,394
effective HSP length: 119
effective length of query: 59
effective length of database: 561,027,566
effective search space: 33100626394
effective search space used: 33100626394
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146567.1


