

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.5 + phase: 2 /pseudo/partial
(263 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
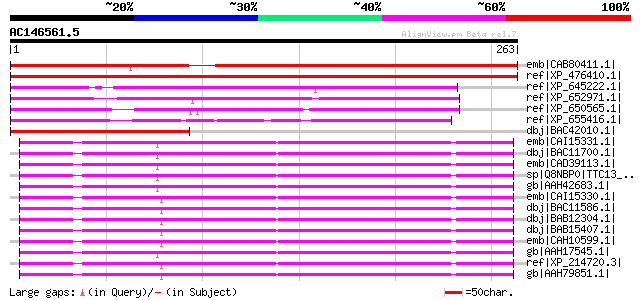
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB80411.1| putative protein [Arabidopsis thaliana] gi|44688... 384 e-105
ref|XP_476410.1| tetratricopeptide repeat(TPR)-containing protei... 349 4e-95
ref|XP_645222.1| hypothetical protein DDB0168736 [Dictyostelium ... 197 3e-49
ref|XP_652971.1| TPR repeat protein [Entamoeba histolytica HM-1:... 164 2e-39
ref|XP_650565.1| TPR repeat protein [Entamoeba histolytica HM-1:... 150 5e-35
ref|XP_655416.1| TPR repeat protein [Entamoeba histolytica HM-1:... 144 2e-33
dbj|BAC42010.1| unknown protein [Arabidopsis thaliana] gi|290290... 138 2e-31
emb|CAI15331.1| tetratricopeptide repeat domain 13 [Homo sapiens... 100 4e-20
dbj|BAC11700.1| unnamed protein product [Homo sapiens] 100 4e-20
emb|CAD39113.1| hypothetical protein [Homo sapiens] 100 4e-20
sp|Q8NBP0|TTC13_HUMAN Tetratricopeptide repeat protein 13 (TPR r... 100 4e-20
gb|AAH42683.1| TTC13 protein [Homo sapiens] 100 4e-20
emb|CAI15330.1| tetratricopeptide repeat domain 13 [Homo sapiens... 100 5e-20
dbj|BAC11586.1| unnamed protein product [Homo sapiens] gi|313777... 100 5e-20
dbj|BAB12304.1| hypothetical protein [Macaca fascicularis] 100 5e-20
dbj|BAB15407.1| unnamed protein product [Homo sapiens] 100 5e-20
emb|CAH10599.1| hypothetical protein [Homo sapiens] 100 5e-20
gb|AAH17545.1| Ttc13 protein [Mus musculus] 100 7e-20
ref|XP_214720.3| PREDICTED: similar to tetratricopeptide repeat ... 99 9e-20
gb|AAH79851.1| Ttc13 protein [Mus musculus] 99 9e-20
>emb|CAB80411.1| putative protein [Arabidopsis thaliana] gi|4468812|emb|CAB38213.1|
putative protein [Arabidopsis thaliana]
gi|7486501|pir||T04740 hypothetical protein F6G17.110 -
Arabidopsis thaliana
Length = 1013
Score = 384 bits (985), Expect = e-105
Identities = 177/265 (66%), Positives = 217/265 (81%), Gaps = 15/265 (5%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
EEFNSGFGSHTPM+LGQAKVVRYFPNYERTL +AK+++K++ V K D++I LS D K+
Sbjct: 761 EEFNSGFGSHTPMVLGQAKVVRYFPNYERTLTLAKSIIKDKLSVRSKKDKVIDLSKDEKI 820
Query: 61 E--EIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTP 118
E +IM A++C +L+ +VGEDFW +TWC+ST EG G+ G+DF+IRTP
Sbjct: 821 EKMQIMRAETCDELHNIVGEDFWVATWCDSTGSEG-------------GRLGYDFSIRTP 867
Query: 119 CTPARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRG 178
CTPARW D+D EM AWEALC AYCGENYGST+ D LE VRDAILRMTYYWYNFMPL+RG
Sbjct: 868 CTPARWSDFDEEMTSAWEALCTAYCGENYGSTELDALETVRDAILRMTYYWYNFMPLARG 927
Query: 179 TAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLKVTTS 238
TA GF V+LGLLLAANMEFT +IP+G Q+DWEAILN++P SFVDSVKSWLYPSLK+ TS
Sbjct: 928 TAVTGFVVLLGLLLAANMEFTETIPKGLQIDWEAILNVEPGSFVDSVKSWLYPSLKINTS 987
Query: 239 WKDYHDVASTFATTGSVVAALSSYD 263
W+D+ +++S F+TTG+VVAALS+Y+
Sbjct: 988 WRDHTEISSAFSTTGAVVAALSTYN 1012
>ref|XP_476410.1| tetratricopeptide repeat(TPR)-containing protein-like [Oryza sativa
(japonica cultivar-group)] gi|34393726|dbj|BAC83208.1|
tetratricopeptide repeat(TPR)-containing protein-like
[Oryza sativa (japonica cultivar-group)]
gi|33147009|dbj|BAC80093.1| tetratricopeptide
repeat(TPR)-containing protein-like [Oryza sativa
(japonica cultivar-group)]
Length = 1011
Score = 349 bits (896), Expect = 4e-95
Identities = 161/263 (61%), Positives = 207/263 (78%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
EEFNSGFGSHTPM+LGQAK++RY+P Y+ L+ AK +M + YV+ D+ I L++ KL
Sbjct: 748 EEFNSGFGSHTPMLLGQAKIIRYYPYYQSVLEAAKNIMLDLKYVNNAEDRAIFLTDIEKL 807
Query: 61 EEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
++I A SCS+LY +VGE +W ST C+S AF+G++LEGTR+T MG+ GFDFAIRTPCT
Sbjct: 808 KKIEVASSCSELYHIVGETYWVSTRCDSIAFQGRRLEGTRITTQNMGKTGFDFAIRTPCT 867
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
P+RWE+YD EM+ AWEA+C AYC + + D D+L+ V+ AILRMTYYWYNFMPLSRG+A
Sbjct: 868 PSRWEEYDEEMSAAWEAICEAYCSDTNPTRDPDMLDAVKAAILRMTYYWYNFMPLSRGSA 927
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLKVTTSWK 240
VG+ V+LGL LAANM+ T SIP G QVDWEAIL+ DP++FVD +K WLYPS+K + + K
Sbjct: 928 VVGYVVLLGLFLAANMDVTASIPHGVQVDWEAILSQDPDTFVDKIKPWLYPSIKTSRNLK 987
Query: 241 DYHDVASTFATTGSVVAALSSYD 263
DY DV+ F+TTGSVVAAL+ D
Sbjct: 988 DYADVSVAFSTTGSVVAALTCVD 1010
>ref|XP_645222.1| hypothetical protein DDB0168736 [Dictyostelium discoideum]
gi|42761489|gb|AAS45307.1| similar to Oryza sativa
(japonica cultivar-group). OJ1513_F02.22 protein
[Dictyostelium discoideum] gi|60473299|gb|EAL71245.1|
hypothetical protein DDB0168736 [Dictyostelium
discoideum]
Length = 1050
Score = 197 bits (500), Expect = 3e-49
Identities = 103/256 (40%), Positives = 153/256 (59%), Gaps = 32/256 (12%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
E+F GFGSHTP+I GQ KVVRY+P +E++ +I K ++ + H +SN L
Sbjct: 771 EQFKEGFGSHTPIITGQCKVVRYYPMFEKSFEIMKLLLPIQ---HPSL-----ISNQTIL 822
Query: 61 EEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+++ A +C DLYK++ +DF+ T C S + E K +EGTR+TL + G++FAIRTP T
Sbjct: 823 DQLESATTCKDLYKLIKKDFFVVTPCYSRSNENKVMEGTRLTLQYIHPEGYEFAIRTPST 882
Query: 121 PARWEDYDAEMAMAWEALCN-AYCGENYGSTDFDVLEN---------------------- 157
P RWE Y E+ + W+ L + + +N ++ D+++N
Sbjct: 883 PYRWEQYSQELDIIWDQLIDKVFIYQNIINSKEDLIDNSSSSSSSSPSPSPSNQESILLD 942
Query: 158 -VRDAILRMTYYWYNFMPLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNL 216
+ D IL +++YWYNFMPLSRG+AA+GF +LGL L+ ++ T SIP+ Q DWEAIL
Sbjct: 943 EISDLILCLSFYWYNFMPLSRGSAALGFTTLLGLFLSLDIAITSSIPKNQQPDWEAILRP 1002
Query: 217 DPNSFVDSVKSWLYPS 232
SF+ +K WLYP+
Sbjct: 1003 TTTSFISVIKPWLYPA 1018
>ref|XP_652971.1| TPR repeat protein [Entamoeba histolytica HM-1:IMSS]
gi|56469881|gb|EAL47585.1| TPR repeat protein [Entamoeba
histolytica HM-1:IMSS]
Length = 917
Score = 164 bits (416), Expect = 2e-39
Identities = 87/240 (36%), Positives = 128/240 (53%), Gaps = 21/240 (8%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
E+F GFGSHTPM +GQ +VVRY+ Y+R +I K ++ E+ + + +
Sbjct: 658 EQFEEGFGSHTPMFIGQTRVVRYYCQYQRAFEIVKKLLPEQLKI-----------KEEWV 706
Query: 61 EEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEG-------KQLEGTRVTLVKMGQHGFDF 113
+++ AK+ D+ ++ DF T C S F+ + EGTR+ + G++F
Sbjct: 707 KKVQTAKNIQDILDIIKSDFHVITPCKSLYFDDPDKPGTKRTYEGTRLVVEIKQPVGYEF 766
Query: 114 AIRTPCTPARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFM 173
AI+TPCT RW+ YD E+ W+ + + + L A+ T+YWYNFM
Sbjct: 767 AIKTPCTMPRWKLYDEELTECWKNIVRLVLTRKGKKGEEEALLR---AMFDFTFYWYNFM 823
Query: 174 PLSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSL 233
PLSRGTAA GF ML +L + N + IP+G Q DWEAIL DP F D + W+ S+
Sbjct: 824 PLSRGTAATGFVFMLAMLASLNYQINKHIPKGMQTDWEAILCDDPKKFEDVMMGWMKDSI 883
>ref|XP_650565.1| TPR repeat protein [Entamoeba histolytica HM-1:IMSS]
gi|56467204|gb|EAL45179.1| TPR repeat protein [Entamoeba
histolytica HM-1:IMSS]
Length = 931
Score = 150 bits (378), Expect = 5e-35
Identities = 83/239 (34%), Positives = 128/239 (52%), Gaps = 20/239 (8%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
E+F GFGSHTPM GQ +V+RYF Y+R I + ++ ++ + + +++
Sbjct: 673 EQFEEGFGSHTPMYTGQGRVIRYFCQYDRAFKIVQELLPKQVQLKPEWVKVVK------- 725
Query: 61 EEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFE----GKQL--EGTRVTLVKMGQHGFDFA 114
AK+ D+ +VV +F +T C S F+ G+++ EGT + G+DFA
Sbjct: 726 ----EAKNIGDILRVVESNFQVTTPCRSDNFDDPKTGERIIYEGTNLVTEMKEPVGYDFA 781
Query: 115 IRTPCTPARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMP 174
I+TPC +RW YD E+ W+ L ++ E + A+ T++WYNFMP
Sbjct: 782 IKTPCKKSRWILYDKELEACWKKLALLILTRKGEKSEE---EKILRAMFSFTFFWYNFMP 838
Query: 175 LSRGTAAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSL 233
L+RGTAA GF M+ +L + + IPQG Q DWEAIL P+ F D + +W+ S+
Sbjct: 839 LTRGTAAGGFVFMMSVLASIGKQIGKHIPQGMQTDWEAILCEYPHKFSDVMYNWMKDSI 897
>ref|XP_655416.1| TPR repeat protein [Entamoeba histolytica HM-1:IMSS]
gi|56472551|gb|EAL50030.1| TPR repeat protein [Entamoeba
histolytica HM-1:IMSS]
Length = 890
Score = 144 bits (364), Expect = 2e-33
Identities = 83/230 (36%), Positives = 131/230 (56%), Gaps = 23/230 (10%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
++F GFGS+T + Q +V RY+P + RT + K + +E S + Q++
Sbjct: 646 QQFEEGFGSNTTLKTAQTRVARYWPMFPRTFGLFKKLFEEESGASKEKKQLV-------- 697
Query: 61 EEIMHAKSCSDLYKVVGED-FWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPC 119
A++ ++ KV+ D F+ +T C + EG +LEGT + + K+ + G F+I+TPC
Sbjct: 698 ---AKAETVDEVQKVLKRDNFFMTTICRNE--EGTRLEGTWLLIEKV-ELGSMFSIKTPC 751
Query: 120 TPARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGT 179
TP R++ +D E+ +AL Y ++ D + DA+LR Y W+NFMPL+RGT
Sbjct: 752 TPKRYKIFDVELK---KALDKYYTVQHSNDQD-----KILDAVLRFCYLWFNFMPLTRGT 803
Query: 180 AAVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWL 229
A G M GLLL+A + G IP+GFQ DWEA+L + N ++ +KSW+
Sbjct: 804 AGCGMYWMFGLLLSAGFKQVGVIPKGFQCDWEALLTDNQNDYITKMKSWI 853
>dbj|BAC42010.1| unknown protein [Arabidopsis thaliana] gi|29029044|gb|AAO64901.1|
At4g37460 [Arabidopsis thaliana]
gi|30690956|ref|NP_195462.2| tetratricopeptide repeat
(TPR)-containing protein [Arabidopsis thaliana]
Length = 883
Score = 138 bits (347), Expect = 2e-31
Identities = 60/93 (64%), Positives = 78/93 (83%)
Query: 1 EEFNSGFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYVHGKTDQIIHLSNDGKL 60
EEFNSGFGSHTPM+LGQAKVVRYFPNYERTL +AK+++K++ V K D++I LS D K+
Sbjct: 788 EEFNSGFGSHTPMVLGQAKVVRYFPNYERTLTLAKSIIKDKLSVRSKKDKVIDLSKDEKI 847
Query: 61 EEIMHAKSCSDLYKVVGEDFWSSTWCNSTAFEG 93
E+IM A++C +L+ +VGEDFW +TWC+ST EG
Sbjct: 848 EKIMRAETCDELHNIVGEDFWVATWCDSTGSEG 880
>emb|CAI15331.1| tetratricopeptide repeat domain 13 [Homo sapiens]
gi|55960678|emb|CAI17216.1| tetratricopeptide repeat
domain 13 [Homo sapiens]
Length = 806
Score = 100 bits (249), Expect = 4e-20
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 543 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 598
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 599 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 658
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 659 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 717
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 718 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 775
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 776 TLPSVSETFPTLRSMIEVLNT 796
>dbj|BAC11700.1| unnamed protein product [Homo sapiens]
Length = 806
Score = 100 bits (249), Expect = 4e-20
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 543 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 598
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 599 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 658
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 659 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 717
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 718 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 775
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 776 TLPSVSETFPTLRSMIEVLNT 796
>emb|CAD39113.1| hypothetical protein [Homo sapiens]
Length = 529
Score = 100 bits (249), Expect = 4e-20
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 266 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 321
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 322 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 381
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 382 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 440
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 441 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 498
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 499 TLPSVSETFPTLRSMIEVLNT 519
>sp|Q8NBP0|TTC13_HUMAN Tetratricopeptide repeat protein 13 (TPR repeat protein 13)
Length = 859
Score = 100 bits (249), Expect = 4e-20
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 596 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 651
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 652 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 711
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 712 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 770
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 771 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 828
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 829 TLPSVSETFPTLRSMIEVLNT 849
>gb|AAH42683.1| TTC13 protein [Homo sapiens]
Length = 502
Score = 100 bits (249), Expect = 4e-20
Identities = 77/261 (29%), Positives = 131/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 239 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 294
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 295 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 354
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 355 ERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 413
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 414 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSYK 471
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 472 TLPSVSETFPTLRSMIEVLNT 492
>emb|CAI15330.1| tetratricopeptide repeat domain 13 [Homo sapiens]
gi|55960679|emb|CAI17217.1| tetratricopeptide repeat
domain 13 [Homo sapiens]
Length = 860
Score = 100 bits (248), Expect = 5e-20
Identities = 77/262 (29%), Positives = 131/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 596 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 651
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 652 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 711
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 712 EERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSS 770
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 771 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSY 828
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 829 KTLPSVSETFPTLRSMIEVLNT 850
>dbj|BAC11586.1| unnamed protein product [Homo sapiens] gi|31377703|ref|NP_078801.2|
tetratricopeptide repeat domain 13 [Homo sapiens]
Length = 860
Score = 100 bits (248), Expect = 5e-20
Identities = 77/262 (29%), Positives = 131/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 596 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 651
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 652 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 711
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 712 EERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSS 770
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 771 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSY 828
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 829 KTLPSVSETFPTLRSMIEVLNT 850
>dbj|BAB12304.1| hypothetical protein [Macaca fascicularis]
Length = 502
Score = 100 bits (248), Expect = 5e-20
Identities = 77/262 (29%), Positives = 131/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 238 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 293
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 294 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 353
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 354 EERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSS 412
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 413 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSY 470
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 471 KTLPSVSETFPTLRSMIEVLNT 492
>dbj|BAB15407.1| unnamed protein product [Homo sapiens]
Length = 308
Score = 100 bits (248), Expect = 5e-20
Identities = 77/262 (29%), Positives = 131/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 44 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 99
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 100 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 159
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 160 EERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSS 218
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 219 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSY 276
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 277 KTLPSVSETFPTLRSMIEVLNT 298
>emb|CAH10599.1| hypothetical protein [Homo sapiens]
Length = 785
Score = 100 bits (248), Expect = 5e-20
Identities = 77/262 (29%), Positives = 131/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 521 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 576
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 577 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 636
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S++F + V + IL + YY+YN MPLSRG++
Sbjct: 637 EERTQLYHAEIDALYKDL-TAKGKVLILSSEFGEADAVCNLILSLVYYFYNLMPLSRGSS 695
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 696 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKVAKSWM--NLKSISPSY 753
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 754 KTLPSVSETFPTLRSMIEVLNT 775
>gb|AAH17545.1| Ttc13 protein [Mus musculus]
Length = 307
Score = 99.8 bits (247), Expect = 7e-20
Identities = 77/261 (29%), Positives = 130/261 (49%), Gaps = 12/261 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 44 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 99
Query: 65 HAKSCSDLYKV---VGEDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCTP 121
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 100 KVEDLLPIMKFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTTE 159
Query: 122 ARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTAA 181
R + Y AE+ ++ L A S +F + V + IL + YY+YN MPLSRG++
Sbjct: 160 ERTQLYHAEIDALYKDL-TAKGKVLTLSAEFGEADAVCNLILSLVYYFYNLMPLSRGSSV 218
Query: 182 VGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSWK 240
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+K
Sbjct: 219 IAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKIAKSWM--NLKSISPSYK 276
Query: 241 DYHDVASTFATTGSVVAALSS 261
V+ TF T S++ L++
Sbjct: 277 TLPSVSETFPTLRSMIEVLNT 297
>ref|XP_214720.3| PREDICTED: similar to tetratricopeptide repeat domain 13 [Rattus
norvegicus]
Length = 738
Score = 99.4 bits (246), Expect = 9e-20
Identities = 77/262 (29%), Positives = 130/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 474 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 529
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 530 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 589
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S +F + V + IL + YY+YN MPLSRG++
Sbjct: 590 EERTQLYHAEIDALYKDL-TAKGKVLTLSAEFGEADAVCNLILSLVYYFYNLMPLSRGSS 648
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 649 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKIAKSWM--NLKSISPSY 706
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 707 KTLPSVSETFPTLRSMIEVLNT 728
>gb|AAH79851.1| Ttc13 protein [Mus musculus]
Length = 585
Score = 99.4 bits (246), Expect = 9e-20
Identities = 77/262 (29%), Positives = 130/262 (49%), Gaps = 13/262 (4%)
Query: 6 GFGSHTPMILGQAKVVRYFPNYERTLDIAKTVMKERSYV-HGKTDQIIHLSNDGKLEEIM 64
GF +H +I GQ +RY +E+ L +K+R V HG + L LE++
Sbjct: 321 GFNNHINLIRGQVINMRYLEYFEKILHF----IKDRILVYHGANNPKGLLEVREALEKVH 376
Query: 65 HAKSCSDLYKVVG----EDFWSSTWCNSTAFEGKQLEGTRVTLVKMGQHGFDFAIRTPCT 120
+ + K + F +T S +GK+ +G +T+ F++ T T
Sbjct: 377 KVEDLLPIMKQFNTKTKDGFTVNTKVPSLKDQGKEYDGFTITITGDKVGNILFSVETQTT 436
Query: 121 PARWEDYDAEMAMAWEALCNAYCGENYGSTDFDVLENVRDAILRMTYYWYNFMPLSRGTA 180
R + Y AE+ ++ L A S +F + V + IL + YY+YN MPLSRG++
Sbjct: 437 EERTQLYHAEIDALYKDL-TAKGKVLTLSAEFGEADAVCNLILSLVYYFYNLMPLSRGSS 495
Query: 181 AVGFAVMLGLLLAANMEFTGSIPQGFQVDWEAILNLDPNSFVDSVKSWLYPSLK-VTTSW 239
+ ++V++G L+A+ E G IP+G VD+EA+ +F KSW+ +LK ++ S+
Sbjct: 496 VIAYSVIVGALMASGKEVAGKIPKGKLVDFEAMTAPGSEAFSKIAKSWM--NLKSISPSY 553
Query: 240 KDYHDVASTFATTGSVVAALSS 261
K V+ TF T S++ L++
Sbjct: 554 KTLPSVSETFPTLRSMIEVLNT 575
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 468,421,574
Number of Sequences: 2540612
Number of extensions: 19210283
Number of successful extensions: 38424
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 27
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 38363
Number of HSP's gapped (non-prelim): 38
length of query: 263
length of database: 863,360,394
effective HSP length: 125
effective length of query: 138
effective length of database: 545,783,894
effective search space: 75318177372
effective search space used: 75318177372
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146561.5


