

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146566.1 + phase: 0
(155 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
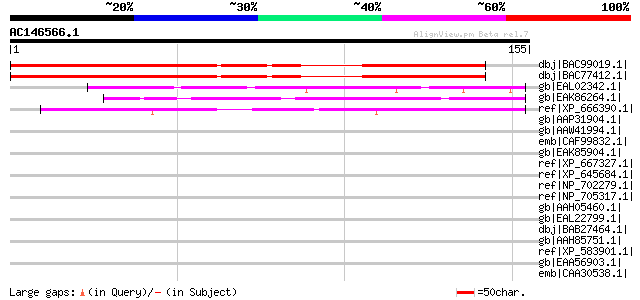
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAC99019.1| makorin ring-zinc-finger protein [Pisum sativum] 167 1e-40
dbj|BAC77412.1| makorin ring-zinc-finger protein [Pisum sativum]... 167 1e-40
gb|EAL02342.1| probable ubiquitin ligase Tom1p fragment [Candida... 47 2e-04
gb|EAK86264.1| hypothetical protein UM04809.1 [Ustilago maydis 5... 46 3e-04
ref|XP_666390.1| hypothetical protein Chro.30151 [Cryptosporidiu... 45 6e-04
gb|AAP31904.1| histidine-rich calcium binding protein [Rattus no... 44 0.002
gb|AAW41994.1| small nucleolar ribonucleoprotein protein, putati... 42 0.005
emb|CAF99832.1| unnamed protein product [Tetraodon nigroviridis] 41 0.011
gb|EAK85904.1| hypothetical protein UM05044.1 [Ustilago maydis 5... 40 0.015
ref|XP_667327.1| CCAAT-box-binding transcription factor [Cryptos... 40 0.015
ref|XP_645684.1| hypothetical protein DDB0202825 [Dictyostelium ... 40 0.015
ref|NP_702279.1| hypothetical protein PF14_0390 [Plasmodium falc... 40 0.019
ref|NP_705317.1| Casein kinase II regulatory subunit, putative [... 40 0.019
gb|AAH05460.1| Nucleolin [Mus musculus] 40 0.019
gb|EAL22799.1| hypothetical protein CNBB0200 [Cryptococcus neofo... 40 0.025
dbj|BAB27464.1| unnamed protein product [Mus musculus] 40 0.025
gb|AAH85751.1| Nucleolin [Rattus norvegicus] 39 0.042
ref|XP_583901.1| PREDICTED: similar to nucleolin-related protein... 39 0.042
gb|EAA56903.1| hypothetical protein MG07258.4 [Magnaporthe grise... 39 0.042
emb|CAA30538.1| nucleolin [Mus musculus] gi|128843|sp|P09405|NUC... 39 0.055
>dbj|BAC99019.1| makorin ring-zinc-finger protein [Pisum sativum]
Length = 411
Score = 167 bits (422), Expect = 1e-40
Identities = 89/142 (62%), Positives = 104/142 (72%), Gaps = 20/142 (14%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M+DMLD+L EVDLSSGEFYSIMRDS+FF+ MDP EMMALSDSLAGGS PCLGPF SDD+
Sbjct: 289 MYDMLDMLSEVDLSSGEFYSIMRDSEFFDEMDPLEMMALSDSLAGGSVPCLGPFGSDDEG 348
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
D+ F V RMAAM EA+ +D FGPDD G DDF PDDF +D
Sbjct: 349 DN-FNVFRMAAMSEALD-NLDDFGPDD------------------FGPDDFDPDDFDELD 388
Query: 121 PMDAALISMMMHSHMDDEEDDE 142
PM+AALISMMMHS++D++ +DE
Sbjct: 389 PMEAALISMMMHSNIDEDSEDE 410
>dbj|BAC77412.1| makorin ring-zinc-finger protein [Pisum sativum]
gi|33468810|dbj|BAC81564.1| makorin RING finger protein
[Pisum sativum]
Length = 461
Score = 167 bits (422), Expect = 1e-40
Identities = 89/142 (62%), Positives = 104/142 (72%), Gaps = 20/142 (14%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M+DMLD+L EVDLSSGEFYSIMRDS+FF+ MDP EMMALSDSLAGGS PCLGPF SDD+
Sbjct: 339 MYDMLDMLSEVDLSSGEFYSIMRDSEFFDEMDPLEMMALSDSLAGGSVPCLGPFGSDDEG 398
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
D+ F V RMAAM EA+ +D FGPDD G DDF PDDF +D
Sbjct: 399 DN-FNVFRMAAMSEALD-NLDDFGPDD------------------FGPDDFDPDDFDELD 438
Query: 121 PMDAALISMMMHSHMDDEEDDE 142
PM+AALISMMMHS++D++ +DE
Sbjct: 439 PMEAALISMMMHSNIDEDSEDE 460
>gb|EAL02342.1| probable ubiquitin ligase Tom1p fragment [Candida albicans SC5314]
gi|46442930|gb|EAL02216.1| probable ubiquitin ligase
Tom1p fragment [Candida albicans SC5314]
Length = 495
Score = 47.0 bits (110), Expect = 2e-04
Identities = 44/143 (30%), Positives = 64/143 (43%), Gaps = 18/143 (12%)
Query: 24 DSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAF 83
D +F++ DP + M + L+G S G D D+DDD + M++ E AI D
Sbjct: 191 DDEFYDAEDPVDAMMTGEDLSGASDD--GDDDDDEDDDGDDDIDDMSS--ELSAIDSDLD 246
Query: 84 GPDD-DEEEFNVFRMAAMQEAMVSGIDDFGPD--DFPGIDPMDAALISMMMHSH------ 134
G D+ D+ E + E ID+ D D ID +D L+S
Sbjct: 247 GGDNADDIEMEIEVDPYDDERGSEDIDEDASDMEDIEIIDDLD--LVSQTDGDDDGDDDD 304
Query: 135 -MDDEEDDEDYEDEE--EYTDDE 154
DDEED ++D+E EY +DE
Sbjct: 305 GRDDEEDSSSWDDDEMSEYDEDE 327
>gb|EAK86264.1| hypothetical protein UM04809.1 [Ustilago maydis 521]
gi|49077026|ref|XP_402424.1| hypothetical protein
UM04809.1 [Ustilago maydis 521]
Length = 608
Score = 46.2 bits (108), Expect = 3e-04
Identities = 40/126 (31%), Positives = 57/126 (44%), Gaps = 11/126 (8%)
Query: 29 EGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDD 88
+G+D +M AL D LAG D DDDD+D EA+ D DDD
Sbjct: 193 DGLDEVDMAAL-DELAGEEEED----DDDDDDEDDEDEENKNDSDEALEHDDD----DDD 243
Query: 89 EEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEE 148
EE + +AA QE + + D + L+ + + DDEEDDE+ E+ E
Sbjct: 244 NEESDSEELAAEQEDEIPRVVPNANGDTLAASIARSGLVPV--EASDDDEEDDEEDEENE 301
Query: 149 EYTDDE 154
+ +DE
Sbjct: 302 QEDEDE 307
Score = 35.0 bits (79), Expect = 0.61
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 12/74 (16%)
Query: 86 DDDE---EEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMM--HSHMDDEED 140
DDDE EE MQ M + DD G+D +D A + + DD++D
Sbjct: 166 DDDESDLEEGQDVSEKGMQRLMAALGDD-------GLDEVDMAALDELAGEEEEDDDDDD 218
Query: 141 DEDYEDEEEYTDDE 154
DED EDEE D +
Sbjct: 219 DEDDEDEENKNDSD 232
>ref|XP_666390.1| hypothetical protein Chro.30151 [Cryptosporidium hominis]
gi|54657377|gb|EAL36160.1| hypothetical protein
Chro.30151 [Cryptosporidium hominis]
Length = 3919
Score = 45.1 bits (105), Expect = 6e-04
Identities = 44/154 (28%), Positives = 66/154 (42%), Gaps = 20/154 (12%)
Query: 10 EVDLSSGEFYSIMRDSDFFEGMDPFEMMALSD-----SLAGGSGPCLGPFDSDDDDDDGF 64
EVDL F D DF + +D S+ SL G G + D DDDD+D
Sbjct: 1535 EVDLDQFGFQFNSSDEDFDDDLDDSLEDDFSEEFDLTSLDGLDGEDIDDNDEDDDDND-- 1592
Query: 65 RVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGI----DDFGPDDFPGID 120
E M ++ DDDE + ++F M V I DD DD G D
Sbjct: 1593 --------VEDMDDDVEDDVEDDDEND-DLFLDETMINDGVIDIEGEDDDLSNDDIVGDD 1643
Query: 121 PMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
+ + + + DD++DD+D +D+++ DD+
Sbjct: 1644 DLTSDVDDVDDDDDDDDDDDDDDDDDDDDDDDDD 1677
>gb|AAP31904.1| histidine-rich calcium binding protein [Rattus norvegicus]
gi|31077132|ref|NP_852034.1| histidine rich calcium
binding protein [Rattus norvegicus]
Length = 755
Score = 43.5 bits (101), Expect = 0.002
Identities = 31/103 (30%), Positives = 45/103 (43%), Gaps = 10/103 (9%)
Query: 52 GPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDF 111
G D DDDD+DG VS Q D DD +EE + R+ ++ D+
Sbjct: 215 GHHDEDDDDEDG--VSTEGEHQAHRYQDHDEEDDDDSDEESHAHRLQGQED---ENDDED 269
Query: 112 GPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G G D H DD++D+E+ E+EEE ++E
Sbjct: 270 GDSSENGHQTQDH-----QGHKEQDDDDDEEEEEEEEEEEEEE 307
>gb|AAW41994.1| small nucleolar ribonucleoprotein protein, putative [Cryptococcus
neoformans var. neoformans JEC21]
gi|58264290|ref|XP_569301.1| small nucleolar
ribonucleoprotein protein, putative [Cryptococcus
neoformans var. neoformans JEC21]
Length = 776
Score = 42.0 bits (97), Expect = 0.005
Identities = 29/87 (33%), Positives = 40/87 (45%), Gaps = 3/87 (3%)
Query: 5 LDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGF 64
L+ E+ SGE M + + E MD FE DS GSG D DDDD++G
Sbjct: 203 LEEFREMLKESGEDVDDMTEEELMERMDEFEGDESDDSEESGSG---SDEDEDDDDEEGD 259
Query: 65 RVSRMAAMQEAMAIGMDAFGPDDDEEE 91
+A+ E + G D+DE+E
Sbjct: 260 EDELLASASEGENDEEVSVGDDEDEDE 286
>emb|CAF99832.1| unnamed protein product [Tetraodon nigroviridis]
Length = 665
Score = 40.8 bits (94), Expect = 0.011
Identities = 45/145 (31%), Positives = 63/145 (43%), Gaps = 15/145 (10%)
Query: 1 MHDMLDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDD 60
M DM ++ G S G+ + R+S G D + S S A GSG G DD+
Sbjct: 1 MADMDELFG----SDGDSDNDQRESGSGSGSDSDQERPPSVSNASGSGS--GSERDQDDE 54
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSG--IDDFGPDDFPG 118
D+ + A+M + + FG DD E+E + + Q + SG D D
Sbjct: 55 DEEDHEAGNASMNKEL------FG-DDSEDEQHSQHSGSDQHSDRSGNQSDASMHSDQGD 107
Query: 119 IDPMDAALISMMMHSHMDDEEDDED 143
DP DA S H D++EDDED
Sbjct: 108 NDPSDAEQHSGSERGHQDEDEDDED 132
>gb|EAK85904.1| hypothetical protein UM05044.1 [Ustilago maydis 521]
gi|49077648|ref|XP_402659.1| hypothetical protein
UM05044.1 [Ustilago maydis 521]
Length = 543
Score = 40.4 bits (93), Expect = 0.015
Identities = 35/125 (28%), Positives = 54/125 (43%), Gaps = 23/125 (18%)
Query: 53 PFDSDDDDDDGFRVS---RMAAMQEAMAIGMDAFGPDDDEEEFNVFR---------MAAM 100
PF+ D+DD DG + R A ++ A G DDDE+ F+V A+
Sbjct: 89 PFNEDEDDSDGEADAGSHRRRADKKRRGDAAGAAGDDDDEDMFDVENDDEKQTSSASKAI 148
Query: 101 QEAMVSGI----------DDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEE-E 149
++A G DFG ++ +D D ++ DD++DD+D D E E
Sbjct: 149 RKAQTRGKYLKLGELEEGQDFGSNNRSTLDAADDEDAKLLRGKGKDDDDDDDDDLDPELE 208
Query: 150 YTDDE 154
DD+
Sbjct: 209 LEDDD 213
>ref|XP_667327.1| CCAAT-box-binding transcription factor [Cryptosporidium hominis]
gi|54658453|gb|EAL37098.1| CCAAT-box-binding
transcription factor [Cryptosporidium hominis]
Length = 1090
Score = 40.4 bits (93), Expect = 0.015
Identities = 25/74 (33%), Positives = 36/74 (47%), Gaps = 5/74 (6%)
Query: 80 MDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDEE 139
+D FG D D + ++ + + + +DDF DDF D +D DD E
Sbjct: 967 VDNFGEDLDNNDDDIDLLDGVN---MDDMDDF--DDFEDFDDLDGESDFAGEEGEEDDSE 1021
Query: 140 DDEDYEDEEEYTDD 153
D+ED EDEE+ DD
Sbjct: 1022 DEEDEEDEEDDEDD 1035
>ref|XP_645684.1| hypothetical protein DDB0202825 [Dictyostelium discoideum]
gi|60473827|gb|EAL71766.1| hypothetical protein
DDB0202825 [Dictyostelium discoideum]
Length = 1461
Score = 40.4 bits (93), Expect = 0.015
Identities = 40/140 (28%), Positives = 64/140 (45%), Gaps = 15/140 (10%)
Query: 17 EFYSIMRDSDFFEGMDPFEMM--ALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQE 74
+ +SI D+ F+ ++ M A +D L G G D D++DDD F+ +++
Sbjct: 801 QIFSIFADNISFQVLEHILTMIKAPTDELFNGLDGN-GDDDEDNEDDDEFKPISPEELKQ 859
Query: 75 AMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSH 134
+ D DD+EEE +E +D DD ++P A + M H
Sbjct: 860 SKENSDDEDEEDDEEEE---------EEEEEEEEED--SDDDNSVNPELVARLKQTMGKH 908
Query: 135 MDDEEDDEDYEDEEEYTDDE 154
+ +EDDED ED E T++E
Sbjct: 909 LLLDEDDED-EDLLEPTEEE 927
>ref|NP_702279.1| hypothetical protein PF14_0390 [Plasmodium falciparum 3D7]
gi|23497460|gb|AAN37003.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 227
Score = 40.0 bits (92), Expect = 0.019
Identities = 31/100 (31%), Positives = 38/100 (38%), Gaps = 40/100 (40%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD D DDDE++ DD D
Sbjct: 119 DEDDDDDD------------------DDDEDDDDEDD--------------EDDDDDDED 146
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
D D MD DD++DDED +DEE+Y DD+
Sbjct: 147 DDDDFDDMD--------EDDDDDDDDDEDDDDEEDYDDDD 178
Score = 37.0 bits (84), Expect = 0.16
Identities = 30/100 (30%), Positives = 38/100 (38%), Gaps = 27/100 (27%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD M E D DDDEE+++ DD D
Sbjct: 137 DEDDDDDDEDDDDDFDDMDEDDDDDDDDDEDDDDEEDYD---------------DDDDDD 181
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
D D + DD+EDD+D D+ EY D+
Sbjct: 182 DDDDEDDDE------------DDDEDDDDENDQNEYAGDD 209
Score = 33.5 bits (75), Expect = 1.8
Identities = 29/101 (28%), Positives = 37/101 (35%), Gaps = 35/101 (34%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D DDDDDD D DDDE++ DD D
Sbjct: 86 DDDDDDDD------------------DDDEDDDDEDD-----------------DDDDDD 110
Query: 115 DFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDEY 155
D D D DD+EDDED +D++E DD++
Sbjct: 111 DDDDDDDDDEDDDDDDDDDEDDDDEDDEDDDDDDEDDDDDF 151
>ref|NP_705317.1| Casein kinase II regulatory subunit, putative [Plasmodium
falciparum 3D7] gi|23615562|emb|CAD52554.1| Casein
kinase II regulatory subunit, putative [Plasmodium
falciparum 3D7]
Length = 385
Score = 40.0 bits (92), Expect = 0.019
Identities = 34/149 (22%), Positives = 66/149 (43%), Gaps = 17/149 (11%)
Query: 9 GEVDLSSGEFY--SIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRV 66
GEV+L+ +FY +++ D E ++ E A +D + + ++ DD+D+D
Sbjct: 21 GEVELTDADFYDLTVINDKIDEEIIEDDEEEADNDDQENDNVQEV--YNIDDEDNDIHND 78
Query: 67 SRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAAL 126
+ Q + + +++EEE +E D+ DD D D
Sbjct: 79 KLLLDQQRDNDVNEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDDDDDDDDDDDDD--- 135
Query: 127 ISMMMHSHMDDEEDDEDYEDEEEYTDDEY 155
D++DD+DY+D++EY +D++
Sbjct: 136 ----------DDDDDDDYDDDDEYDEDDF 154
Score = 33.9 bits (76), Expect = 1.4
Identities = 31/111 (27%), Positives = 47/111 (41%), Gaps = 34/111 (30%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDD--DEEEFNV--------FRMAAMQEAM 104
D DDDDDD D + DD DE++FN F +
Sbjct: 133 DDDDDDDD------------------DDYDDDDEYDEDDFNEATVSWIEWFCQLKQNLFL 174
Query: 105 VSGIDDFGPDDFPGID-----PMDAALISMMM-HSHMDDEEDDEDYEDEEE 149
V +DF D+F I P L+ +++ DD++DD+DY+DE++
Sbjct: 175 VEVDEDFIRDEFNLIGLQTKVPHFKKLLKIILDEDDDDDDDDDDDYDDEDD 225
>gb|AAH05460.1| Nucleolin [Mus musculus]
Length = 707
Score = 40.0 bits (92), Expect = 0.019
Identities = 27/77 (35%), Positives = 36/77 (46%), Gaps = 1/77 (1%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A D DE+E + ++ D+F P G+ P AA + DDE
Sbjct: 137 GKNAKKEDSDEDEDEEDEEDSDEDEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDE 196
Query: 139 -EDDEDYEDEEEYTDDE 154
EDDE+ EDEEE D E
Sbjct: 197 DEDDEEEEDEEEEDDSE 213
Score = 35.4 bits (80), Expect = 0.47
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 8/108 (7%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPD-------DDEEEFNVFRMAAMQEAMVSG 107
D DD+++D F + ++ A A D DDEEE + +E ++
Sbjct: 160 DEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDEDEDDEEEEDEEEEDDSEEEVMEI 219
Query: 108 IDDFGPDDFPGIDPMDAALISMMMHSHMDDEED-DEDYEDEEEYTDDE 154
G + PM A ++ +DE+D DED E+E++ DDE
Sbjct: 220 TTAKGKKTPAKVVPMKAKSVAEEEDEEEEDEDDEDEDDEEEDDEDDDE 267
>gb|EAL22799.1| hypothetical protein CNBB0200 [Cryptococcus neoformans var.
neoformans B-3501A]
Length = 777
Score = 39.7 bits (91), Expect = 0.025
Identities = 29/93 (31%), Positives = 42/93 (44%), Gaps = 9/93 (9%)
Query: 5 LDVLGEVDLSSGEFYSIMRDSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGF 64
L+ E+ SGE M + + E MD FE DS GSG D DDDD++G
Sbjct: 203 LEEFREMLKESGEDVDDMTEEELMERMDEFEGDESDDSEESGSG---SDEDEDDDDEEGD 259
Query: 65 RVSRMAAM------QEAMAIGMDAFGPDDDEEE 91
+ A E +++G D ++DE+E
Sbjct: 260 EDDELLASASEGENDEEVSVGDDEDEDEEDEDE 292
>dbj|BAB27464.1| unnamed protein product [Mus musculus]
Length = 444
Score = 39.7 bits (91), Expect = 0.025
Identities = 22/75 (29%), Positives = 37/75 (49%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A + DE+E + ++ G D+F P G+ P AA + DDE
Sbjct: 137 GKNAKKEESDEDEDEEDEDDSDEDEDDEGEDEFEPPIVKGVKPAKAAPAAPASEDEGDDE 196
Query: 139 EDDEDYEDEEEYTDD 153
++D++ +D+EE DD
Sbjct: 197 DEDDEEDDDEEEEDD 211
Score = 34.7 bits (78), Expect = 0.80
Identities = 28/107 (26%), Positives = 46/107 (42%), Gaps = 11/107 (10%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPD-------DDEEEFNVFRMAAMQEAMVSG 107
D DD+ +D F + ++ A A D DDEE+ + +E ++
Sbjct: 160 DEDDEGEDEFEPPIVKGVKPAKAAPAAPASEDEGDDEDEDDEEDDDEEEEDDSEEEVMEI 219
Query: 108 IDDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G + PM A ++ DDEE+DED EDE++ +D+
Sbjct: 220 TTAKGKKTPAKVVPMKAKSVA----EEEDDEEEDEDDEDEDDEEEDD 262
>gb|AAH85751.1| Nucleolin [Rattus norvegicus]
Length = 714
Score = 38.9 bits (89), Expect = 0.042
Identities = 30/105 (28%), Positives = 48/105 (45%), Gaps = 6/105 (5%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
D D+D++D F + ++ A A D+DEE+ + + DD +
Sbjct: 158 DEDEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDEEDDDDDDEEEEEEDD-SEE 216
Query: 115 DFPGIDPMD-----AALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
+ I P A ++ + S ++EEDDED EDEEE D+E
Sbjct: 217 EVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEE 261
Score = 31.6 bits (70), Expect = 6.7
Identities = 29/133 (21%), Positives = 51/133 (37%), Gaps = 24/133 (18%)
Query: 24 DSDFFEGMDPFEMMALSDSLAGGSGPCLGPFDSDDDDDDGFRVSRMAAMQEAMAIGMDAF 83
D D + +P + + + A + P D +DDDD+
Sbjct: 160 DEDEEDEFEPPVVKGVKPAKAAPAAPASEDEDEEDDDDE--------------------- 198
Query: 84 GPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE--EDD 141
DDD+++ +E ++ G + P+ A ++ DDE E+D
Sbjct: 199 -EDDDDDDEEEEEEDDSEEEVMEITPAKGKKTPAKVVPVKAKSVAEEEEDDEDDEDEEED 257
Query: 142 EDYEDEEEYTDDE 154
ED EDEE+ D++
Sbjct: 258 EDEEDEEDDEDED 270
>ref|XP_583901.1| PREDICTED: similar to nucleolin-related protein, partial [Bos
taurus]
Length = 675
Score = 38.9 bits (89), Expect = 0.042
Identities = 28/108 (25%), Positives = 48/108 (43%), Gaps = 8/108 (7%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPD 114
DS+++DDD A + A A G D+D+E+ + +E D+ +
Sbjct: 107 DSEEEDDDDEEEDEPAVRKAAAAFGAAPATDDEDDEDDDDDDDEEEEEDEEDEEDEDDSE 166
Query: 115 DFP--------GIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
+ P P+ A + + +DEEDD+D +DEEE +D+
Sbjct: 167 EEPMEIVPAKGKKAPVKAVPVKAKSTAEDEDEEDDDDDDDEEEEEEDD 214
Score = 32.7 bits (73), Expect = 3.0
Identities = 24/76 (31%), Positives = 35/76 (45%), Gaps = 10/76 (13%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A D DEE+ ++ DD +D P + AA + DDE
Sbjct: 92 GKNAKKEDSDEED---------EDDSEEEDDDDEEEDEPAVRKAAAAFGAAPATDDEDDE 142
Query: 139 EDDEDYEDEEEYTDDE 154
+DD+D +DEEE D+E
Sbjct: 143 DDDDD-DDEEEEEDEE 157
>gb|EAA56903.1| hypothetical protein MG07258.4 [Magnaporthe grisea 70-15]
gi|39971885|ref|XP_367333.1| hypothetical protein
MG07258.4 [Magnaporthe grisea 70-15]
Length = 755
Score = 38.9 bits (89), Expect = 0.042
Identities = 29/93 (31%), Positives = 44/93 (47%), Gaps = 18/93 (19%)
Query: 61 DDGFRVSRMAAMQEAMAIGMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGID 120
+DGF V + +E+ A G A G DDDE VSG+ + + GI
Sbjct: 164 EDGFEVGSDDSEEESDASG-SASGSDDDE---------------VSGVSEDESELENGIQ 207
Query: 121 PMDAALISMMMHSHMDDEEDDEDYEDEEEYTDD 153
D+ + M+D D++DYE+EE+Y +D
Sbjct: 208 GSDSE--QNVTDGDMEDMGDEDDYEEEEDYEED 238
>emb|CAA30538.1| nucleolin [Mus musculus] gi|128843|sp|P09405|NUCL_MOUSE Nucleolin
(Protein C23) gi|12802527|gb|AAK07920.1| nucleolin [Mus
musculus] gi|26350437|dbj|BAC38858.1| unnamed protein
product [Mus musculus]
Length = 707
Score = 38.5 bits (88), Expect = 0.055
Identities = 22/75 (29%), Positives = 36/75 (47%)
Query: 79 GMDAFGPDDDEEEFNVFRMAAMQEAMVSGIDDFGPDDFPGIDPMDAALISMMMHSHMDDE 138
G +A D DE+E + ++ D+F P G+ P AA + DDE
Sbjct: 137 GKNAKKEDSDEDEDEEDEDDSDEDEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDE 196
Query: 139 EDDEDYEDEEEYTDD 153
++D++ +D+EE DD
Sbjct: 197 DEDDEEDDDEEEEDD 211
Score = 35.8 bits (81), Expect = 0.36
Identities = 28/107 (26%), Positives = 47/107 (43%), Gaps = 11/107 (10%)
Query: 55 DSDDDDDDGFRVSRMAAMQEAMAIGMDAFGPD-------DDEEEFNVFRMAAMQEAMVSG 107
D DD+++D F + ++ A A D DDEE+ + +E ++
Sbjct: 160 DEDDEEEDEFEPPIVKGVKPAKAAPAAPASEDEEDDEDEDDEEDDDEEEEDDSEEEVMEI 219
Query: 108 IDDFGPDDFPGIDPMDAALISMMMHSHMDDEEDDEDYEDEEEYTDDE 154
G + PM A ++ DDEE+DED EDE++ +D+
Sbjct: 220 TTAKGKKTPAKVVPMKAKSVA----EEEDDEEEDEDDEDEDDEEEDD 262
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 296,961,976
Number of Sequences: 2540612
Number of extensions: 13454838
Number of successful extensions: 153733
Number of sequences better than 10.0: 749
Number of HSP's better than 10.0 without gapping: 311
Number of HSP's successfully gapped in prelim test: 463
Number of HSP's that attempted gapping in prelim test: 137600
Number of HSP's gapped (non-prelim): 6106
length of query: 155
length of database: 863,360,394
effective HSP length: 117
effective length of query: 38
effective length of database: 566,108,790
effective search space: 21512134020
effective search space used: 21512134020
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146566.1


