

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146561.3 + phase: 0
(262 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
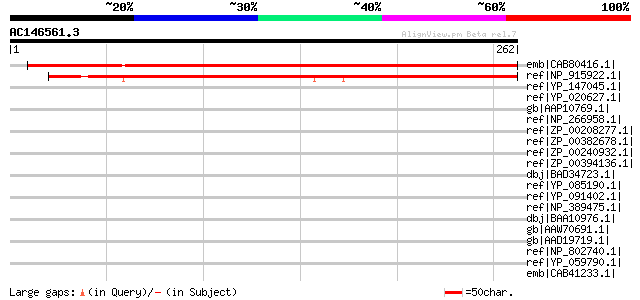
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB80416.1| putative protein [Arabidopsis thaliana] gi|44688... 370 e-101
ref|NP_915922.1| P0468B07.18 [Oryza sativa (japonica cultivar-gr... 317 2e-85
ref|YP_147045.1| dsRNA-specific ribonuclease [Geobacillus kausto... 44 0.005
ref|YP_020627.1| ribonuclease iii [Bacillus anthracis str. 'Ames... 42 0.017
gb|AAP10769.1| Ribonuclease III [Bacillus cereus ATCC 14579] gi|... 42 0.017
ref|NP_266958.1| ribonuclease III [Lactococcus lactis subsp. lac... 42 0.017
ref|ZP_00208277.1| COG0571: dsRNA-specific ribonuclease [Magneto... 42 0.017
ref|ZP_00382678.1| COG0571: dsRNA-specific ribonuclease [Lactoco... 42 0.017
ref|ZP_00240932.1| ribonuclease III [Bacillus cereus G9241] gi|4... 42 0.017
ref|ZP_00394136.1| COG0571: dsRNA-specific ribonuclease [Bacillu... 42 0.017
dbj|BAD34723.1| Dicer-related RNase III protein Dcr2p [Tetrahyme... 42 0.023
ref|YP_085190.1| ribonuclease III [Bacillus cereus E33L] gi|5197... 42 0.023
ref|YP_091402.1| Rnc [Bacillus licheniformis ATCC 14580] gi|5234... 41 0.030
ref|NP_389475.1| ribonuclease III [Bacillus subtilis subsp. subt... 41 0.030
dbj|BAA10976.1| ORF3 [Bacillus subtilis] 41 0.030
gb|AAW70691.1| dsRNA-specific ribonuclease [Wolbachia endosymbio... 41 0.039
gb|AAD19719.1| ribonuclease III; RNaseIII [Zymomonas mobilis] 41 0.039
ref|NP_802740.1| putative ribonuclease III [Streptococcus pyogen... 40 0.050
ref|YP_059790.1| Ribonuclease III [Streptococcus pyogenes MGAS10... 40 0.050
emb|CAB41233.1| SPCC188.13c [Schizosaccharomyces pombe] 39 0.15
>emb|CAB80416.1| putative protein [Arabidopsis thaliana] gi|4468817|emb|CAB38218.1|
putative protein [Arabidopsis thaliana]
gi|15235580|ref|NP_195467.1| ribonuclease III family
protein [Arabidopsis thaliana] gi|7486504|pir||T04745
hypothetical protein F6G17.160 - Arabidopsis thaliana
Length = 537
Score = 370 bits (949), Expect = e-101
Identities = 185/253 (73%), Positives = 208/253 (82%), Gaps = 1/253 (0%)
Query: 10 SPKPTNFSFSSSFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRNNNPQRLLKELAE 69
S SFSSS S FP + TKSR L + A T+ PR++ PQRLLKELA+
Sbjct: 17 SSSAPEISFSSSISQFPSKTQSILTKSRFQNLRICASVTAETQGLPRDS-PQRLLKELAQ 75
Query: 70 RKKITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSLRLNNADKL 129
RK T PK K PP+R+IL+PPLDDKKLAERFLNSPQLSLKSFPLLSSCLPS +LNNADK
Sbjct: 76 RKTATGPKKKVPPKRFILRPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSSKLNNADKT 135
Query: 130 WIDEYLLEAKQALGYSLEPSETLEDDNPAKQFDTLLYLAFQHPSCERTKQKHVRSGHSRL 189
WIDEYLLE KQALGYSLEPSE+L DDNPAK FDTLLYLAFQHPSC+R + +HV++GHSRL
Sbjct: 136 WIDEYLLEVKQALGYSLEPSESLGDDNPAKHFDTLLYLAFQHPSCDRARARHVKNGHSRL 195
Query: 190 FFLGQYVLELALAEFFLQRYPRELPGPMRERVYGLIGKENLPKWIKAASLQNLIFPYDNM 249
+FLGQYVLELAL EFFLQRYPRE PGPMRERV+ LIGK LPKWIKAASLQNLIFPYD+M
Sbjct: 196 WFLGQYVLELALTEFFLQRYPRESPGPMRERVFALIGKRYLPKWIKAASLQNLIFPYDDM 255
Query: 250 DKIVRRDREGPVK 262
DK++R++RE PVK
Sbjct: 256 DKLIRKEREPPVK 268
Score = 43.9 bits (102), Expect = 0.005
Identities = 37/144 (25%), Positives = 72/144 (49%), Gaps = 7/144 (4%)
Query: 104 PQLSLKSFP-LLSSCLPSLRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQFD 162
P+ +L + P L +C+P +W + + Q LGY L ++ +++ A+ +
Sbjct: 338 PEDALFAHPRLFRACVPPGMHRFRGNIWDFDSKPKVMQTLGYPLTMNDRIKEITEARNIE 397
Query: 163 TLL--YLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGP-MRE 219
L L F HPS + K +H R RL ++GQ + ++A+AE L ++ + PG ++E
Sbjct: 398 LGLGLQLCFLHPS--KHKFEHPRFCFERLEYVGQKIQDIAMAERLLMKH-LDAPGKWLQE 454
Query: 220 RVYGLIGKENLPKWIKAASLQNLI 243
+ L+ + ++++ L N I
Sbjct: 455 KHRRLLMNKFCGRYLREKRLHNFI 478
>ref|NP_915922.1| P0468B07.18 [Oryza sativa (japonica cultivar-group)]
gi|20160696|dbj|BAB89639.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 533
Score = 317 bits (812), Expect = 2e-85
Identities = 163/256 (63%), Positives = 194/256 (75%), Gaps = 17/256 (6%)
Query: 21 SFSPFPIQILIKNTKSRNYPLHVLAVAIDPTKDFPRN--------NNPQRLLKELAERKK 72
+ P P+ + + R + VLAVA D T P + N+P RLL+ELAERKK
Sbjct: 12 ALGPLPLPLPAARRRRR---VRVLAVAADHTPPPPPSPSSPPEPANSPSRLLRELAERKK 68
Query: 73 ITNPKSKSPPRRYILKPPLDDKKLAERFLNSPQLSLKSFPLLSSCLPSLRLNNADKLWID 132
+PK K PPRR+ILKPPLDD++L +RFL+SPQLSLK+ PLLSSCLPS L+ AD+ W+D
Sbjct: 69 AVSPKKKHPPRRFILKPPLDDERLTQRFLSSPQLSLKALPLLSSCLPSAPLSAADRTWMD 128
Query: 133 EYLLEAKQALGYSLEPSETLEDDN----PAKQFDTLLYLAFQH--PSCERTKQKHVRSGH 186
EYLLEAKQALGY L PSETL D + PA+ FD LLYLAFQH SCERT+ +HVRSGH
Sbjct: 129 EYLLEAKQALGYPLAPSETLGDGDDDGCPARHFDVLLYLAFQHLDTSCERTRTRHVRSGH 188
Query: 187 SRLFFLGQYVLELALAEFFLQRYPRELPGPMRERVYGLIGKENLPKWIKAASLQNLIFPY 246
SRL+FLGQYVLELA EFFLQRYPRE PGPMRERV+ LIGK +PKWIKAASL NL+FPY
Sbjct: 189 SRLWFLGQYVLELAFCEFFLQRYPRESPGPMRERVFALIGKRAIPKWIKAASLHNLVFPY 248
Query: 247 DNMDKIVRRDREGPVK 262
D++DK++R+DRE P K
Sbjct: 249 DDLDKMIRKDREPPAK 264
Score = 36.2 bits (82), Expect = 0.95
Identities = 26/99 (26%), Positives = 46/99 (46%), Gaps = 4/99 (4%)
Query: 113 LLSSCLPSLRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQFDTLL--YLAFQ 170
L +C+P +W + + LGY L ++ + + A+ + L L F
Sbjct: 344 LFRACVPPGMHRFRGNIWDFDNRPKVMNTLGYPLPMNDRIPEITEARNIELGLGLQLCFL 403
Query: 171 HPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRY 209
HPS + K +H R RL ++GQ + +L +AE L ++
Sbjct: 404 HPS--KHKFEHPRFCLERLEYVGQKIQDLVMAERLLMKH 440
>ref|YP_147045.1| dsRNA-specific ribonuclease [Geobacillus kaustophilus HTA426]
gi|56379569|dbj|BAD75477.1| dsRNA-specific ribonuclease
[Geobacillus kaustophilus HTA426]
Length = 246
Score = 43.9 bits (102), Expect = 0.005
Identities = 23/82 (28%), Positives = 42/82 (51%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S ++ + + RL FLG VLEL ++++ Q++P G + +
Sbjct: 32 EKLLIQAFTHSSYVNEHRRRLHEDNERLEFLGDAVLELTVSQYLFQKFPHMSEGQLTKLR 91
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L K+ A S L+
Sbjct: 92 AAIVCEPSLVKFANALSFGELV 113
>ref|YP_020627.1| ribonuclease iii [Bacillus anthracis str. 'Ames Ancestor']
gi|30263852|ref|NP_846229.1| ribonuclease III [Bacillus
anthracis str. Ames] gi|49478414|ref|YP_037910.1|
ribonuclease III [Bacillus thuringiensis serovar
konkukian str. 97-27] gi|49186699|ref|YP_029951.1|
ribonuclease III [Bacillus anthracis str. Sterne]
gi|30258496|gb|AAP27715.1| ribonuclease III [Bacillus
anthracis str. Ames] gi|49329970|gb|AAT60616.1|
ribonuclease III [Bacillus thuringiensis serovar
konkukian str. 97-27] gi|47504426|gb|AAT33102.1|
ribonuclease III [Bacillus anthracis str. 'Ames
Ancestor'] gi|49180626|gb|AAT56002.1| ribonuclease III
[Bacillus anthracis str. Sterne]
gi|38372420|sp|Q81WI8|RNC_BACAN Ribonuclease III (RNase
III)
Length = 245
Score = 42.0 bits (97), Expect = 0.017
Identities = 23/82 (28%), Positives = 41/82 (49%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S +K + RL FLG VLEL ++++ Q+YP G + +
Sbjct: 33 EKLLIQAFTHSSYVNEHRKKPHEDNERLEFLGDAVLELTVSQYLFQKYPTMSEGELTKLR 92
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L ++ S +L+
Sbjct: 93 AAIVCEPSLVRFANELSFGSLV 114
>gb|AAP10769.1| Ribonuclease III [Bacillus cereus ATCC 14579]
gi|30021937|ref|NP_833568.1| Ribonuclease III [Bacillus
cereus ATCC 14579] gi|38372411|sp|Q819V8|RNC_BACCR
Ribonuclease III (RNase III)
Length = 245
Score = 42.0 bits (97), Expect = 0.017
Identities = 23/82 (28%), Positives = 41/82 (49%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S +K + RL FLG VLEL ++++ Q+YP G + +
Sbjct: 33 EKLLIQAFTHSSYVNEHRKKPHEDNERLEFLGDAVLELTVSQYLFQKYPTMSEGELTKLR 92
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L ++ S +L+
Sbjct: 93 AAIVCEPSLVRFANELSFGSLV 114
>ref|NP_266958.1| ribonuclease III [Lactococcus lactis subsp. lactis Il1403]
gi|12723723|gb|AAK04900.1| ribonuclease III (EC
3.1.26.3) [Lactococcus lactis subsp. lactis Il1403]
gi|13878708|sp|Q9CHD0|RNC_LACLA Ribonuclease III (RNase
III)
Length = 231
Score = 42.0 bits (97), Expect = 0.017
Identities = 24/97 (24%), Positives = 46/97 (46%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S ++ + + RL FLG L L ++++ + YP +L G + +
Sbjct: 19 EDLLKTAFTHSSFTNEERLPKIANNERLEFLGDVALSLVISDYLYRTYPEKLEGELSKMR 78
Query: 222 YGLIGKENLPKWIKAASLQNLIFPYDNMDKIVRRDRE 258
++ E+L + ++ + +K+ RDRE
Sbjct: 79 SSIVRTESLANFSRSCGFGEFLRLGHGEEKMGGRDRE 115
>ref|ZP_00208277.1| COG0571: dsRNA-specific ribonuclease [Magnetospirillum
magnetotacticum MS-1]
Length = 230
Score = 42.0 bits (97), Expect = 0.017
Identities = 23/65 (35%), Positives = 33/65 (50%)
Query: 164 LLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERVYG 223
LL A HPS ++ ++ + RL FLG VL L +AE R+P E G + R
Sbjct: 21 LLSQALTHPSTQQHRRSQGSDPYERLEFLGDRVLGLVVAEMLFLRFPDEAEGALARRHAA 80
Query: 224 LIGKE 228
L+ +E
Sbjct: 81 LVRRE 85
>ref|ZP_00382678.1| COG0571: dsRNA-specific ribonuclease [Lactococcus lactis subsp.
cremoris SK11]
Length = 231
Score = 42.0 bits (97), Expect = 0.017
Identities = 24/97 (24%), Positives = 46/97 (46%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S ++ + + RL FLG L L ++++ + YP +L G + +
Sbjct: 19 EDLLKTAFTHSSFTNEERLPKIANNERLEFLGDVALSLVISDYLYRTYPEKLEGELSKMR 78
Query: 222 YGLIGKENLPKWIKAASLQNLIFPYDNMDKIVRRDRE 258
++ E+L + ++ + +K+ RDRE
Sbjct: 79 SSIVRTESLANFSRSCGFGEFLRLGHGEEKMGGRDRE 115
>ref|ZP_00240932.1| ribonuclease III [Bacillus cereus G9241] gi|47553047|gb|EAL11449.1|
ribonuclease III [Bacillus cereus G9241]
gi|42782941|ref|NP_980188.1| ribonuclease III [Bacillus
cereus ATCC 10987] gi|42738868|gb|AAS42796.1|
ribonuclease III [Bacillus cereus ATCC 10987]
Length = 245
Score = 42.0 bits (97), Expect = 0.017
Identities = 23/82 (28%), Positives = 41/82 (49%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S +K + RL FLG VLEL ++++ Q+YP G + +
Sbjct: 33 EKLLIQAFTHSSYVNEHRKKPHEDNERLEFLGDAVLELTVSQYLFQKYPTMSEGELTKLR 92
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L ++ S +L+
Sbjct: 93 AAIVCEPSLVRFANELSFGSLV 114
>ref|ZP_00394136.1| COG0571: dsRNA-specific ribonuclease [Bacillus anthracis str.
A2012]
Length = 262
Score = 42.0 bits (97), Expect = 0.017
Identities = 23/82 (28%), Positives = 41/82 (49%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S +K + RL FLG VLEL ++++ Q+YP G + +
Sbjct: 50 EKLLIQAFTHSSYVNEHRKKPHEDNERLEFLGDAVLELTVSQYLFQKYPTMSEGELTKLR 109
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L ++ S +L+
Sbjct: 110 AAIVCEPSLVRFANELSFGSLV 131
>dbj|BAD34723.1| Dicer-related RNase III protein Dcr2p [Tetrahymena thermophila]
Length = 1838
Score = 41.6 bits (96), Expect = 0.023
Identities = 32/124 (25%), Positives = 55/124 (43%), Gaps = 9/124 (7%)
Query: 107 SLKSFPLLSSCLPSLRLNNADKLWIDEYLLEAKQALGYSLEPSETLEDDNPAKQFDTLLY 166
+L S L +PSL+ N +KL ++ K+ G + E + + +N K
Sbjct: 1552 TLPSIESLCIQIPSLKNTNLNKLHQINNAIQKKENQG-TFEKIDQISKENQNK------- 1603
Query: 167 LAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERVYGLIG 226
+Q S E K+ + RL FLG VL+L + E+ ++P PG + + L+
Sbjct: 1604 -IYQKDSNEWENLKNYNLCNQRLEFLGDAVLDLIIVEYLFNKFPNSDPGELTQMKTSLVQ 1662
Query: 227 KENL 230
+ L
Sbjct: 1663 NKTL 1666
>ref|YP_085190.1| ribonuclease III [Bacillus cereus E33L] gi|51975103|gb|AAU16653.1|
ribonuclease III [Bacillus cereus E33L]
Length = 245
Score = 41.6 bits (96), Expect = 0.023
Identities = 23/82 (28%), Positives = 40/82 (48%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
+ LL AF H S +K + RL FLG VLEL ++++ Q+YP G + +
Sbjct: 33 EKLLIQAFTHSSYVNEHRKKPHEDNERLEFLGDAVLELTVSQYLFQKYPTMSEGELTKLR 92
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
++ + +L ++ S L+
Sbjct: 93 AAIVCEPSLVRFANELSFGGLV 114
>ref|YP_091402.1| Rnc [Bacillus licheniformis ATCC 14580] gi|52348075|gb|AAU40709.1|
Rnc [Bacillus licheniformis DSM 13]
Length = 249
Score = 41.2 bits (95), Expect = 0.030
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 160 QFDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRE 219
Q + LLY AF H S +K + RL FLG VLEL +++F +YP G + +
Sbjct: 32 QNEKLLYQAFTHSSYVNEHRKKPYEDNERLEFLGDAVLELTISQFLYAKYPAMSEGDLTK 91
Query: 220 RVYGLIGKENLPKWIKAASLQNLI 243
++ + +L S L+
Sbjct: 92 LRAAIVCEPSLVSLAHELSFGELV 115
>ref|NP_389475.1| ribonuclease III [Bacillus subtilis subsp. subtilis str. 168]
gi|2633965|emb|CAB13466.1| ribonuclease III [Bacillus
subtilis subsp. subtilis str. 168]
gi|7531275|sp|P51833|RNC_BACSU Ribonuclease III (RNase
III)
Length = 249
Score = 41.2 bits (95), Expect = 0.030
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 160 QFDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRE 219
Q + LLY AF H S +K + RL FLG VLEL ++ F +YP G + +
Sbjct: 32 QNEKLLYQAFTHSSYVNEHRKKPYEDNERLEFLGDAVLELTISRFLFAKYPAMSEGDLTK 91
Query: 220 RVYGLIGKENLPKWIKAASLQNLI 243
++ + +L S +L+
Sbjct: 92 LRAAIVCEPSLVSLAHELSFGDLV 115
>dbj|BAA10976.1| ORF3 [Bacillus subtilis]
Length = 266
Score = 41.2 bits (95), Expect = 0.030
Identities = 25/84 (29%), Positives = 39/84 (45%)
Query: 160 QFDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRE 219
Q + LLY AF H S +K + RL FLG VLEL ++ F +YP G + +
Sbjct: 32 QNEKLLYQAFTHSSYVNEHRKKPYEDNERLEFLGDAVLELTISRFLFPKYPAMSEGDLTK 91
Query: 220 RVYGLIGKENLPKWIKAASLQNLI 243
++ + +L S +L+
Sbjct: 92 LRAAIVCEPSLVSLAHELSFGDLV 115
>gb|AAW70691.1| dsRNA-specific ribonuclease [Wolbachia endosymbiont strain TRS of
Brugia malayi] gi|58584360|ref|YP_197933.1|
dsRNA-specific ribonuclease [Wolbachia endosymbiont
strain TRS of Brugia malayi]
Length = 243
Score = 40.8 bits (94), Expect = 0.039
Identities = 22/82 (26%), Positives = 36/82 (43%)
Query: 162 DTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERV 221
D +L A HPS + K+ + RL FLG +L + ++ + +P+E G + R
Sbjct: 29 DAILEEALTHPSLNKRNSKNQIENYERLEFLGDSILNMIVSAILFRLFPKEKEGALARRK 88
Query: 222 YGLIGKENLPKWIKAASLQNLI 243
L+ + K L N I
Sbjct: 89 TDLVCGNTIANVAKEIKLGNFI 110
>gb|AAD19719.1| ribonuclease III; RNaseIII [Zymomonas mobilis]
Length = 222
Score = 40.8 bits (94), Expect = 0.039
Identities = 27/93 (29%), Positives = 41/93 (44%), Gaps = 16/93 (17%)
Query: 152 LEDDNPAKQFDTLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPR 211
+E P+ + L A HPS H S + RL FLG VL L +A + +PR
Sbjct: 10 IEKFRPSAKDPALFLRAMTHPS-------HGNSDYQRLEFLGDRVLGLVIANWLYDLFPR 62
Query: 212 ELPGPMRERVYGLIGKEN---------LPKWIK 235
E G + R+ L+ + LP+W++
Sbjct: 63 EPEGKLSRRLNSLVSGASCASIARIVGLPQWLR 95
>ref|NP_802740.1| putative ribonuclease III [Streptococcus pyogenes SSI-1]
gi|21904099|gb|AAM78982.1| putative ribonuclease III
[Streptococcus pyogenes MGAS315]
gi|19747784|gb|AAL97288.1| putative ribonuclease III
[Streptococcus pyogenes MGAS8232]
gi|19745653|ref|NP_606789.1| putative ribonuclease III
[Streptococcus pyogenes MGAS8232]
gi|13621744|gb|AAK33526.1| putative ribonuclease III
[Streptococcus pyogenes M1 GAS]
gi|21909911|ref|NP_664179.1| putative ribonuclease III
[Streptococcus pyogenes MGAS315]
gi|54039245|sp|P66672|RNC_STRP8 Ribonuclease III (RNase
III) gi|54039244|sp|P66671|RNC_STRP3 Ribonuclease III
(RNase III) gi|54041616|sp|P66670|RNC_STRPY Ribonuclease
III (RNase III) gi|28811641|dbj|BAC64573.1| putative
ribonuclease III [Streptococcus pyogenes SSI-1]
gi|15674631|ref|NP_268805.1| putative ribonuclease III
[Streptococcus pyogenes M1 GAS]
Length = 230
Score = 40.4 bits (93), Expect = 0.050
Identities = 25/81 (30%), Positives = 41/81 (49%)
Query: 163 TLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERVY 222
TLL AF H S + S + RL FLG VL+L ++E+ +YP++ G M +
Sbjct: 20 TLLETAFTHTSYANEHRLLNVSHNERLEFLGDAVLQLIISEYLFAKYPKKTEGDMSKLRS 79
Query: 223 GLIGKENLPKWIKAASLQNLI 243
++ +E+L + + S I
Sbjct: 80 MIVREESLAGFSRFCSFDAYI 100
>ref|YP_059790.1| Ribonuclease III [Streptococcus pyogenes MGAS10394]
gi|50902892|gb|AAT86607.1| Ribonuclease III
[Streptococcus pyogenes MGAS10394]
gi|56808931|ref|ZP_00366639.1| COG0571: dsRNA-specific
ribonuclease [Streptococcus pyogenes M49 591]
Length = 230
Score = 40.4 bits (93), Expect = 0.050
Identities = 25/81 (30%), Positives = 41/81 (49%)
Query: 163 TLLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRERVY 222
TLL AF H S + S + RL FLG VL+L ++E+ +YP++ G M +
Sbjct: 20 TLLETAFTHTSYANEHRLLNVSHNERLEFLGDAVLQLIISEYLFAKYPKKTEGDMSKLRS 79
Query: 223 GLIGKENLPKWIKAASLQNLI 243
++ +E+L + + S I
Sbjct: 80 MIVREESLAGFSRFCSFDAYI 100
>emb|CAB41233.1| SPCC188.13c [Schizosaccharomyces pombe]
Length = 653
Score = 38.9 bits (89), Expect = 0.15
Identities = 28/102 (27%), Positives = 52/102 (50%), Gaps = 13/102 (12%)
Query: 164 LLYLAFQHPSCERTKQKHVRSGHSRLFFLGQYVLELALAEFFLQRYPRELPGPMRE-RVY 222
LL+LAF HPS Q+ + + +L FLG VL+ + ++ ++YP G + + + +
Sbjct: 378 LLHLAFIHPSM--MSQQGIYENYQQLEFLGDAVLDYIIVQYLYKKYPNATSGELTDYKSF 435
Query: 223 GLIGKE--------NLPKWI--KAASLQNLIFPYDNMDKIVR 254
+ K NL K+I ++A++ + IF Y + + R
Sbjct: 436 YVCNKSLSYIGFVLNLHKYIQHESAAMCDAIFEYQELIEAFR 477
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.137 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,262,505
Number of Sequences: 2540612
Number of extensions: 19937600
Number of successful extensions: 48837
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 56
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 48737
Number of HSP's gapped (non-prelim): 137
length of query: 262
length of database: 863,360,394
effective HSP length: 125
effective length of query: 137
effective length of database: 545,783,894
effective search space: 74772393478
effective search space used: 74772393478
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC146561.3


