

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146563.10 + phase: 0
(231 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
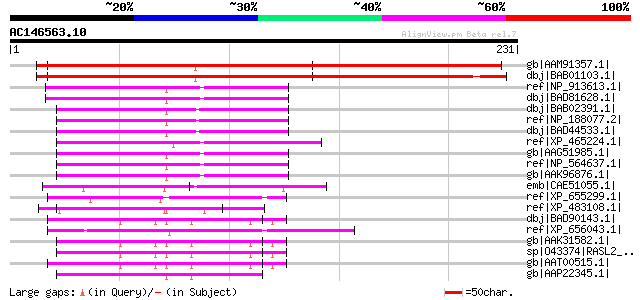
gb|AAM91357.1| At3g18370/MYF24_8 [Arabidopsis thaliana] gi|15983... 280 2e-74
dbj|BAB01103.1| unnamed protein product [Arabidopsis thaliana] 266 3e-70
ref|NP_913613.1| B1066G12.11 [Oryza sativa (japonica cultivar-gr... 81 2e-14
dbj|BAD81628.1| C2 domain-containing protein-like [Oryza sativa ... 81 2e-14
dbj|BAB02391.1| unnamed protein product [Arabidopsis thaliana] 78 2e-13
ref|NP_188077.2| C2 domain-containing protein [Arabidopsis thali... 78 2e-13
dbj|BAD44533.1| hypothetical protein [Arabidopsis thaliana] 78 2e-13
ref|XP_465224.1| C2 domain-containing protein-like [Oryza sativa... 77 4e-13
gb|AAG51985.1| hypothetical protein; 75132-72058 [Arabidopsis th... 74 3e-12
ref|NP_564637.1| C2 domain-containing protein [Arabidopsis thali... 74 3e-12
gb|AAK96876.1| Unknown protein [Arabidopsis thaliana] 74 3e-12
emb|CAE51055.1| novel protein similar to mouse and human membran... 69 1e-10
ref|XP_655299.1| C2 domain protein, putative [Entamoeba histolyt... 67 4e-10
ref|XP_483108.1| C2 domain/GRAM domain-containing protein-like [... 66 7e-10
dbj|BAD90143.1| mKIAA0538 protein [Mus musculus] 66 7e-10
ref|XP_656043.1| C2 domain protein, putative [Entamoeba histolyt... 66 7e-10
gb|AAK31582.1| calcium-promoted Ras inactivator [Homo sapiens] 64 3e-09
sp|O43374|RASL2_HUMAN Ras GTPase-activating protein 4 (RasGAP-ac... 64 3e-09
gb|AAT00515.1| Ca2+ promoted Ras inactivator [Mus musculus] gi|5... 64 3e-09
gb|AAP22345.1| unknown [Homo sapiens] 64 3e-09
>gb|AAM91357.1| At3g18370/MYF24_8 [Arabidopsis thaliana] gi|15983787|gb|AAL10490.1|
AT3g18370/MYF24_8 [Arabidopsis thaliana]
gi|18401863|ref|NP_566607.1| C2 domain-containing
protein [Arabidopsis thaliana]
Length = 815
Score = 280 bits (716), Expect = 2e-74
Identities = 138/212 (65%), Positives = 171/212 (80%)
Query: 13 SGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD 72
S G IELVL+E RDLVAAD+RGTSDPYVRV YG K+RTKVIYKTL P+WNQT+EFPDD
Sbjct: 602 SSKGLIELVLVEARDLVAADIRGTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDD 661
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPE 132
GS L L+VKD+N LLPTSSIG CVVEYQ L PN+ ADKWI LQGVK GE+H+++TRKV E
Sbjct: 662 GSSLELHVKDYNTLLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRVTRKVTE 721
Query: 133 MQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQEGYV 192
+Q+R S +K + Q+KQ+MIKF++ I+DG+LEGL+ L ELE+LED QE Y+
Sbjct: 722 IQRRASAGPGTPFNKALLLSNQMKQVMIKFQNLIDDGDLEGLAEALEELESLEDEQEQYL 781
Query: 193 AQLETEQMLLLSKIKELGQEIINSSPSPSLSR 224
QL+TEQ LL++KIK+LG+EI+NSSP+ + SR
Sbjct: 782 LQLQTEQSLLINKIKDLGKEILNSSPAQAPSR 813
Score = 57.4 bits (137), Expect = 3e-07
Identities = 37/122 (30%), Positives = 61/122 (49%), Gaps = 3/122 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
I + ++ G++LV+ D G D V++ YG ++TK++ WNQ EF +
Sbjct: 483 IIVTVLAGKNLVSKDKSGKCDASVKLQYGKIIQKTKIV-NAAECVWNQKFEFEELAGEEY 541
Query: 78 LYVKDH-NALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQKR 136
L VK + +L T +IG + Q + ++M W+PL+ V GEI + I PE +
Sbjct: 542 LKVKCYREEMLGTDNIGTATLSLQGINNSEM-HIWVPLEDVNSGEIELLIEALDPEYSEA 600
Query: 137 QS 138
S
Sbjct: 601 DS 602
>dbj|BAB01103.1| unnamed protein product [Arabidopsis thaliana]
Length = 786
Score = 266 bits (680), Expect = 3e-70
Identities = 134/214 (62%), Positives = 165/214 (76%), Gaps = 3/214 (1%)
Query: 13 SGNGWIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDD 72
S G IELVL+E RDLVAAD+RGTSDPYVRV YG K+RTKVIYKTL P+WNQT+EFPDD
Sbjct: 572 SSKGLIELVLVEARDLVAADIRGTSDPYVRVQYGEKKQRTKVIYKTLQPKWNQTMEFPDD 631
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPE 132
GS L L+VKD+N LLPTSSIG CVVEYQ L PN+ ADKWI LQGVK GE+H+++TRKV E
Sbjct: 632 GSSLELHVKDYNTLLPTSSIGNCVVEYQGLKPNETADKWIILQGVKHGEVHVRVTRKVTE 691
Query: 133 MQKRQSMDSEPSLSKLHQIPTQIKQMMIKFRSQIEDGNLEGLSTTLSELETLEDTQEGYV 192
+Q+R S +K + Q+KQ+MIKF++ I+DG+LEGL+ L ELE+LED QE Y+
Sbjct: 692 IQRRASAGPGTPFNKALLLSNQMKQVMIKFQNLIDDGDLEGLAEALEELESLEDEQEQYL 751
Query: 193 AQLETEQMLLLSKIKELGQEIINSSPSPSLSRRI 226
QL+TEQ LL++KIK+LG S S +RR+
Sbjct: 752 LQLQTEQSLLINKIKDLGS---GSGTGSSYNRRL 782
Score = 57.4 bits (137), Expect = 3e-07
Identities = 37/122 (30%), Positives = 61/122 (49%), Gaps = 3/122 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDGSPLM 77
I + ++ G++LV+ D G D V++ YG ++TK++ WNQ EF +
Sbjct: 453 IIVTVLAGKNLVSKDKSGKCDASVKLQYGKIIQKTKIV-NAAECVWNQKFEFEELAGEEY 511
Query: 78 LYVKDH-NALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQKR 136
L VK + +L T +IG + Q + ++M W+PL+ V GEI + I PE +
Sbjct: 512 LKVKCYREEMLGTDNIGTATLSLQGINNSEM-HIWVPLEDVNSGEIELLIEALDPEYSEA 570
Query: 137 QS 138
S
Sbjct: 571 DS 572
>ref|NP_913613.1| B1066G12.11 [Oryza sativa (japonica cultivar-group)]
Length = 852
Score = 81.3 bits (199), Expect = 2e-14
Identities = 41/115 (35%), Positives = 68/115 (58%), Gaps = 5/115 (4%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DD 72
+++L ++EG D+ +D+ G SDPYV+ G FK +T++ KTL+P+W + + P +
Sbjct: 464 YVKLEILEGSDMKPSDMNGLSDPYVKGRLGPFKFQTQIQKKTLSPKWFEEFKIPITSWES 523
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ L + V D + + S+G C ++ L Q DKWI L+ VK+G IH+ IT
Sbjct: 524 LNELAMEVCDKDHMF-DDSLGTCTIDIHELRGGQRHDKWISLKNVKKGRIHLAIT 577
>dbj|BAD81628.1| C2 domain-containing protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 674
Score = 81.3 bits (199), Expect = 2e-14
Identities = 41/115 (35%), Positives = 68/115 (58%), Gaps = 5/115 (4%)
Query: 17 WIELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DD 72
+++L ++EG D+ +D+ G SDPYV+ G FK +T++ KTL+P+W + + P +
Sbjct: 286 YVKLEILEGSDMKPSDMNGLSDPYVKGRLGPFKFQTQIQKKTLSPKWFEEFKIPITSWES 345
Query: 73 GSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ L + V D + + S+G C ++ L Q DKWI L+ VK+G IH+ IT
Sbjct: 346 LNELAMEVCDKDHMF-DDSLGTCTIDIHELRGGQRHDKWISLKNVKKGRIHLAIT 399
>dbj|BAB02391.1| unnamed protein product [Arabidopsis thaliana]
Length = 660
Score = 78.2 bits (191), Expect = 2e-13
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
++E D+ +DL G +DPYV+ G ++ +TK+++KTL P+W + + P D + L
Sbjct: 212 VVEACDVKPSDLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSANILN 271
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V+D + S+G+C V Q D W+PLQ +K G +H+ IT
Sbjct: 272 IEVQDKDR-FSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAIT 320
>ref|NP_188077.2| C2 domain-containing protein [Arabidopsis thaliana]
Length = 737
Score = 78.2 bits (191), Expect = 2e-13
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
++E D+ +DL G +DPYV+ G ++ +TK+++KTL P+W + + P D + L
Sbjct: 289 VVEACDVKPSDLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSANILN 348
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V+D + S+G+C V Q D W+PLQ +K G +H+ IT
Sbjct: 349 IEVQDKDR-FSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAIT 397
>dbj|BAD44533.1| hypothetical protein [Arabidopsis thaliana]
Length = 692
Score = 78.2 bits (191), Expect = 2e-13
Identities = 37/110 (33%), Positives = 62/110 (55%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
++E D+ +DL G +DPYV+ G ++ +TK+++KTL P+W + + P D + L
Sbjct: 244 VVEACDVKPSDLNGLADPYVKGQLGAYRFKTKILWKTLAPKWQEEFKIPICTWDSANILN 303
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V+D + S+G+C V Q D W+PLQ +K G +H+ IT
Sbjct: 304 IEVQDKDR-FSDDSLGDCSVNIAEFRGGQRNDMWLPLQNIKMGRLHLAIT 352
>ref|XP_465224.1| C2 domain-containing protein-like [Oryza sativa (japonica
cultivar-group)] gi|46391036|dbj|BAD15979.1| C2
domain-containing protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 718
Score = 77.0 bits (188), Expect = 4e-13
Identities = 41/125 (32%), Positives = 68/125 (53%), Gaps = 5/125 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPDDG----SPLM 77
++EG D+ +D G +DPYV+ H G ++ +TK+ KTL P+W + + P + L
Sbjct: 291 ILEGADMKPSDPNGLADPYVKGHLGPYRFQTKIHKKTLNPKWMEEFKIPVTSWAALNLLS 350
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQKRQ 137
L V+D + + ++G+C + +L Q D WI L+ +K G IHI +T E +K
Sbjct: 351 LQVRDKDPIF-DDTLGDCSISINKLRGGQRHDIWIALKNIKTGRIHIAVTVLEDENEKVP 409
Query: 138 SMDSE 142
+ D E
Sbjct: 410 NDDDE 414
>gb|AAG51985.1| hypothetical protein; 75132-72058 [Arabidopsis thaliana]
gi|25405650|pir||A96576 hypothetical protein F22G10.28
[imported] - Arabidopsis thaliana
Length = 706
Score = 73.9 bits (180), Expect = 3e-12
Identities = 38/110 (34%), Positives = 61/110 (54%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
+ E DL +DL G +DPYV+ G ++ +TK+ KTL+P+W++ + P D S L
Sbjct: 243 VFEASDLKPSDLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPSILN 302
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V D + + ++GEC V + Q D W+ LQ +K G +H+ IT
Sbjct: 303 IEVGDKDRFV-DDTLGECSVNIEEFRGGQRNDMWLSLQNIKMGRLHLAIT 351
>ref|NP_564637.1| C2 domain-containing protein [Arabidopsis thaliana]
Length = 751
Score = 73.9 bits (180), Expect = 3e-12
Identities = 38/110 (34%), Positives = 61/110 (54%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
+ E DL +DL G +DPYV+ G ++ +TK+ KTL+P+W++ + P D S L
Sbjct: 288 VFEASDLKPSDLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPSILN 347
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V D + + ++GEC V + Q D W+ LQ +K G +H+ IT
Sbjct: 348 IEVGDKDRFV-DDTLGECSVNIEEFRGGQRNDMWLSLQNIKMGRLHLAIT 396
>gb|AAK96876.1| Unknown protein [Arabidopsis thaliana]
Length = 751
Score = 73.9 bits (180), Expect = 3e-12
Identities = 38/110 (34%), Positives = 61/110 (54%), Gaps = 5/110 (4%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP----DDGSPLM 77
+ E DL +DL G +DPYV+ G ++ +TK+ KTL+P+W++ + P D S L
Sbjct: 288 VFEASDLKPSDLNGLADPYVKGKLGAYRFKTKIQKKTLSPKWHEEFKIPIFTWDSPSILN 347
Query: 78 LYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQIT 127
+ V D + + ++GEC V + Q D W+ LQ +K G +H+ IT
Sbjct: 348 IEVGDKDRFV-DDTLGECSVNIEEFRGGQRNDMWLSLQNIKMGRLHLAIT 396
>emb|CAE51055.1| novel protein similar to mouse and human membrane bound C2 domain
containing protein (MBC2) [Danio rerio]
Length = 1076
Score = 68.9 bits (167), Expect = 1e-10
Identities = 42/143 (29%), Positives = 71/143 (49%), Gaps = 15/143 (10%)
Query: 16 GWIELVLIEGRDLVAAD------LRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G + L+L+E +DLVA D ++G SDPYV++H G+ ++ VI + L P WN+ E
Sbjct: 810 GVLRLILLEAQDLVAKDGLMGGMVKGKSDPYVKIHIGDTTFKSHVIKENLNPTWNEMYEL 869
Query: 70 ---PDDGSPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIH--- 123
S +++ V D + + +G + Q + +Q+ D+W L VK G +H
Sbjct: 870 VLTSSSSSEVLVEVFDKD-MDKDDFLGRMKISLQEIIQSQITDRWFSLSDVKHGRVHLIL 928
Query: 124 --IQITRKVPEMQKRQSMDSEPS 144
+ K +QK + S+ S
Sbjct: 929 EWLNTVTKPDPLQKAVQLQSDHS 951
Score = 50.8 bits (120), Expect = 3e-05
Identities = 26/73 (35%), Positives = 41/73 (55%), Gaps = 6/73 (8%)
Query: 16 GWIELVLIEGRDLVAAD------LRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF 69
G + + L+EG++LVA D ++G SDPYV++ G ++ VI + L P WN+ E
Sbjct: 582 GLLRIHLVEGQNLVAKDNLMGGMVKGKSDPYVKIQIGGETFKSHVIKENLNPTWNEMYEL 641
Query: 70 PDDGSPLMLYVKD 82
P L ++ KD
Sbjct: 642 PGQELTLEVFDKD 654
Score = 42.0 bits (97), Expect = 0.014
Identities = 37/159 (23%), Positives = 73/159 (45%), Gaps = 30/159 (18%)
Query: 16 GWIELVLIEGRDLVAAD------LRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTL-- 67
G + + L+E +L A D + G SDPY V G ++ + TL+P+W +
Sbjct: 314 GVVRIHLLEADNLAAKDNYVKGVMAGMSDPYAIVRVGPQTFKSHHLDNTLSPKWGEVYEV 373
Query: 68 ---EFPDDGSPLMLYVK--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEI 122
E P + ++ K DH+ L + + +V+ +++ D+W L+ + G +
Sbjct: 374 VVHEVPGQELEVEVFDKDPDHDDFLGRTKLDLGIVK-----KSKIVDEWFNLKDTQTGRV 428
Query: 123 HIQI--------TRKVPEMQKRQSMDSEPSLSKLHQIPT 153
H+++ T ++ E+ KR +E +SK + P+
Sbjct: 429 HLKLEWLTLETHTERLKEVLKR----NESVVSKAAEPPS 463
>ref|XP_655299.1| C2 domain protein, putative [Entamoeba histolytica HM-1:IMSS]
gi|56472424|gb|EAL49910.1| C2 domain protein, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 188
Score = 67.0 bits (162), Expect = 4e-10
Identities = 42/113 (37%), Positives = 61/113 (53%), Gaps = 8/113 (7%)
Query: 18 IELVLIEGRDLVAADLRG-TSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTL---EFPDDG 73
IEL +IEGR+L AD G +SDPY ++ ++T+ +YKT P WN++ FP G
Sbjct: 3 IELKVIEGRNLKGADFGGLSSDPYCKIITRQCTQQTQTVYKTRNPSWNKSFIMDVFP--G 60
Query: 74 SPLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQI 126
+ V D++ S+G + P Q+ D W+ L K+GEIHIQI
Sbjct: 61 EDIKFEVYDYDTFGKNDSLGSTHYKVLNGFPGQVVDTWLGLS--KKGEIHIQI 111
>ref|XP_483108.1| C2 domain/GRAM domain-containing protein-like [Oryza sativa
(japonica cultivar-group)] gi|42408774|dbj|BAD10009.1|
C2 domain/GRAM domain-containing protein-like [Oryza
sativa (japonica cultivar-group)]
Length = 1081
Score = 66.2 bits (160), Expect = 7e-10
Identities = 39/108 (36%), Positives = 57/108 (52%), Gaps = 5/108 (4%)
Query: 14 GNGWIELV-LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEF--- 69
G+GW+ V LIEG L D G SDPYV K + + ++TL PQWN EF
Sbjct: 585 GDGWLLTVALIEGTKLAPVDATGFSDPYVVFTCNGKSKTSSIKFQTLEPQWNDIFEFDAM 644
Query: 70 PDDGSPLMLYVKDHNALL-PTSSIGECVVEYQRLPPNQMADKWIPLQG 116
D S + ++V D + +S+G + + + +++AD WIPLQG
Sbjct: 645 DDPPSVMNVHVYDFDGPFDEVTSLGHAEINFVKSNLSELADVWIPLQG 692
Score = 52.4 bits (124), Expect = 1e-05
Identities = 30/78 (38%), Positives = 42/78 (53%), Gaps = 2/78 (2%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP--DDGSPLMLY 79
+IE R+L A D G SDPYV++ G + +TKV+ K L P W+Q F D L LY
Sbjct: 7 VIEARNLRAMDSNGFSDPYVKLQLGKQRFKTKVVKKNLNPAWDQEFSFSVGDVRDVLKLY 66
Query: 80 VKDHNALLPTSSIGECVV 97
V D + + +G+ V
Sbjct: 67 VYDEDMIGIDDFLGQVKV 84
>dbj|BAD90143.1| mKIAA0538 protein [Mus musculus]
Length = 826
Score = 66.2 bits (160), Expect = 7e-10
Identities = 36/101 (35%), Positives = 54/101 (52%), Gaps = 3/101 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPL 76
+ ++E RDL D G SDP+VRVHY + T V+ K+ P+WN+T +F + G+
Sbjct: 159 LRCAVLEARDLAPKDRNGASDPFVRVHYNGRTQETSVVKKSCYPRWNETFDFELEKGASE 218
Query: 77 MLYVK--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
L V+ D + + +G+ VV QRL Q + W LQ
Sbjct: 219 ALLVEAWDWDLVSQNDFLGKVVVNVQRLCSAQQEEGWFRLQ 259
Score = 48.1 bits (113), Expect = 2e-04
Identities = 35/118 (29%), Positives = 56/118 (46%), Gaps = 9/118 (7%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFK-KRTKVIYKTLTPQWNQ--TLEFPDDGS 74
+ + ++EG++L A D+ G+SDPY V N RT ++KTL P W + + P
Sbjct: 31 LSIRIVEGKNLPAKDITGSSDPYCIVKVDNEPIIRTATVWKTLCPFWGEDYQVHLPPTFH 90
Query: 75 PLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGVK-----RGEIHIQI 126
+ YV D +AL IG+ + L + W L V +GEIH+++
Sbjct: 91 TVAFYVMDEDALSRDDVIGKVCLTRDALASHPKGFSGWTHLVEVDPNEEVQGEIHLRL 148
>ref|XP_656043.1| C2 domain protein, putative [Entamoeba histolytica HM-1:IMSS]
gi|56473220|gb|EAL50659.1| C2 domain protein, putative
[Entamoeba histolytica HM-1:IMSS]
Length = 389
Score = 66.2 bits (160), Expect = 7e-10
Identities = 45/142 (31%), Positives = 69/142 (47%), Gaps = 5/142 (3%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFPD--DGSP 75
I L ++ G+ L A DLR +SDPYV V G +++TK + K L P W + EF + G+
Sbjct: 29 IRLTVVSGKQLKAMDLR-SSDPYVIVSVGIEQRKTKTVMKNLNPTWGDSFEFYNVTPGTM 87
Query: 76 LMLYVKDHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQGVKRGEIHIQITRKVPEMQK 135
V D++ ++G + Q+LPP QMA +PL +G I IQ T
Sbjct: 88 ATFTVMDYDKRGKDDNMGNASLVIQKLPPGQMATNELPLS--TKGSICIQYTIISSSSST 145
Query: 136 RQSMDSEPSLSKLHQIPTQIKQ 157
+ S ++ Q P+ +Q
Sbjct: 146 KTSQQCGTKPTQQQQYPSYPQQ 167
>gb|AAK31582.1| calcium-promoted Ras inactivator [Homo sapiens]
Length = 803
Score = 64.3 bits (155), Expect = 3e-09
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPLMLYV 80
++E RDL D GTSDP+VRV Y + T ++ K+ P+WN+T EF +G+ L V
Sbjct: 139 VLEARDLAPKDRNGTSDPFVRVRYKGRTRETSIVKKSCYPRWNETFEFELQEGAMEALCV 198
Query: 81 K--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
+ D + + +G+ V++ QRL Q + W LQ
Sbjct: 199 EAWDWDLVSRNDFLGKVVIDVQRLRVVQQEEGWFRLQ 235
Score = 47.4 bits (111), Expect = 3e-04
Identities = 34/114 (29%), Positives = 54/114 (46%), Gaps = 9/114 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFK-KRTKVIYKTLTPQWNQ--TLEFPDDGSPLML 78
++EG++L A D+ G+SDPY V N RT ++KTL P W + + P +
Sbjct: 11 IVEGKNLPAKDITGSSDPYCIVKVDNEPIIRTATVWKTLCPFWGEEYQVHLPPTFHAVAF 70
Query: 79 YVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGVK-----RGEIHIQI 126
YV D +AL IG+ + + + W L V +GEIH+++
Sbjct: 71 YVMDEDALSRDDVIGKVCLTRDTIASHPKGFSGWAHLTEVDPDEEVQGEIHLRL 124
>sp|O43374|RASL2_HUMAN Ras GTPase-activating protein 4 (RasGAP-activating-like protein 2)
(Calcium-promoted Ras inactivator)
Length = 803
Score = 64.3 bits (155), Expect = 3e-09
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPLMLYV 80
++E RDL D GTSDP+VRV Y + T ++ K+ P+WN+T EF +G+ L V
Sbjct: 139 VLEARDLAPKDRNGTSDPFVRVRYKGRTRETSIVKKSCYPRWNETFEFELQEGAMEALCV 198
Query: 81 K--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
+ D + + +G+ V++ QRL Q + W LQ
Sbjct: 199 EAWDWDLVSRNDFLGKVVIDVQRLRVVQQEEGWFRLQ 235
Score = 47.4 bits (111), Expect = 3e-04
Identities = 34/114 (29%), Positives = 54/114 (46%), Gaps = 9/114 (7%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFK-KRTKVIYKTLTPQWNQ--TLEFPDDGSPLML 78
++EG++L A D+ G+SDPY V N RT ++KTL P W + + P +
Sbjct: 11 IVEGKNLPAKDITGSSDPYCIVKVDNEPIIRTATVWKTLCPFWGEEYQVHLPPTFHAVAF 70
Query: 79 YVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGVK-----RGEIHIQI 126
YV D +AL IG+ + + + W L V +GEIH+++
Sbjct: 71 YVMDEDALSRDDVIGKVCLTRDTIASHPKGFSGWAHLTEVDPDEEVQGEIHLRL 124
>gb|AAT00515.1| Ca2+ promoted Ras inactivator [Mus musculus]
gi|51711391|ref|XP_355650.2| PREDICTED: Ca2+promoted Ras
inactivator [Mus musculus] gi|34785418|gb|AAH57460.1|
Rasa4 protein [Mus musculus]
Length = 802
Score = 64.3 bits (155), Expect = 3e-09
Identities = 35/101 (34%), Positives = 53/101 (51%), Gaps = 3/101 (2%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPL 76
+ ++E RDL D G SDP+VRVHY + T V+ K+ P+WN+T +F + G+
Sbjct: 135 LRCAVLEARDLAPKDRNGASDPFVRVHYNGRTQETSVVKKSCYPRWNETFDFELEKGASE 194
Query: 77 MLYVK--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
L V+ D + + +G+ V QRL Q + W LQ
Sbjct: 195 ALLVEAWDWDLVSRNDFLGKVAVNVQRLCSAQQEEGWFRLQ 235
Score = 48.1 bits (113), Expect = 2e-04
Identities = 35/118 (29%), Positives = 56/118 (46%), Gaps = 9/118 (7%)
Query: 18 IELVLIEGRDLVAADLRGTSDPYVRVHYGNFK-KRTKVIYKTLTPQWNQ--TLEFPDDGS 74
+ + ++EG++L A D+ G+SDPY V N RT ++KTL P W + + P
Sbjct: 7 LSIRIVEGKNLPAKDITGSSDPYCIVKVDNEPIIRTATVWKTLCPFWGEDYQVHLPPTFH 66
Query: 75 PLMLYVKDHNALLPTSSIGECVVEYQRLPPNQMA-DKWIPLQGVK-----RGEIHIQI 126
+ YV D +AL IG+ + L + W L V +GEIH+++
Sbjct: 67 TVAFYVMDEDALSRDDVIGKVCLTRDALASHPKGFSGWTHLVEVDPNEEVQGEIHLRL 124
>gb|AAP22345.1| unknown [Homo sapiens]
Length = 724
Score = 64.3 bits (155), Expect = 3e-09
Identities = 35/97 (36%), Positives = 54/97 (55%), Gaps = 3/97 (3%)
Query: 22 LIEGRDLVAADLRGTSDPYVRVHYGNFKKRTKVIYKTLTPQWNQTLEFP-DDGSPLMLYV 80
++E RDL D GTSDP+VRV Y + T ++ K+ P+WN+T EF +G+ L V
Sbjct: 162 VLEARDLAPKDRNGTSDPFVRVRYKGRTRETSIVKKSCYPRWNETFEFELQEGAMEALCV 221
Query: 81 K--DHNALLPTSSIGECVVEYQRLPPNQMADKWIPLQ 115
+ D + + +G+ V++ QRL Q + W LQ
Sbjct: 222 EAWDWDLVSRNDFLGKVVIDVQRLRVVQQEEGWFRLQ 258
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.314 0.133 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 390,847,271
Number of Sequences: 2540612
Number of extensions: 15647759
Number of successful extensions: 36727
Number of sequences better than 10.0: 1232
Number of HSP's better than 10.0 without gapping: 458
Number of HSP's successfully gapped in prelim test: 774
Number of HSP's that attempted gapping in prelim test: 34836
Number of HSP's gapped (non-prelim): 2021
length of query: 231
length of database: 863,360,394
effective HSP length: 124
effective length of query: 107
effective length of database: 548,324,506
effective search space: 58670722142
effective search space used: 58670722142
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC146563.10


