

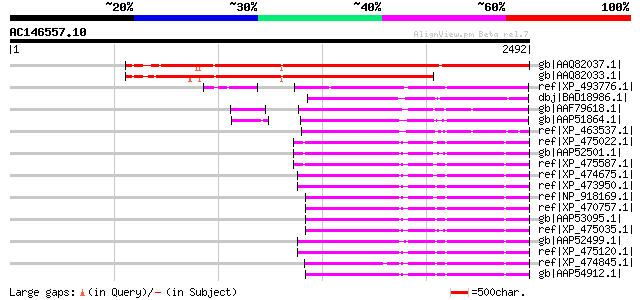
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146557.10 - phase: 0
(2492 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum] 2205 0.0
gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum] 1514 0.0
ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica... 905 0.0
dbj|BAD18986.1| GAG-POL precursor [Vitis vinifera] 768 0.0
gb|AAF79618.1| F5M15.26 [Arabidopsis thaliana] gi|25402907|pir||... 763 0.0
gb|AAP51864.1| putative retroelement pol polyprotein [Oryza sati... 752 0.0
ref|XP_463537.1| putative polyprotein [Oryza sativa (japonica cu... 701 0.0
ref|XP_475022.1| OSJNBb0093G06.3 [Oryza sativa (japonica cultiva... 699 0.0
gb|AAP52501.1| putative gag-pol precursor [Oryza sativa (japonic... 698 0.0
ref|XP_475587.1| putative polyprotein [Oryza sativa (japonica cu... 698 0.0
ref|XP_474675.1| OSJNBa0027O01.4 [Oryza sativa (japonica cultiva... 696 0.0
ref|XP_473950.1| OSJNBa0053K19.16 [Oryza sativa (japonica cultiv... 696 0.0
ref|NP_918169.1| putative gag-pol polyprotein [Oryza sativa (jap... 693 0.0
ref|XP_470757.1| putative gag-pol precursor [Oryza sativa] gi|18... 693 0.0
gb|AAP53095.1| putative retroelement [Oryza sativa (japonica cul... 692 0.0
ref|XP_475035.1| OSJNBa0042F21.5 [Oryza sativa (japonica cultiva... 691 0.0
gb|AAP52499.1| putative gag-pol precursor [Oryza sativa (japonic... 691 0.0
ref|XP_475120.1| putative polyprotein [Oryza sativa (japonica cu... 690 0.0
ref|XP_474845.1| OSJNBa0035O13.3 [Oryza sativa (japonica cultiva... 685 0.0
gb|AAP54912.1| gag-pol precursor [Oryza sativa (japonica cultiva... 682 0.0
>gb|AAQ82037.1| gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 2205 bits (5714), Expect = 0.0
Identities = 1137/2019 (56%), Positives = 1436/2019 (70%), Gaps = 139/2019 (6%)
Query: 557 YVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKAESSVNASKRYGNGHHKKKETEVG 616
+++RM +FS++V G R E ++ G I + ++SK+ G +++E E
Sbjct: 300 FMDRMGSCPFGSFSDVVICGERTESLIKTGKI----QDVGSSSSKKPFAGAPRRREGETN 355
Query: 617 MVSAGAGQAMATVAPINAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQ 676
V Q A +P P +
Sbjct: 356 AVQHRRDQNRIEYRQAAAVTIPAPQP---------------------------------R 382
Query: 677 MKQQLPVQQQQQQQQQQYTRP-------TFPPIPMLYAELLPTLLQRG---HCTTRQGKP 726
+QQ VQQ QQQQQQ+ +P F +PM YAELLP LL+ G CT
Sbjct: 383 QQQQQRVQQPQQQQQQRPYQPRQRMPDRRFDSLPMSYAELLPELLRLGLVELCT----MA 438
Query: 727 PPDPLPPRFRSDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLP 786
PP LPP + ++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P
Sbjct: 439 PPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINFA-PVPNVVNNPMP 497
Query: 787 NHAA--VNMIEVCEEAPRLD-VRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRDPLGCH 843
H VN IE E +D V +V T L+ + +L ++S CLGC GC
Sbjct: 498 QHGGHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAESENGCD 557
Query: 844 AVQDDIQSLMNDNLLTVS-------DVCVIVPVF-----HDPPVKSVPLKKNAEPLVIRL 891
++ IQ +M++ L S V I F H V S P N P+ I +
Sbjct: 558 QLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGHGQRVVSAP-ATNGTPVTIPV 616
Query: 892 P------------GPIPYVSDKAVPYKY-----------------NATMIENGV--EVPL 920
P G + +AVP+KY N T + G+ VP
Sbjct: 617 PVTISAPTTIVASGRRAVENSRAVPWKYDNAYRSNRRVESQTKPVNQTPVTIGLANRVPA 676
Query: 921 ASFATVSNIAEGTSAALRSGKVRPPLFQKKVATPTIPPVEEATPTV-VSPIATDVNQSGK 979
V N+ G RSG++ P + + + V P+ + +
Sbjct: 677 TVGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGS- 734
Query: 980 SIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHKNTLLKVLEQAYVDHE 1039
+ +++E +++IK+SDYKIVDQL QTPSKIS+LSLLL SEAH+N LLK+L AYV E
Sbjct: 735 --FEKDVEEFMKMIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQE 792
Query: 1040 VTVDRFGGIVGNITACNNLWFSEEELPEAGKSHNLALHISVNCKSDMISNVLVDTGSSLN 1099
++V++ G++ N++ + + F+ +LP G++HN ALHI++ CK ++S+VLVDTGSSLN
Sbjct: 793 ISVNQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLN 852
Query: 1100 VMPKTTLDQLSYRGTPLRRSTFLVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINA 1159
V+PK L ++ G L S +V+AFD S+++V GE+ LP+ IGPE F + F VMDI
Sbjct: 853 VLPKQILKKIDVEGFVLTPSDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQP 912
Query: 1160 SYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTIHGEEAYLVSQLLSFSCIEA-GSAE 1218
+YSCLLGRPWIH AGAV+STLHQKLK+V NG++VT+ GEE LVS L SF +E G
Sbjct: 913 AYSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIH 972
Query: 1219 GT---AFQGLTIEG---AEPKEAGAAMASLKDAQRAIQEGQTAGWGKVIRLCENKRKEGL 1272
T F+ + +E AE ++ G ++ S K A+ + G+ GWGK++ L + K G+
Sbjct: 973 ETLCQVFETVALEKVAYAEQRKPGVSITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGV 1032
Query: 1273 GFSPSSRVSSG-----VFHSAGFVNAISEEATGS-----------GLRPVFVTPGGIATD 1316
G+ P +G F SAG +N ATGS +RP PGG +
Sbjct: 1033 GYEPLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINN 1090
Query: 1317 WDAIDIPSIMHVSELNHNKPVEHSNPTVPPSFDFPVYEAKD--EEGDGIPYEITRLLEQE 1374
W A ++ + ++E + + + FD P+Y+A++ EE +P E+ RLL+QE
Sbjct: 1091 WTAEEVVQVTLLTESDLMAFTMNGSVIAQYDFDNPIYQAEEESEEDCELPAELVRLLKQE 1150
Query: 1375 KKAIQPHQEEIELVNIGTEENKREIKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMPGL 1434
++ IQPHQEE+E+VN+GTE+ REIKIGA LE+ VKR++I++LREY +IFAWSY+DMPGL
Sbjct: 1151 ERVIQPHQEELEVVNLGTEDATREIKIGAALEDSVKRRLIEMLREYVEIFAWSYQDMPGL 1210
Query: 1435 DPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVP 1494
D IV HR+P + CP V+QKLRRT PDMA KIK EVQKQ DAGFL YP W+ANIVP
Sbjct: 1211 DTDIVVHRLPLREGCPSVKQKLRRTSPDMATKIKEEVQKQWDAGFLAVTSYPPWMANIVP 1270
Query: 1495 VPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSP 1554
VPKKDGKVRMCVD+RDLN+ASPKD+FPLPHIDVLVDNTAQS VFSFMDGFSGYNQIKM+P
Sbjct: 1271 VPKKDGKVRMCVDYRDLNRASPKDDFPLPHIDVLVDNTAQSSVFSFMDGFSGYNQIKMAP 1330
Query: 1555 EDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTD 1614
ED EKT+FITPWGTFCYKVMPFGL NAGATYQR MTTLFHDM HKE+EVYVDDMI KS
Sbjct: 1331 EDMEKTTFITPWGTFCYKVMPFGLKNAGATYQRAMTTLFHDMMHKEIEVYVDDMIAKSQT 1390
Query: 1615 EEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMP 1674
EE+H+ L K+F+RLRK+KLRLNPNKCTFGVRSGKLLGFIVS+KGIEVDP KV+AI+EMP
Sbjct: 1391 EEEHLVNLQKLFDRLRKFKLRLNPNKCTFGVRSGKLLGFIVSEKGIEVDPAKVKAIQEMP 1450
Query: 1675 APRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYLL 1734
P+TEKQVRGFLGRLNYI+RFISH+TATC PIFKLLRKNQ + WND+CQ+AFD IK+YL
Sbjct: 1451 EPKTEKQVRGFLGRLNYIARFISHLTATCEPIFKLLRKNQAIKWNDDCQKAFDKIKEYLQ 1510
Query: 1735 EPPILVPPVEGRPLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLE 1794
+PPIL+PPV GRPLIMYL+V + SMGCVLG+ DE+G+KEHAIYYLSKKFTDCETRY++LE
Sbjct: 1511 KPPILIPPVPGRPLIMYLSVTENSMGCVLGRHDESGRKEHAIYYLSKKFTDCETRYSLLE 1570
Query: 1795 KTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKT 1854
KTCCALAWAA+RLR Y++NHTT LIS+MDP+KYIFEK A+TG++ARWQM+L+EYDI + +
Sbjct: 1571 KTCCALAWAARRLRQYMLNHTTLLISKMDPVKYIFEKPALTGRVARWQMILTEYDIQYTS 1630
Query: 1855 QKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVF 1914
QKAIKGSIL+D+LA QP++DYQP+ F+FPDE+IMYLK KDC+EPL+EEGPDP+ KW L+F
Sbjct: 1631 QKAIKGSILSDYLAEQPIEDYQPMMFEFPDEDIMYLKMKDCKEPLVEEGPDPDDKWTLMF 1690
Query: 1915 DGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDI 1974
DGAVN G G+GAV+++P+G H+PF+AR+ F+ TNN AEYEACI GIEEAID+RIK LDI
Sbjct: 1691 DGAVNMNGNGVGAVLINPKGAHMPFSARLTFDVTNNEAEYEACIMGIEEAIDLRIKTLDI 1750
Query: 1975 YGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSS 2034
+GDSALV+NQ+ G+W T+ LIPYRDY RR+LT+F KV+L+H+PRDENQMADALATLSS
Sbjct: 1751 FGDSALVVNQVNGDWNTNQPHLIPYRDYTRRILTFFKKVKLYHVPRDENQMADALATLSS 1810
Query: 2035 MFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSREYPPG 2094
M +VN WN VP + V RLERP++VFA V V+D KPWYYDIK FL ++EYP G
Sbjct: 1811 MIKVNWWNHVPHVAVNRLERPAYVFAAESV-------VIDEKPWYYDIKNFLKTQEYPEG 1863
Query: 2095 ASKQDKKTLRRLASRFLLD-GDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATG 2153
ASK DKKTLRRLA F L+ D+LYKRN+DMVLLRC+D EA+ LM +VH+G+FGTHA G
Sbjct: 1864 ASKNDKKTLRRLAGSFYLNQDDVLYKRNFDMVLLRCMDRPEADMLMQEVHEGSFGTHAGG 1923
Query: 2154 HTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSMWGID 2213
H M++KLLRAGYYWM ME DC++YARKCHKCQIYAD++HVPP LNVM+SPWPF+MWGID
Sbjct: 1924 HAMAKKLLRAGYYWMTMESDCFKYARKCHKCQIYADRVHVPPSPLNVMNSPWPFAMWGID 1983
Query: 2214 MIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIIT 2273
MIG+IEP ASNGHRFILVAIDYFTKWVEAASY N+TKQVV +FIK IICRYGVP +IIT
Sbjct: 1984 MIGKIEPTASNGHRFILVAIDYFTKWVEAASYANITKQVVTRFIKKEIICRYGVPERIIT 2043
Query: 2274 DNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVTTYKDWHEM 2333
DNG+NLNN +++ LC++FKIEHHNSSPYRP+MNGAVEAANKNIK+IV+KMV TYKDWHEM
Sbjct: 2044 DNGSNLNNKMMKELCKDFKIEHHNSSPYRPKMNGAVEAANKNIKKIVRKMVVTYKDWHEM 2103
Query: 2334 LPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQ 2393
LP+ALHGYRT+VR+STGATP+SLVYGMEAVLP+EVEIPSLRV+++ KL EAEW ++R+++
Sbjct: 2104 LPFALHGYRTSVRTSTGATPYSLVYGMEAVLPVEVEIPSLRVLLDVKLDEAEWIRTRFNE 2163
Query: 2394 LNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPDPRGKWAPNY 2453
L+LIEE+R+ + GQ YQ RMK AFD+KVRPR +++G+LVLKR + D RGKW PNY
Sbjct: 2164 LSLIEERRLAVVCHGQLYQRRMKRAFDQKVRPRSYQIGDLVLKRILPPGTDNRGKWTPNY 2223
Query: 2454 EGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
EGPYVVKK FSGGAL+LT MDG + P+PVN+D+VKKYFA
Sbjct: 2224 EGPYVVKKVFSGGALMLTTMDGEDFPSPVNSDVVKKYFA 2262
>gb|AAQ82033.1| gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 1514 bits (3921), Expect = 0.0
Identities = 814/1550 (52%), Positives = 1046/1550 (66%), Gaps = 108/1550 (6%)
Query: 557 YVERMIIAAPNNFSEMVTMGTRLEEAVRDGIIVFEKAESSVNASKRYGNGHHKKKETEVG 616
+++RM +FS++V G R E ++ G I ++ ++SK+ G +++E E
Sbjct: 301 FMDRMGSCPFVSFSDVVICGERTESLIKTGKI----QDAGSSSSKKPFAGAPRRREGETN 356
Query: 617 MVSAGAGQAMATVAPINAAQMPPSYPYMPYSQHPFFPPFYHQYPLPPGQPQVPVNAIAQQ 676
V Q + + A +P P P Q Q V QQ
Sbjct: 357 AVQYRRDQNRSQRCQVAAVTIPA--------------------PQPRQQQQQRVQQPQQQ 396
Query: 677 MKQQLPVQQQQQQQQQQYTRPTFPPIPMLYAELLPTLLQRGHCTTRQGKPPPDPLPPRFR 736
+QQ P Q +Q+ ++ F +PM YAELLP LL+ G R PP LPP +
Sbjct: 397 QQQQRPYQPRQRMPDRR-----FDSLPMSYAELLPELLRLGMVELRT-MAPPTVLPPGYD 450
Query: 737 SDLKCDFHQGALGHDVEGCYALKYIVKKLIDQGKLTFENNVPHVLDNPLPNHAA--VNMI 794
++++CDFH GA GH E C AL++ V+ LID + F VP+V++NP+P H VN I
Sbjct: 451 ANVRCDFHSGAPGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQHGGHRVNNI 509
Query: 795 EVCE-EAPRLDVRNVATPLVPLHIKLCKASLFSHDHAKCLGCLRDPLGCHAVQDDIQSLM 853
E E E ++V +V T L+ + +L ++S CLGC GC ++ IQ +M
Sbjct: 510 EGKEAEDLVVNVDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAESENGCDQLRTGIQGMM 569
Query: 854 NDNLL----------TVSDVCVI-------------VPVFHDPPVK-SVPLKKNAEPLVI 889
++ L TVS + + P + PV SVP+ +A P I
Sbjct: 570 DEGCLQFSRAVKDRGTVSTITIYFKPSEGRGQRVVSAPATNGTPVTISVPVTISA-PTTI 628
Query: 890 RLPGPIPYVSDKAVPYKY-----------------NATMIENGV--EVPLASFATVSNIA 930
G + +AVP+KY N + G+ VP V N+
Sbjct: 629 AASGRRAVENSRAVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVG 688
Query: 931 EGTSAALRSGKVRPPLFQKKVATPTIPPVEEATPTVVSPIATDVNQSGKSIEDSNLDEIL 990
G RSG++ P + + + V G +D ++E +
Sbjct: 689 -GPGGFTRSGRLFAPQPLRDNNAEALAKAKGKQAVVEEEPVQKEAPEGSFEKD--VEEFM 745
Query: 991 RIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHKNTLLKVLEQAYVDHEVTVDRFGGIVG 1050
+IIK+SDYKIVDQL QTPSKIS+LSLLL SEAH+N LLK+L AYV E++V++ G++
Sbjct: 746 KIIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQLEGVMA 805
Query: 1051 NITACNNLWFSEEELPEAGKSHNLALHISVNCKSDMISNVLVDTGSSLNVMPKTTLDQLS 1110
N++ + + F+ +L G++HN ALHI++ CK ++S+VLVDTGSSLNV+PK L ++
Sbjct: 806 NVSTRHGVGFTNLDLTPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKID 865
Query: 1111 YRGTPLRRSTFLVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWI 1170
G L S +V+AFDGS+++V GE+ LP+ IGPE F + F VMDI +YSCLLGRPWI
Sbjct: 866 VEGFVLTPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWI 925
Query: 1171 HDAGAVTSTLHQKLKFVKNGKLVTIHGEEAYLVSQLLSFSCIEA-GSAEGT---AFQGLT 1226
H AGAV+STLHQKLK+V NG++VT+ GEE LVS L SF +E G T AF+ +
Sbjct: 926 HAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVA 985
Query: 1227 IEG---AEPKEAGAAMASLKDAQRAIQEGQTAGWGKVIRLCENKRKEGLGFSPSSRVSSG 1283
+E AE ++ GA++ S K A+ + G+ GWGK++ L + K G+G+ P +G
Sbjct: 986 LEKVAYAEQRKPGASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGYEPLQAEQNG 1045
Query: 1284 -----VFHSAGFVNAISEEATGSG-----------LRPVFVTPGGIATDWDAIDIPSIMH 1327
F SAG +N ATGS +RP PGG +W A ++ +
Sbjct: 1046 QAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRPC--APGGSINNWTAEEVVQVTL 1103
Query: 1328 VSELNHNKPVEHSNPTVPPSFDFPVYEAKDEEGDG--IPYEITRLLEQEKKAIQPHQEEI 1385
++E + +++ F P+Y+A++E + +P E+ RLL QE++ IQPHQEE+
Sbjct: 1104 LTESDLMAFTMNNSVMAQYDFGNPIYQAEEESEEDCELPAELVRLLRQEERVIQPHQEEL 1163
Query: 1386 ELVNIGTEENKREIKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPT 1445
E+VN+GTE+ KREIKIGA LE+ VKR++I++LREY +IFAWSY+DMPGLD IV HR+P
Sbjct: 1164 EVVNLGTEDAKREIKIGAALEDSVKRRLIEMLREYVEIFAWSYQDMPGLDTDIVVHRLPL 1223
Query: 1446 KPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMC 1505
+ CP V+QKLRRT PDMA KIK EVQKQ DAGFL YP WVANIVPVPKKDGKVRMC
Sbjct: 1224 REGCPSVKQKLRRTSPDMATKIKEEVQKQWDAGFLAVTSYPPWVANIVPVPKKDGKVRMC 1283
Query: 1506 VDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP 1565
VD+RDLN+ASPKD+FPLPHIDVLVDNTAQS VFSFMDGFSGYNQIKM+PED EKT+FITP
Sbjct: 1284 VDYRDLNRASPKDDFPLPHIDVLVDNTAQSSVFSFMDGFSGYNQIKMAPEDMEKTTFITP 1343
Query: 1566 WGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQHVEYLTKM 1625
WGTF YKVMPFGL NAGATYQR MTTLFHDM HKE+EVYVDDMI KS EE+H+ L K+
Sbjct: 1344 WGTFYYKVMPFGLKNAGATYQRAMTTLFHDMMHKEIEVYVDDMIAKSQTEEEHLVNLQKL 1403
Query: 1626 FERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPRTEKQVRGF 1685
F+RLRK+KLRLNPNKCTFGVRSGKLLGFIVS+KGIEVDP KV+AI+EMP P+TEKQVRGF
Sbjct: 1404 FDRLRKFKLRLNPNKCTFGVRSGKLLGFIVSEKGIEVDPAKVKAIQEMPEPKTEKQVRGF 1463
Query: 1686 LGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEG 1745
LGRLNYI+RFISH+TATC PIFKLLRKNQ + WND+CQ+AFD IK+YL +PPIL PPV G
Sbjct: 1464 LGRLNYIARFISHLTATCEPIFKLLRKNQAIKWNDDCQKAFDKIKEYLQKPPILTPPVPG 1523
Query: 1746 RPLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAK 1805
RPLIMYL+V + SMGCVLGQ DE+G+KEHAIYYLSKKFTDCETRY++LEKTCCALAWAA+
Sbjct: 1524 RPLIMYLSVTENSMGCVLGQHDESGRKEHAIYYLSKKFTDCETRYSLLEKTCCALAWAAR 1583
Query: 1806 RLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKTQKAIKGSILAD 1865
RLR Y++NHTT LIS+MDP+KYIFEK A+TG++ARWQM+L+EYDI + +QKAIKGSIL+D
Sbjct: 1584 RLRQYMLNHTTLLISKMDPVKYIFEKPALTGRVARWQMILTEYDIQYTSQKAIKGSILSD 1643
Query: 1866 HLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGI 1925
+LA QP++DYQP+ F+FPDE+IMYLK KDCEEPL+EEGPDP+ KW L+FDGAVN G G+
Sbjct: 1644 YLAEQPIEDYQPMMFEFPDEDIMYLKVKDCEEPLVEEGPDPDDKWTLMFDGAVNMNGNGV 1703
Query: 1926 GAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQI 1985
GAV+++P+G HIPF+AR+ F+ TNN AEYEACI GIEEAID+RIK LDIYGDSALV+NQ+
Sbjct: 1704 GAVLINPKGAHIPFSARLTFDVTNNEAEYEACIMGIEEAIDLRIKTLDIYGDSALVVNQV 1763
Query: 1986 KGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSM 2035
G+W T+ LIPYRDY RR+LT+F KV L+H+PRDENQMADALATLSSM
Sbjct: 1764 NGDWNTNQPHLIPYRDYTRRILTFFKKVRLYHVPRDENQMADALATLSSM 1813
>ref|XP_493776.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9927274|dbj|BAB08213.2| Similar to Arabidopsis
thaliana chromosome II BAC F26H6; putative retroelement
pol polyprotein (AC006920) [Oryza sativa (japonica
cultivar-group)]
Length = 2876
Score = 905 bits (2339), Expect = 0.0
Identities = 478/1140 (41%), Positives = 688/1140 (59%), Gaps = 37/1140 (3%)
Query: 1366 EITRLLEQEKKAIQPHQEEIELVNIGTEENKREIKIGATLEEGVKRKIIQLLREYPDIFA 1425
++ L+Q QP +E+ +N+GTE++ R I + L E + L E+ D FA
Sbjct: 1759 DVAGALKQLDDGGQPTIDELVEMNLGTEDDPRPIFVSGMLTEEEREDYRSFLMEFRDCFA 1818
Query: 1426 WSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEY 1485
W+Y++MPGLD + H++ P+ PV+Q RR P+ ++ +EV + I+ GF+ ++Y
Sbjct: 1819 WTYKEMPGLDSRVATHKLAIDPQFRPVKQPPRRLRPEFQDQVIAEVDRLINVGFIKEIQY 1878
Query: 1486 PEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFS 1545
P W+ANIVPV KK+G+VR+CVDFRDLN+A PKD+FPLP +++VD+T S
Sbjct: 1879 PRWLANIVPVEKKNGQVRVCVDFRDLNRACPKDDFPLPITEMVVDSTTG------YGALS 1932
Query: 1546 GYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYV 1605
GYNQIKM D T+F TP G F Y VMPFGL NAGATYQR M + D+ H VE YV
Sbjct: 1933 GYNQIKMDLLDAFDTAFRTPKGNFYYTVMPFGLKNAGATYQRAMQFVLDDLIHHSVECYV 1992
Query: 1606 DDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPD 1665
DDM+VK+ D E H E L +FERLR+++L++NP KC F V+SG LGF++ +GIE++P
Sbjct: 1993 DDMVVKTKDHEHHQEDLRIVFERLRRHQLKMNPLKCAFAVQSGVFLGFVIRHRGIEIEPK 2052
Query: 1666 KVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEA 1725
K++AI MP P+ K +R G+L YI RFIS+++ P KL++K P VW++ECQ
Sbjct: 2053 KIKAILNMPPPQELKDLRKLQGKLAYIRRFISNLSGRIQPFSKLMKKGTPFVWDEECQNG 2112
Query: 1726 FDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTD 1785
FDSIK+YLL PP+L PV+GRPLI+Y+A S+G +L Q ++ G KE A YYLS+
Sbjct: 2113 FDSIKRYLLNPPVLAAPVKGRPLILYIATQPASIGALLAQHNDEG-KEVACYYLSRTMVG 2171
Query: 1786 CETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLL 1845
E Y+ +EK C AL +A K+LRHY++ H LI+R DPI+Y+ + +TG++ +W +L+
Sbjct: 2172 AEQNYSPIEKLCLALIFALKKLRHYMLAHQIQLIARADPIRYVLSQPVLTGRLGKWALLM 2231
Query: 1846 SEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPD 1905
EYDI F QKAIKG LA+ LA P+ D P+ + PDEEI + ++
Sbjct: 2232 MEYDITFVPQKAIKGQALAEFLATHPMPDDSPLIANLPDEEIFTAELQE----------- 2280
Query: 1906 PNSKWGLVFDGA----VNAYG-----KGIGAVIVSPQGHHIPFTARIL-FECTNNMAEYE 1955
+W L FDGA +N G G G V +PQG I + +L EC+NN AEYE
Sbjct: 2281 ---QWELYFDGASRKDINPDGTPRRRAGAGLVFKTPQGGVIYHSFSLLKEECSNNEAEYE 2337
Query: 1956 ACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVEL 2015
A IFG+ A+ M ++ L +GDS L+I QI +E +L+PY ARRL+ F +E+
Sbjct: 2338 ALIFGLLLALSMEVRSLRAHGDSRLIIRQINNIYEVRKPELVPYYTVARRLMDKFEHIEV 2397
Query: 2016 HHIPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDY 2075
H+PR +N ADALA L++ N ++ +R P+ + I + ++ N +
Sbjct: 2398 IHVPRSKNAPADALAKLAAALVFQGDNPAQIVVEERWLLPAVLELIPEEVNIIITNSAEE 2457
Query: 2076 KPWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDM-VLLRCVDEHE 2134
+ W + P +++ L+R ++ +LYKR+Y VLLRCVD E
Sbjct: 2458 EDWRQPFLDYFKHGSLP--EDPVERRQLQRRLPSYIYKAGVLYKRSYGQEVLLRCVDRSE 2515
Query: 2135 AEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVP 2194
A +++ +VH G G H +G M + GYYW + DC + A+ CH CQI+ + H P
Sbjct: 2516 ANRVLQEVHHGVCGGHQSGPKMYHSIRLVGYYWPGIMADCLKTAKTCHGCQIHDNFKHQP 2575
Query: 2195 PHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVA 2254
P L+ WPF WGID+IG I P +S GHRFIL A DYF+KW EA V V
Sbjct: 2576 PAPLHPTVPSWPFDAWGIDVIGLINPPSSRGHRFILTATDYFSKWAEAVPLREVKSSDVI 2635
Query: 2255 KFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANK 2314
F++ +II R+GVP +I +DN + + E++KI+ + S+ Y PQ NG EA NK
Sbjct: 2636 NFLERHIIYRFGVPHRITSDNAKAFKSQKIYRFMEKYKIKWNYSTGYYPQANGMAEAFNK 2695
Query: 2315 NIKRIVQKMVTTY-KDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL 2373
+ +I++K V + +DWH+ L AL YR TVR+ T ATP+SLVYG EAVLPLE+++PSL
Sbjct: 2696 TLGKILKKTVDKHRRDWHDRLYEALWAYRVTVRTPTQATPYSLVYGNEAVLPLEIQLPSL 2755
Query: 2374 RVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGE- 2432
RV + +L++ E + R+ +L+ +EE+R+ A+ + Y+ M A+DK V+ R F+ GE
Sbjct: 2756 RVAIHDELTKDEQIRLRFQELDAVEEERLGALQNLELYRQNMVRAYDKLVKQRVFRKGEL 2815
Query: 2433 -LVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYF 2491
LVL+R I +GK+ P +EGPYV+++A+ GGA L G + P+N +KKYF
Sbjct: 2816 VLVLRRPIVVTHKMKGKFEPKWEGPYVIEQAYDGGAYQLIDHQGSQPMPPINGRFLKKYF 2875
Score = 113 bits (283), Expect = 7e-23
Identities = 80/258 (31%), Positives = 125/258 (48%), Gaps = 17/258 (6%)
Query: 931 EGTSAALRSGKVRPPLFQKKVATPTIP-PVEEATPTVVSPIATDVNQSGKSIEDSNLDEI 989
E + +LR GK P + KV P + P ++A+P +P A + K
Sbjct: 1252 EEVNISLRGGKTLPDPHKSKV--PNVDKPAKKASPPGEAPEAPETKTGSKEKP------- 1302
Query: 990 LRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHKNTLLKVLEQAYVDHEVTVDRFGGIV 1049
DYK++ L + P+ +SV L+ + L+K L+ V +EV + + +
Sbjct: 1303 -----AVDYKVLAHLKRIPALLSVYDALMMVPDLREALIKALQAPEV-YEVDMAKHR-LY 1355
Query: 1050 GNITACNNLWFSEEELPEAGKSHNLALHISVNCKSDMISNVLVDTGSSLNVMPKTTLDQL 1109
N N + F++E+ G HN L+I N S + +L+D GS++N++P +L +
Sbjct: 1356 DNPLFVNEITFADEDNIIKGGDHNRPLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRA 1415
Query: 1110 SYRGTPLRRSTFLVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPW 1169
+ L ++ FD K LG I + I + +F V F V++ N SYS LLGRPW
Sbjct: 1416 GFTTKDLEPIDVVICGFDNQGKPTLGAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPW 1475
Query: 1170 IHDAGAVTSTLHQKLKFV 1187
IH V STLHQ LKF+
Sbjct: 1476 IHKYRVVPSTLHQCLKFL 1493
>dbj|BAD18986.1| GAG-POL precursor [Vitis vinifera]
Length = 1027
Score = 768 bits (1982), Expect = 0.0
Identities = 420/1063 (39%), Positives = 616/1063 (57%), Gaps = 39/1063 (3%)
Query: 1431 MPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVA 1490
M G+ P I HR+ PVRQ++RR HPD I++E+ K ++AGF+ V YP+W+A
Sbjct: 1 MKGIHPSITSHRLNVVSTARPVRQRIRRFHPDRQRVIRNEIDKLLEAGFIREVSYPDWLA 60
Query: 1491 NIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQI 1550
N+V VPKK+GK R+CVD+ +LN A PKD+FPLP ID +VD+T+ + SF+D FSGY+QI
Sbjct: 61 NVVVVPKKEGKWRVCVDYTNLNNACPKDSFPLPRIDQIVDSTSGQGMLSFLDAFSGYHQI 120
Query: 1551 KMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIV 1610
MSP+D EK +FITP +CYKVMPFGL NAGATYQR MT +F + VEVY+DD++V
Sbjct: 121 PMSPDDEEKIAFITPHDLYCYKVMPFGLKNAGATYQRLMTKIFKPLIGHSVEVYIDDIVV 180
Query: 1611 KSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAI 1670
KS EQH+ +L ++F LR+Y ++LNP+KC FGV + K LGF+VSQ+GIEV PD+V+A+
Sbjct: 181 KSKTREQHILHLQEVFYLLRRYGMKLNPSKCAFGVSARKFLGFMVSQRGIEVSPDQVKAV 240
Query: 1671 REMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIK 1730
E P PR +K+++ G+L + RFI+ P F +RK W D CQ A + IK
Sbjct: 241 METPPPRNKKELQRLTGKLVALGRFIARFIDELRPFFLAIRKAGTHGWTDNCQNALERIK 300
Query: 1731 KYLLEPPILVPPVEGRPLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRY 1790
YL++PPIL P+ L MYLAV + ++ VL + + K++ IYY+S+ D ETRY
Sbjct: 301 HYLMQPPILSSPIPKEKLYMYLAVSEWAISAVL-FRCPSPKEQKPIYYVSRALADVETRY 359
Query: 1791 TMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDI 1850
+ +E AL AA++LR Y H ++ P++ I K +TG++ +W + LSE+ I
Sbjct: 360 SKMELISLALRSAAQKLRPYFQAHPV-IVLTDQPLRNILHKPDLTGRMLQWAIELSEFGI 418
Query: 1851 VFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKW 1910
F+ + ++KG ++AD + +E+ +P EG W
Sbjct: 419 EFQPRLSMKGQVMADFV----------LEYS--------------RKPGQHEGSRKKEWW 454
Query: 1911 GLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIK 1970
L DGA + G G+G ++ SP G H+ R+ F +NN AEYEA + G++ A+ + +
Sbjct: 455 TLRVDGASRSSGSGVGLLLQSPTGEHLEQAIRLGFSASNNEAEYEAILSGLDLALALSVS 514
Query: 1971 HLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALA 2030
L I+ DS LV+ ++ E+E A++ Y R L FT+ + I R +N+ ADALA
Sbjct: 515 KLRIFSDSQLVVKHVQEEYEAKDARMARYLAKVRNTLQQFTEWTIEKIKRADNRRADALA 574
Query: 2031 TLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSRE 2090
+++ + +P I VQ S + + D + W DI +++ +
Sbjct: 575 GIAASLSIKEAILLP-IHVQTNPSVSEI----SICSTTEAPQADDQEWMNDITEYIRTGT 629
Query: 2091 YPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTH 2150
PG KQ K +R A+RF L G LYKR++ LRC+ EA+ ++ ++H+G +G H
Sbjct: 630 L-PGDPKQAHK-VRVQAARFTLIGGHLYKRSFTGPYLRCLGHSEAQYVLAELHEGIYGNH 687
Query: 2151 ATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSMW 2210
+ G +++ + GYYW M+ + Y ++C KCQ YA H+P L +S PWPF+ W
Sbjct: 688 SGGRSLAHRAHSQGYYWPTMKKEAAAYVKRCDKCQRYAPIPHMPSTTLKSISGPWPFAQW 747
Query: 2211 GIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSK 2270
G+D++ R P A +F+LVA DYF+KWVEA +Y + + V KF+ NIICR+G+P
Sbjct: 748 GMDIV-RPLPTAPAQKKFLLVATDYFSKWVEAEAYASTKDKDVTKFVWKNIICRFGIPQT 806
Query: 2271 IITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVTTYK-D 2329
II DNG ++ + C E I + S+P PQ NG EA NK + ++K + K
Sbjct: 807 IIADNGPQFDSIAFRNFCSELNIRNSYSTPRYPQSNGQAEATNKTLITALKKRLEQAKGK 866
Query: 2330 WHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQS 2389
W E LP L YRTT TG TPF+L YGM+AV+P+E+ +P++ AK S+A
Sbjct: 867 WVEELPGVLWAYRTTPGRPTGNTPFALAYGMDAVIPIEIGLPTIWT-NAAKQSDANMQLG 925
Query: 2390 RYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPD-PRGK 2448
R L+ +E R A R YQ R +++KVRPR K G LVL++ + GK
Sbjct: 926 R--NLDWTDEVRESASIRMADYQQRASAHYNRKVRPRSLKNGTLVLRKFFENTTEVGAGK 983
Query: 2449 WAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYF 2491
+ N+EGPY+V KA GA L +DG L P N +K+Y+
Sbjct: 984 FQANWEGPYIVSKASDNGAYHLQKLDGTPLLRPWNVSNLKQYY 1026
>gb|AAF79618.1| F5M15.26 [Arabidopsis thaliana] gi|25402907|pir||H86337 protein
F5M15.26 [imported] - Arabidopsis thaliana
Length = 1838
Score = 763 bits (1969), Expect = 0.0
Identities = 432/1117 (38%), Positives = 632/1117 (55%), Gaps = 49/1117 (4%)
Query: 1386 ELVNIGTEENKREIKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPT 1445
E+VNI + R + +GA + ++ ++I LL+ FAWS EDM G+DP I H +
Sbjct: 759 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 818
Query: 1446 KPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMC 1505
P PV+QK R+ P+ A + EV+K + AG ++ V+YPEW+AN V V KK+GK R+C
Sbjct: 819 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 878
Query: 1506 VDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITP 1565
VD+ DLNKA PKD++PLPHID LV+ T+ + + SFMD FSGYNQI M +D+EKTSF+T
Sbjct: 879 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 938
Query: 1566 WGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQHVEYLTKM 1625
GT+CYKVM FGL NAGATYQR + + D + VEVY+DDM+VKS E HVE+L+K
Sbjct: 939 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 998
Query: 1626 FERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPRTEKQVRGF 1685
F+ L Y ++LNP KCTFGV SG+ LG++V+++GIE +P ++RAI E+P+PR ++V+
Sbjct: 999 FDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNAREVQRL 1058
Query: 1686 LGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEG 1745
GR+ ++RFIS T C P + LL++ W+ + +EAF+ +K YL PPILV P G
Sbjct: 1059 TGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPILVKPEVG 1118
Query: 1746 RPLIMYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAK 1805
L +Y+AV D ++ VL ++D ++ I+Y SK + ETRY ++EK A+ +A+
Sbjct: 1119 ETLYLYIAVSDHAVSSVLVREDR--GEQRPIFYTSKSLVEAETRYPVIEKAALAVVTSAR 1176
Query: 1806 RLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKTQKAIKGSILAD 1865
+LR Y +HT +++ P++ + +G++ +W + LSEYDI F+ + A+K +LAD
Sbjct: 1177 KLRPYFQSHTIAVLTD-QPLRVALHSPSQSGRMTKWAVELSEYDIDFRPRPAMKSQVLAD 1235
Query: 1866 HLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGI 1925
L PL + EE W L DG+ +A G GI
Sbjct: 1236 FLIELPLQSAERAVSGNRGEE-----------------------WSLYVDGSSSARGSGI 1272
Query: 1926 GAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQI 1985
G +VSP + + R+ F TNN+AEYE I G+ A M+I + + DS L+ Q+
Sbjct: 1273 GIRLVSPTAEVLEQSFRLRFVATNNVAEYEVLIAGLRLAAGMQITTIHAFTDSQLIAGQL 1332
Query: 1986 KGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDV- 2044
GE+E + K+ Y + + F +L IPR +N ADALA L+ + +D+
Sbjct: 1333 SGEYEAKNEKMDAYLKIVQLMTKDFENFKLSKIPRGDNAPADALAALA----LTSDSDLR 1388
Query: 2045 PLIKVQRLERPS-----HVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSREYPPGASKQD 2099
+I V+ +++PS V + + + D W +I+ +L G D
Sbjct: 1389 RIIPVESIDKPSIDSTDAVEIVNTIRSSNAPDPADPTDWRVEIRDYL-----SDGTLPSD 1443
Query: 2100 KKTLRRL---ASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTM 2156
K T RRL A+++ L + L K + +L C+ E ++M + H+G G H+ G +
Sbjct: 1444 KWTARRLRIKAAKYTLMKEHLLKVSAFGAMLNCLHGTEINEIMKETHEGAAGNHSGGRAL 1503
Query: 2157 SRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSMWGIDMIG 2216
+ KL + G+YW M DC + KC +CQ +A IH P L +P+PF W +D++G
Sbjct: 1504 ALKLKKLGFYWPTMISDCKTFTAKCEQCQRHAPTIHQPTELLRAGVAPYPFMRWAMDIVG 1563
Query: 2217 RIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNG 2276
+ AS RFILV DYFTKWVEA SY + V F+ IICR+G+P +IITDNG
Sbjct: 1564 PM--PASRQKRFILVMTDYFTKWVEAESYATIRANDVQNFVWKFIICRHGLPYEIITDNG 1621
Query: 2277 TNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVTTYKD-WHEMLP 2335
+ + + C +KI + S+P PQ NG EA NK I ++K + K W + L
Sbjct: 1622 SQFISLSFENFCASWKIRLNKSTPRYPQGNGQAEATNKTILSGLKKRLDEKKGAWADELD 1681
Query: 2336 YALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLN 2395
L YRTT RS+T TPF+ YGMEA+ P EV SLR M K E + D+L+
Sbjct: 1682 GVLWSYRTTPRSATDQTPFAHAYGMEAMAPAEVGYSSLRRSMMVKNPELN-DRMMLDRLD 1740
Query: 2396 LIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPD-PRGKWAPNYE 2454
+EE R A+ R Q+YQ +++KV R F VG+LVL++ + GK N+E
Sbjct: 1741 DLEEIRNAALCRIQNYQLAAAKHYNQKVHNRHFDVGDLVLRKVFENTAEINAGKLGANWE 1800
Query: 2455 GPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYF 2491
G Y V K G L M G +P N+ +K+Y+
Sbjct: 1801 GSYQVSKIVRPGDYELLTMSGTAVPRTWNSMHLKRYY 1837
Score = 73.6 bits (179), Expect = 8e-11
Identities = 48/176 (27%), Positives = 82/176 (46%), Gaps = 7/176 (3%)
Query: 1060 FSEEELPEAGKSHNLALHISVNCKSDMISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRS 1119
F++ +L HN L + + ++ VL+DTGSS++++ K L ++ ++
Sbjct: 602 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 661
Query: 1120 TFLVKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTST 1179
+ + FDG +G I LPI +G V F V+ A Y+ +LG PWIH A+ ST
Sbjct: 662 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 721
Query: 1180 LHQKLKFVKNGKLVTIHG-------EEAYLVSQLLSFSCIEAGSAEGTAFQGLTIE 1228
HQ +KF + + T+ +Y S+L + ++ T G+ E
Sbjct: 722 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 777
>gb|AAP51864.1| putative retroelement pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|37530550|ref|NP_919577.1| putative
retroelement pol polyprotein [Oryza sativa (japonica
cultivar-group)] gi|18581535|gb|AAK52540.2| Putative
retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 752 bits (1941), Expect = 0.0
Identities = 419/1105 (37%), Positives = 632/1105 (56%), Gaps = 66/1105 (5%)
Query: 1394 ENKREIKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVR 1453
++ EI I + +G + +I+LL+E+ D F W Y +MPG IVEHR+P KP P +
Sbjct: 528 DDLEEIDISPAIGQG-RHLLIELLKEFRDCFTWEYYEMPGHSRSIVEHRLPLKPGVRPHQ 586
Query: 1454 QKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNK 1513
Q RR DM +K+E+++ DAGF+ Y EWV++IVPV KK+GKVR+C+DFR LNK
Sbjct: 587 QLPRRCKADMLEPVKAEIKRLYDAGFIRPCRYAEWVSSIVPVIKKNGKVRVCIDFRYLNK 646
Query: 1514 ASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPW--GTFCY 1571
A+PKD +P+ D LVD + K+ SFMDG +GYNQI M+ ED KT+F P G F +
Sbjct: 647 ATPKDEYPMLVADQLVDAASGHKILSFMDGNAGYNQIFMAEEDIHKTTFRCPGAIGLFEW 706
Query: 1572 KVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRK 1631
V+ F L +AGATYQR M ++HD+ VEVY+DD++VKS + E H+ L K+FER RK
Sbjct: 707 VVITFVLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIADLRKVFERTRK 766
Query: 1632 YKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPRTEKQVRGFLGRLNY 1691
Y L++NP KC FGV +G+ LGF+V ++GIEV + AI+++ P + +++ +G++N+
Sbjct: 767 YGLKMNPTKCAFGVSAGQFLGFLVHERGIEVTQRSINAIKKIKPPEDKTELQEMIGKINF 826
Query: 1692 ISRFISHMTATCGPIFKLLR--KNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEGRPLI 1749
+ RFIS ++ P LLR +Q W E Q+A D+IK+YL PP+L+PP +G P
Sbjct: 827 VRRFISILSGKLEPFTPLLRLKADQQFAWGAEQQKALDNIKEYLSSPPVLIPPQKGIPFR 886
Query: 1750 MYLAVFDESMGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKRLRH 1809
+YL+ D+S+ VL Q+ E +KE ++YLS++ + ETRY+ +EK C L ++ RLRH
Sbjct: 887 LYLSAGDKSISSVLIQELE--RKERVVFYLSRRLLNAETRYSPVEKLCLCLYFSCTRLRH 944
Query: 1810 YLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKTQKAIKGSILADHLAY 1869
YL+++ +I + D +KY+ + G+I +W L+E+D+ +++ KAIKG +A+ +
Sbjct: 945 YLLSNECTVICKADVVKYMLSAPILKGRIGKWIFSLTEFDLRYESPKAIKGQAIANFIV- 1003
Query: 1870 QPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVI 1929
D+ D+ I G W L FDG+V +G GIG VI
Sbjct: 1004 -----------DYRDDSI---------------GSVEVVPWTLFFDGSVCTHGCGIGLVI 1037
Query: 1930 VSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEW 1989
+SP+G F I TNN AEYEA + G++ ++ ++I GDS LVI+Q+ GE+
Sbjct: 1038 ISPRGACFEFAYTIKPYATNNQAEYEAVLKGLQLLKEVEADTIEIMGDSLLVISQLAGEY 1097
Query: 1990 ETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFRVNHWNDVPLIKV 2049
E + LI Y + + L+ F V L H+ R++N A+ LA +S +++ +IK
Sbjct: 1098 ECKNDTLIVYNEKCQELMREFRLVTLKHVSREQNIEANDLAQGASGYKL-------MIKH 1150
Query: 2050 QRLERPSHVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSREYPPGASKQDKKTLRRLASR 2109
++E V+ W Y + Q+L S+ + LR A +
Sbjct: 1151 VQIEVA----------------VITADDWRYYVYQYLQD------PSQSASRKLRYKALK 1188
Query: 2110 FLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMA 2169
++L D LY R D VLL+C+ +A+ + +VH+G GTH + H M L AGY+W
Sbjct: 1189 YILLDDELYYRTIDGVLLKCLSTDQAKVAIGEVHEGICGTHQSAHKMKWLLRCAGYFWPT 1248
Query: 2170 MEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFI 2229
M DC++Y + C CQ + P A+N + PWPF WGIDMIG I P +S GH+FI
Sbjct: 1249 MLEDCFRYYKGCQDCQKFGAIQRAPASAMNPIIKPWPFRGWGIDMIGMINPPSSKGHKFI 1308
Query: 2230 LVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCE 2289
LVA DYFTKWVEA V +F++ +II R G+P I TD G+ ++ +
Sbjct: 1309 LVATDYFTKWVEAIPLKKVDSGDAIQFVQEHIIYRVGIPQTITTDQGSIFVSDEFIQFAD 1368
Query: 2290 EFKIEHHNSSPYRPQMNGAVEAANKNIKRIVQKMVTTY-KDWHEMLPYALHGYRTTVRSS 2348
I+ NSSPY Q NG EA+NK++ +++++ ++ Y + WH L AL YR S
Sbjct: 1369 SMGIKLLNSSPYYAQANGQAEASNKSLIKLIKRKISDYPRQWHTRLAEALWSYRMACHGS 1428
Query: 2349 TGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARG 2408
P+ LVY +A+LP EV I S R ++ L+ E+ D+ + + R+ A+A+
Sbjct: 1429 IQVPPYKLVYRHKAILPWEVRIGSRRTELQNDLTADEYSNLMADEREDLVQSRLRALAKV 1488
Query: 2409 QSYQARMKTAFDKKVRPREFKVGELVLK--RRISQQPDPRGKWAPNYEGPYVVKKAFSGG 2466
+ R+ ++KKV P++F GEL+ K I + GKW+PN+EGP+ + K S G
Sbjct: 1489 IKDKERVTRHYNKKVVPKDFSEGELIWKLILPIGTRDSKFGKWSPNWEGPFQIHKVVSKG 1548
Query: 2467 ALILTHMDGVELPNPVNADIVKKYF 2491
A +L +DG +N +KKY+
Sbjct: 1549 AYMLQGLDGEVYDRALNGKYLKKYY 1573
Score = 77.4 bits (189), Expect = 6e-12
Identities = 50/179 (27%), Positives = 90/179 (49%), Gaps = 3/179 (1%)
Query: 1064 ELPEAGKSHNLA-LHISVNCKSDMISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFL 1122
E PE ++ +L L+I+ +S ++VD G+++N+MP T +L L ++ +
Sbjct: 351 EKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMV 410
Query: 1123 VKAFDGSRKNVLGEIDLPITIGPENFLVTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQ 1182
+K F G+ +G +++ +T+G + TF V+D SYS LLGR WIH + ST+HQ
Sbjct: 411 LKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTIHQ 470
Query: 1183 KLKFVKNGKLVTIHGEEAYLVSQLLSFSCIEAGSAEGTAFQGLTIEGAEPKEAGAAMAS 1241
L G + I ++ L + S+ C E + T++ + K+ M++
Sbjct: 471 CL-IQWQGDKIEIVPADSQLKMENPSY-CFEGVMEGSNVYTKDTVDDLDDKQGQGLMSA 527
>ref|XP_463537.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 2001
Score = 701 bits (1809), Expect = 0.0
Identities = 395/1098 (35%), Positives = 618/1098 (55%), Gaps = 56/1098 (5%)
Query: 1399 IKIGATLEEGVKRKIIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRR 1458
I IG LE+ ++ +I+++++E +FAWS +++ G+D ++EH + K P +QKLRR
Sbjct: 956 ILIGENLEKHIEEEILKVVKENMAVFAWSPDELQGVDRSLIEHNLAIKSGYKPKKQKLRR 1015
Query: 1459 THPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 1518
D K E++K + A + V +PEW+AN V V K +GK RMC+DF DLNKA PKD
Sbjct: 1016 MSTDRQQAAKIELEKLLKAKVIREVMHPEWLANPVLVKKANGKWRMCIDFTDLNKACPKD 1075
Query: 1519 NFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGL 1578
+FPLP ID LVD TA ++ SF+D +SGY+Q+ M ED EKTSFITP+GT+C+ MPFGL
Sbjct: 1076 DFPLPRIDQLVDATAGCELMSFLDAYSGYHQVFMVKEDEEKTSFITPFGTYCFIRMPFGL 1135
Query: 1579 INAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNP 1638
NAGAT+ R + + + VE Y+DD++VKS H + L + FE LRK ++LNP
Sbjct: 1136 KNAGATFARLIGKVLAKQLGRNVEAYIDDIVVKSKQAFTHGKDLQETFENLRKCSVKLNP 1195
Query: 1639 NKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISH 1698
KC FGVR+GKLLGF+VS++GIE +PDK+ AI +M P+ ++V+ GR+ +SRF+S
Sbjct: 1196 EKCVFGVRAGKLLGFLVSKRGIEANPDKIAAIHQMEPPKNTREVQRLTGRMASLSRFLSK 1255
Query: 1699 MTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDES 1758
P FK LR W ECQ+AFD +KKYL E P L P +G+PL+MY+A +
Sbjct: 1256 SAEKGLPFFKTLRGANTFEWTAECQQAFDDLKKYLHEMPTLASPPKGQPLLMYVAATPAT 1315
Query: 1759 MGCVLGQQDETGKKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWL 1818
+ VL Q++E ++ +Y++S+ +TRY+ +EK A+ A+++LRHY ++H +
Sbjct: 1316 VSAVLVQEEE--NRQVPVYFVSEALQGPKTRYSEVEKLIYAIVMASRKLRHYFLSHDITI 1373
Query: 1819 ISRMDPIKYIFEKAAVTGKIARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPI 1878
S PI + V G+IA+W M L +D+ + ++ AIK +LAD +A ++ P
Sbjct: 1374 PSAY-PIGEVLTNKEVAGRIAKWAMELLPFDLKYISRTAIKSQVLADFVA-----EWTPN 1427
Query: 1879 EFDFPDEEIMYLKSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIP 1938
E +E+ + W + DGA NA G G AV+ +P +
Sbjct: 1428 E--------------------VEQQEEVKKPWIVFSDGACNAAGAGAAAVVKTPMKQTLK 1467
Query: 1939 FTARILFECTNNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIP 1998
++ +++F TNN AEYE + + +A + + L + DS LV +E +
Sbjct: 1468 YSVQLVFPSTNNTAEYEGVLLAMRKARALGARRLIVKTDSKLVAGHFSKSFEAKEETMAK 1527
Query: 1999 YRDYARRLLTYFTKVELHHIPRDENQMADALATLSSMFR--VNHWNDVPLIKVQRLERPS 2056
Y + AR +F + + I R+EN AD LA ++ + N + D+ + +PS
Sbjct: 1528 YLEEARLNEKHFLGITVKAITREENGEADELAKAAATGQPLENSFFDI-------ITQPS 1580
Query: 2057 HVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDI 2116
+ I + G+ W I ++L+S + P +++ K ++ ++ ++ +
Sbjct: 1581 YEKKEVACIQREGD-------WREPILKYLVSAQLP--EKEEEAKRIQLMSKKYKVVEGQ 1631
Query: 2117 LYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQ 2176
LYK LL+CV E +++ ++H+G G H +++ K++R G YW + D +
Sbjct: 1632 LYKSGVTAPLLKCVTREEGMKMVVEIHEGLCGAHQAPWSVASKVIRQGIYWPTIMKDTEK 1691
Query: 2177 YARKCHKCQIYADKIHVPPHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYF 2236
Y + C CQ + PP L + WPF WGID++G + P+A RF++VAI+YF
Sbjct: 1692 YIKTCKACQKFGPMTKAPPKELQPIPPVWPFYRWGIDIVGPL-PRAKGDLRFVIVAIEYF 1750
Query: 2237 TKWVEAASYTNVTKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHH 2296
++W+EA + +T V KF+ NIICR+G+P +I+ DNG + Q +C+ ++ +
Sbjct: 1751 SRWIEAEAVARITSAAVQKFVWKNIICRFGIPKEIVCDNGKQFESGKFQDMCKGLNLQIN 1810
Query: 2297 NSSPYRPQMNGAVEAANKNIKRIVQKMV--TTYKDWHEMLPYALHGYRTTVRSSTGATPF 2354
+S PQ NG VE AN I ++K + + W E L L RTTV STG TPF
Sbjct: 1811 FASVGHPQTNGVVERANGKIMEAIKKRLEGSAKGKWPEDLLSVLWALRTTVVRSTGMTPF 1870
Query: 2355 SLVYGMEAVLPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQAR 2414
LVYG EA+ P EV S R+I + K E R L +++E R++A+ + SY
Sbjct: 1871 RLVYGDEAMTPSEVGAHSPRMIFDQKDEE-----GREITLEMLDEIRVEALEKMASYTEG 1925
Query: 2415 MKTAFDKKVRPREFKVGELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMD 2474
K+ +++KV+ R + G+LVLK+ +++ GK +EGP++VKK GA L ++D
Sbjct: 1926 TKSYYNQKVKTRPIEEGDLVLKKVLNEV--AVGKLESKWEGPFIVKKKTETGAFKLAYLD 1983
Query: 2475 GVELPNPVNADIVKKYFA 2492
G EL + NA +KK++A
Sbjct: 1984 GEELKHTWNAVSLKKFYA 2001
Score = 44.3 bits (103), Expect = 0.054
Identities = 31/132 (23%), Positives = 60/132 (44%), Gaps = 4/132 (3%)
Query: 1072 HNLALHISVNCKSDMISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKAFDGSRK 1131
H L +SV + VL+D GSS +V+ ++ L + ++ F G +
Sbjct: 772 HQDPLVVSVEIAQCEVQRVLIDGGSSADVLFYDAFKKMQIPEDRLTNAGVPLQGFGGQQV 831
Query: 1132 NVLGEIDLPITIGPENFL----VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFV 1187
+ +G+I L + G + + F V+D+ Y+ +LGR I+ A+ + +K
Sbjct: 832 HAIGKISLQVVFGKGTNVRKEEIVFDVVDMPYQYNAILGRSTINIFEAIIHHNYICMKLP 891
Query: 1188 KNGKLVTIHGEE 1199
++T+ GE+
Sbjct: 892 GLRGVITVRGEQ 903
>ref|XP_475022.1| OSJNBb0093G06.3 [Oryza sativa (japonica cultivar-group)]
gi|38347562|emb|CAE04995.2| OSJNBb0093G06.3 [Oryza sativa
(japonica cultivar-group)]
Length = 1986
Score = 699 bits (1805), Expect = 0.0
Identities = 407/1139 (35%), Positives = 613/1139 (53%), Gaps = 53/1139 (4%)
Query: 1364 PYEITRLLEQEKKAIQPHQEEIELVNIGTEENKRE-IKIGATLEEGVKRKIIQLLREYPD 1422
P + L K+A+ +E ++ + RE I++ AT E +I L+ D
Sbjct: 891 PRGVLSLRSDIKQAVTCDKESCDMAQTREMASAREEIRLAATTESA----LITFLQNNKD 946
Query: 1423 IFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMT 1482
IFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 947 IFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKE 1006
Query: 1483 VEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMD 1542
V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D
Sbjct: 1007 VLHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLD 1066
Query: 1543 GFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVE 1602
+SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + VE
Sbjct: 1067 CYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1126
Query: 1603 VYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEV 1662
YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1127 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1186
Query: 1663 DPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDEC 1722
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1187 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDNFQWGPEA 1246
Query: 1723 QEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYY 1778
Q+AF+ K+ L +PP+L P PL++Y++ + + VL +++E G K + IY+
Sbjct: 1247 QKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYF 1306
Query: 1779 LSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKI 1838
+S+ D +TRY ++K + ++L HY H+ +++ P+ + G+I
Sbjct: 1307 VSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDVLHNREANGRI 1365
Query: 1839 ARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEP 1898
A+W + L DI FK + +IK LAD +A ++ + D P E+I +
Sbjct: 1366 AKWALELMSLDISFKPRTSIKSQALADFVA-----EWTECQEDTPVEKIEH--------- 1411
Query: 1899 LIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACI 1958
W + FDG+ G G G V++SP G + + I F ++N+AEYEA +
Sbjct: 1412 -----------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALL 1460
Query: 1959 FGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHI 2018
G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H+
Sbjct: 1461 HGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHV 1520
Query: 2019 PRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPW 2078
R N+ AD LA S R +DV V+ L P+ + V+ W
Sbjct: 1521 LRHNNEAADRLANFGSK-REAAPSDV---FVEHLYTPTVPHKDTTQDADTHDVVMVEADW 1576
Query: 2079 YYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQL 2138
+FL S+E P K + + + R + +++ LYK++ +L RCV E QL
Sbjct: 1577 REPFIRFLSSQELP--QDKDEAERISRRSKLYVMHESELYKKSPSGILQRCVSLEEGRQL 1634
Query: 2139 MHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHAL 2198
+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P L
Sbjct: 1635 LKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQEL 1694
Query: 2199 NVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIK 2258
+ WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T F
Sbjct: 1695 QTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF- 1752
Query: 2259 NNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-- 2316
NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG VE AN I
Sbjct: 1753 INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQ 1812
Query: 2317 ---KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL 2373
R+ ++ W + LP L RTT +TG +PF LVYG EA+LP EVE SL
Sbjct: 1813 GIKARVFDRLKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESL 1872
Query: 2374 RVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGEL 2433
R E + + R D L+ +EE R A+ + Y ++ ++ VR R F VG+L
Sbjct: 1873 RF---RNFREERYEEDRVDDLHRLEEAREAALIQSARYLQGLRRYHNRNVRSRAFLVGDL 1929
Query: 2434 VLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
VL++ Q R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1930 VLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 1986
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 784 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 843
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 844 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 887
>gb|AAP52501.1| putative gag-pol precursor [Oryza sativa (japonica cultivar-group)]
gi|37531824|ref|NP_920214.1| putative gag-pol precursor
[Oryza sativa (japonica cultivar-group)]
gi|22128685|gb|AAM92798.1| putative gag-pol precursor
[Oryza sativa (japonica cultivar-group)]
Length = 1986
Score = 698 bits (1801), Expect = 0.0
Identities = 410/1141 (35%), Positives = 615/1141 (52%), Gaps = 57/1141 (4%)
Query: 1364 PYEITRLLEQEKKAIQPHQEEIELVNIGTEENKRE-IKIGATLEEGVKRKIIQLLREYPD 1422
P + L K+A+ +E ++ + RE I++ AT E +I L+ D
Sbjct: 891 PRGVLTLRSDIKQAVTCDKESCDMAQTREIASAREEIRLAATTESA----LITFLQNNKD 946
Query: 1423 IFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMT 1482
IFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 947 IFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKE 1006
Query: 1483 VEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMD 1542
V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D
Sbjct: 1007 VLHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLD 1066
Query: 1543 GFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVE 1602
+SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + VE
Sbjct: 1067 CYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1126
Query: 1603 VYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEV 1662
YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1127 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1186
Query: 1663 DPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDEC 1722
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1187 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDNFQWGPEA 1246
Query: 1723 QEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYY 1778
Q+AF+ K+ L +PP+L P PL++Y++ + + VL +++E G K + IY+
Sbjct: 1247 QKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYF 1306
Query: 1779 LSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKI 1838
+S+ D + RY ++K + ++L HY H+ +++ P+ + G+I
Sbjct: 1307 VSEVLADSKARYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDVLHNREANGRI 1365
Query: 1839 ARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEP 1898
A+W + L DI FK + +IK LAD +A ++ + D P E+I +
Sbjct: 1366 AKWALELMSLDISFKPRTSIKSQALADFVA-----EWTECQEDTPVEKIEH--------- 1411
Query: 1899 LIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACI 1958
W + FDG+ G G G V++SP G + + I F ++N+AEYEA +
Sbjct: 1412 -----------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALL 1460
Query: 1959 FGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHI 2018
G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H+
Sbjct: 1461 HGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHV 1520
Query: 2019 PRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPS--HVFAIGDVIDQAGENVVDYK 2076
R N+ AD LA S R +DV V+ L P+ H D D +V+
Sbjct: 1521 LRHNNEAADRLANFGSK-RETAPSDV---FVEHLYTPTVPHKDTTQDA-DTRNIAMVE-A 1574
Query: 2077 PWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAE 2136
W +FL S+E P K + + + R + ++L LYK++ +L RCV E
Sbjct: 1575 DWREPFIRFLTSQELP--QDKDEAERISRRSKLYVLHESELYKKSPSGILQRCVSLEEGR 1632
Query: 2137 QLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPH 2196
QL+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P
Sbjct: 1633 QLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQ 1692
Query: 2197 ALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKF 2256
L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T F
Sbjct: 1693 ELQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDF 1751
Query: 2257 IKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI 2316
NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG VE AN I
Sbjct: 1752 F-INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMI 1810
Query: 2317 -----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIP 2371
R+ ++ W + LP L RTT +TG +PF LVYG EA+LP EVE
Sbjct: 1811 LQGIKARVFDRLKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFE 1870
Query: 2372 SLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVG 2431
SLR E + + R D L+ +EE R A+ + Y ++ ++ VR R F VG
Sbjct: 1871 SLRF---RNFREERYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRAFLVG 1927
Query: 2432 ELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYF 2491
+LVL++ Q R K +P +EGP+++ + G+ L DG + N N + +++++
Sbjct: 1928 DLVLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFY 1985
Query: 2492 A 2492
A
Sbjct: 1986 A 1986
Score = 47.4 bits (111), Expect = 0.006
Identities = 33/117 (28%), Positives = 55/117 (46%), Gaps = 5/117 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTF-LVKAFDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 784 LRRTLIDGGSALNILFAKTLDDMQIPRSELKSSNAPFHGVIPGLSATPLGQITLPVTFGT 843
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTIHGE 1198
ENF ++F+V D +Y +LG P + AV + +K ++T+ +
Sbjct: 844 RENFRTENISFEVADFETAYHAILGHPALAKFMAVPHYTYMMMKMPGPRGVLTLRSD 900
>ref|XP_475587.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|46391119|gb|AAS90646.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1991
Score = 698 bits (1801), Expect = 0.0
Identities = 405/1139 (35%), Positives = 613/1139 (53%), Gaps = 53/1139 (4%)
Query: 1364 PYEITRLLEQEKKAIQPHQEEIELVNIGTEENKRE-IKIGATLEEGVKRKIIQLLREYPD 1422
P + L K+A+ +E ++ + RE I++ AT E +I L+ D
Sbjct: 896 PRGVLTLRSDIKQAVTCDKESCDMAQTREMASAREEIRLAATTESA----LITFLQNNKD 951
Query: 1423 IFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMT 1482
IFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 952 IFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKE 1011
Query: 1483 VEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMD 1542
V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D
Sbjct: 1012 VLHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLD 1071
Query: 1543 GFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVE 1602
+SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + VE
Sbjct: 1072 CYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1131
Query: 1603 VYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEV 1662
YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1132 AYVDDVVVKTKQKDDLILDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1191
Query: 1663 DPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDEC 1722
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1192 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDNFQWGPEA 1251
Query: 1723 QEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYY 1778
Q+AF+ K+ L +PP+L P PL++Y++ + + VL +++E G K + IY+
Sbjct: 1252 QKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYF 1311
Query: 1779 LSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKI 1838
+S+ D +TRY ++K + ++L HY H+ +++ P+ + G+I
Sbjct: 1312 VSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDVLHNREANGRI 1370
Query: 1839 ARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEP 1898
A+W + L DI FK + +IK LAD +A ++ + D P E+I +
Sbjct: 1371 AKWALELMSLDISFKPRTSIKSQALADFVA-----EWTECQEDTPVEKIEH--------- 1416
Query: 1899 LIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACI 1958
W + FDG+ G G G V++SP G + + I F ++N+AEYEA +
Sbjct: 1417 -----------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALL 1465
Query: 1959 FGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHI 2018
G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H+
Sbjct: 1466 HGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHV 1525
Query: 2019 PRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPW 2078
R N+ AD LA S R +DV V+ L P+ + + + W
Sbjct: 1526 LRHNNEAADRLANFGSK-REAAPSDV---FVEHLYSPTVPHKDATQVADTHDIAMVEADW 1581
Query: 2079 YYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQL 2138
+ +FL S+E P K + + + R + +++ LYK++ +L RCV E QL
Sbjct: 1582 REPLIRFLTSQELP--QYKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQL 1639
Query: 2139 MHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHAL 2198
+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P L
Sbjct: 1640 LKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQEL 1699
Query: 2199 NVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIK 2258
+ WPF++WG+DM+G + KA G+ + +AID F+KW+EA +T F
Sbjct: 1700 QTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFMAIDKFSKWIEAKPVVTITADNARDFF- 1757
Query: 2259 NNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-- 2316
NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG VE AN I
Sbjct: 1758 INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQ 1817
Query: 2317 ---KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSL 2373
R+ ++ W LP L RTT +TG +PF LVYG EA+LP EVE SL
Sbjct: 1818 RIKARVFDRLKPYAGKWVSQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESL 1877
Query: 2374 RVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGEL 2433
R E + + R D L+ +EE R A+ + Y ++ ++ VR R F VG+L
Sbjct: 1878 RF---RNFREERYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRAFLVGDL 1934
Query: 2434 VLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
VL++ Q R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1935 VLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHIRRFYA 1991
Score = 49.3 bits (116), Expect = 0.002
Identities = 34/117 (29%), Positives = 56/117 (47%), Gaps = 5/117 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 789 LRRTLIDGGSALNILFAKTLDDMQIPRSELKSSNAPFHGVIPGLSATPLGQITLPVTFGT 848
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTIHGE 1198
ENF ++F+V D +Y +LGRP + AV + +K ++T+ +
Sbjct: 849 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMKMPGPRGVLTLRSD 905
>ref|XP_474675.1| OSJNBa0027O01.4 [Oryza sativa (japonica cultivar-group)]
gi|38346188|emb|CAD39529.2| OSJNBa0027O01.4 [Oryza sativa
(japonica cultivar-group)]
Length = 2013
Score = 696 bits (1797), Expect = 0.0
Identities = 410/1128 (36%), Positives = 606/1128 (53%), Gaps = 56/1128 (4%)
Query: 1382 QEEIELVNIGTEENK-REIKIGATLEEGVKRKIIQLLREYP-------DIFAWSYEDMPG 1433
+E+I L E + KI + E K K I L P DIFAW DMPG
Sbjct: 925 REDIRLAAATASEGEVPATKISKSGESEAKTKKIPLDPSDPTKTANNKDIFAWKPSDMPG 984
Query: 1434 LDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIV 1493
+ ++EH + K + P++Q+LRR D IK E+ K + AGF+ V +P+W+AN V
Sbjct: 985 IPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPV 1044
Query: 1494 PVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMS 1553
V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D +SGY+QI++
Sbjct: 1045 LVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLK 1104
Query: 1554 PEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKST 1613
D KTSFITP+G +CY MPFGL NAGATYQR + F + VE YVDD++VK+
Sbjct: 1105 ESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTK 1164
Query: 1614 DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREM 1673
++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+ +P+KV AI M
Sbjct: 1165 QKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNM 1224
Query: 1674 PAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYL 1733
P T+K V+ G + +SRF+S + P FKLL+K W E Q+AF+ KK L
Sbjct: 1225 KPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFQWGPEAQKAFEDFKKLL 1284
Query: 1734 LEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYYLSKKFTDCETR 1789
EPP+L P PL++Y++ + + VL +++E G K + IY++S+ D +TR
Sbjct: 1285 TEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTR 1344
Query: 1790 YTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYD 1849
Y ++K + ++L HY H+ +++ P+ I V G+IA+W + L D
Sbjct: 1345 YPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREVNGRIAKWALELMSLD 1403
Query: 1850 IVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSK 1909
I FK + +IK LAD +A ++ + D P E + +
Sbjct: 1404 ISFKPRISIKSQALADFVA-----EWTECQEDTPAENMEH-------------------- 1438
Query: 1910 WGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRI 1969
W + FDG+ G G G V++SP G + + I F ++N+AEYEA + G+ AI + I
Sbjct: 1439 WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGI 1498
Query: 1970 KHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADAL 2029
K L + GDS LV+NQ+ EW ++ YR R+L F +EL H+ R N+ AD L
Sbjct: 1499 KRLIVRGDSQLVVNQVMKEWSYLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRL 1558
Query: 2030 ATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSR 2089
A S R +DV V+ L P+ + + + W + +FL S+
Sbjct: 1559 ANFGSK-REAAPSDV---FVEHLYTPTVPHKDTTQVAGTHDAAMVEVDWREPLIRFLTSQ 1614
Query: 2090 EYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGT 2149
E P K + + + R + ++L LYK++ +L RCV E QL+ D+H G G
Sbjct: 1615 ELP--QDKDEAERISRRSKLYVLHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGN 1672
Query: 2150 HATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSM 2209
HA T+ K R G++W D + R C CQ +A +IH+P L + WPF++
Sbjct: 1673 HAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAV 1732
Query: 2210 WGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPS 2269
WG+DM+G + KA G+ + VAID F+KW+EA +T F NI+ R+GVP+
Sbjct: 1733 WGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF-INIVHRFGVPN 1790
Query: 2270 KIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-----KRIVQKMV 2324
+IITDNGT V + CE+F I+ +S P NG VE AN I R+ ++
Sbjct: 1791 RIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLK 1850
Query: 2325 TTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEA 2384
W + LP L RTT +TG +PF LVYG EA+LP EVE SLR E
Sbjct: 1851 PYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESLRF---RNFREE 1907
Query: 2385 EWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPD 2444
+ + R D L+ +EE R A+ R Y ++ ++ VR R F VG+LVL++ Q
Sbjct: 1908 RYEEDRVDDLHRLEEVREAALIRSARYLQGLRRYHNRNVRSRAFLVGDLVLRK--IQTTR 1965
Query: 2445 PRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1966 DRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 2013
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 785 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 844
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 845 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 888
>ref|XP_473950.1| OSJNBa0053K19.16 [Oryza sativa (japonica cultivar-group)]
gi|38344177|emb|CAE03508.2| OSJNBa0053K19.16 [Oryza
sativa (japonica cultivar-group)]
Length = 2010
Score = 696 bits (1795), Expect = 0.0
Identities = 409/1130 (36%), Positives = 606/1130 (53%), Gaps = 60/1130 (5%)
Query: 1382 QEEIELVNIGTEENKREI-KIGATLEEGVKRKIIQLLREYP-------DIFAWSYEDMPG 1433
+E+I L E + KI + E K K I L P DIFAW DMPG
Sbjct: 922 REDIRLAAATASEGEEPATKISKSGESEAKTKKIPLDPSDPAKTANNKDIFAWKPSDMPG 981
Query: 1434 LDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIV 1493
+ ++EH + K + P++Q+LRR D IK E+ K + AGF+ V +P+W+AN V
Sbjct: 982 IPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPV 1041
Query: 1494 PVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMS 1553
V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D +SGY+QI++
Sbjct: 1042 LVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGRELLSFLDCYSGYHQIRLK 1101
Query: 1554 PEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKST 1613
D KTSFITP+G +CY MPFGL NAGATYQR + F + VE YVDD++VK+
Sbjct: 1102 ESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTK 1161
Query: 1614 DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREM 1673
++ + L + F +R ++++LNP KCTFGV SGKL+GF+VS +GI+ +P+KV AI M
Sbjct: 1162 QKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLVGFMVSHRGIQANPEKVTAILNM 1221
Query: 1674 PAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYL 1733
P T+K V+ G + +SRF+S + P FKLL+K W E Q+AF+ KK L
Sbjct: 1222 KPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDNFQWGPEAQKAFEDFKKLL 1281
Query: 1734 LEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYYLSKKFTDCETR 1789
EPP+L P PL++Y++ + + VL +++E G K + IY++S+ D +TR
Sbjct: 1282 TEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTR 1341
Query: 1790 YTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYD 1849
Y ++K + ++L HY H+ +++ P+ I G+IA+W + L D
Sbjct: 1342 YPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGRIAKWALELMSLD 1400
Query: 1850 IVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSK 1909
I FK + +IK LAD +A ++ + D P E + +
Sbjct: 1401 ISFKPRISIKSQALADFVA-----EWTECQEDTPAENMEH-------------------- 1435
Query: 1910 WGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRI 1969
W + FDG+ G G G V++SP G + + I F ++N+AEYEA + G+ AI + I
Sbjct: 1436 WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGI 1495
Query: 1970 KHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADAL 2029
K L + GDS LV+NQ+ EW ++ YR R+L F +EL H+ R N+ AD L
Sbjct: 1496 KRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRL 1555
Query: 2030 ATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDY--KPWYYDIKQFLL 2087
A S V + + V+ L P+ D AG + V W + +FL
Sbjct: 1556 ANFGSKREVAPSD----VFVEHLYTPT--VPHKDTTQVAGTHDVAMVETDWREPLIRFLT 1609
Query: 2088 SREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTF 2147
S+E P K + + + R + +++ LYK++ +L RCV E QL+ D+H G
Sbjct: 1610 SQELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLQDIHSGIC 1667
Query: 2148 GTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPF 2207
G HA T+ K R G++W D + R C CQ +A +IH+P L + WPF
Sbjct: 1668 GNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPF 1727
Query: 2208 SMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGV 2267
++WG+DM+G + KA G+ + VAID F+KW+EA +T F NI+ R+GV
Sbjct: 1728 AVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF-INIVHRFGV 1785
Query: 2268 PSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-----KRIVQK 2322
P++IITDNGT V + CE+F I+ +S P NG VE AN I R+ +
Sbjct: 1786 PNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDR 1845
Query: 2323 MVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLS 2382
+ W + LP L RTT +TG +PF LVYG EA+LP EVE SLR
Sbjct: 1846 LKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESLRF---RNFR 1902
Query: 2383 EAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQ 2442
E + + R D L+ +EE R A+ R Y ++ ++ VR R F VG+LVL++ Q
Sbjct: 1903 EERYEEDRVDDLHRLEEVREAALIRSARYLQGLRRYHNRNVRSRAFLVGDLVLRK--IQT 1960
Query: 2443 PDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1961 TRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 2010
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 782 LRRTLIDGGSALNILFAKTLDDMQIPHSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 841
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 842 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 885
>ref|NP_918169.1| putative gag-pol polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 2012
Score = 693 bits (1789), Expect = 0.0
Identities = 397/1080 (36%), Positives = 589/1080 (53%), Gaps = 48/1080 (4%)
Query: 1422 DIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLM 1481
DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 972 DIFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIK 1031
Query: 1482 TVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 1541
V +P+W+AN V V KK G+ MCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+
Sbjct: 1032 EVLHPDWLANPVLVRKKTGQWLMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFL 1091
Query: 1542 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEV 1601
D +SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + V
Sbjct: 1092 DCYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNV 1151
Query: 1602 EVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 1661
E YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1152 EAYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQ 1211
Query: 1662 VDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDE 1721
+P+KV AI M +P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1212 ANPEKVTAILNMKSPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDSFRWGPE 1271
Query: 1722 CQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIY 1777
Q+AF+ KK L EPP+L P PL++Y++ + + VL +++E G K + IY
Sbjct: 1272 AQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIY 1331
Query: 1778 YLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGK 1837
++S+ D +TRY ++K + ++L HY H+ +++ P+ I G+
Sbjct: 1332 FVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGR 1390
Query: 1838 IARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEE 1897
IA+W + L DI FK + +IK LAD +A ++ + D P E + +
Sbjct: 1391 IAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAENMEH-------- 1437
Query: 1898 PLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEAC 1957
W + FDG+ G G G V++SP G + + I F ++N+AEYEA
Sbjct: 1438 ------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEAL 1485
Query: 1958 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 2017
+ G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H
Sbjct: 1486 LHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSH 1545
Query: 2018 IPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKP 2077
+ R N+ AD LA S R +DV V+ L P+ A + +
Sbjct: 1546 VLRHNNEAADRLANFGSK-REAAPSDV---FVEHLYSPTVPHKDATQAAGAHDVAMVEAD 1601
Query: 2078 WYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQ 2137
W + +FL S+E P K + + + R + ++L LYK++ +L RCV E Q
Sbjct: 1602 WREPLIRFLTSQELP--QDKDEAERISRRSKLYVLHEAELYKKSPSGILQRCVSLEEGRQ 1659
Query: 2138 LMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHA 2197
L+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P
Sbjct: 1660 LLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQE 1719
Query: 2198 LNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFI 2257
L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T F
Sbjct: 1720 LQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF 1778
Query: 2258 KNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI- 2316
NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG VE AN I
Sbjct: 1779 -INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMIL 1837
Query: 2317 ----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPS 2372
R+ ++ W + LP L RTT +TG +PF LVYG EA+LP EVE S
Sbjct: 1838 QGIKARVFDRLKPYAGKWVQQLPSVLWSLRTTSSRATGQSPFFLVYGAEAMLPSEVEFES 1897
Query: 2373 LRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGE 2432
LR E + + R D L+ +EE R A+ R Y ++ ++ VR R F VG+
Sbjct: 1898 LRF---RNFREERYEEDRVDDLHRLEEAREAALIRSARYLQGLRRYHNRNVRSRAFLVGD 1954
Query: 2433 LVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
LVL++ Q R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1955 LVLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 2012
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 784 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 843
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 844 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 887
>ref|XP_470757.1| putative gag-pol precursor [Oryza sativa] gi|18071370|gb|AAL58229.1|
putative gag-pol precursor [Oryza sativa]
Length = 2026
Score = 693 bits (1788), Expect = 0.0
Identities = 397/1082 (36%), Positives = 589/1082 (53%), Gaps = 52/1082 (4%)
Query: 1422 DIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLM 1481
DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 977 DIFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIK 1036
Query: 1482 TVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 1541
V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+
Sbjct: 1037 EVHHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFL 1096
Query: 1542 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEV 1601
D +SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + V
Sbjct: 1097 DCYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNV 1156
Query: 1602 EVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 1661
E YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1157 EAYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQ 1216
Query: 1662 VDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDE 1721
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1217 ANPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFQWGPE 1276
Query: 1722 CQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIY 1777
Q+AF+ KK L EPP+L P PL++Y++ + + VL +++E G K + IY
Sbjct: 1277 AQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIY 1336
Query: 1778 YLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGK 1837
++S+ D +TRY ++K + ++L HY H+ +++ P+ I G+
Sbjct: 1337 FVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGR 1395
Query: 1838 IARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEE 1897
IA+W + L DI FK + +IK LAD +A ++ + D P E++ +
Sbjct: 1396 IAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAEKMEH-------- 1442
Query: 1898 PLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEAC 1957
W + FDG+ G G G V++SP G + + I F ++N+AEYEA
Sbjct: 1443 ------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEAL 1490
Query: 1958 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 2017
+ G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H
Sbjct: 1491 LHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSH 1550
Query: 2018 IPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDY-- 2075
+ R N+ AD LA S V + + V+ L P+ D AG + V
Sbjct: 1551 VLRHNNEAADRLANFGSKREVAPSD----VFVEHLYTPT--VPHKDTTQVAGTHDVAMVE 1604
Query: 2076 KPWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEA 2135
W + +FL S+E P K + + + R + +++ LYK++ +L RCV E
Sbjct: 1605 ADWREPLIRFLTSQELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEG 1662
Query: 2136 EQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPP 2195
QL+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P
Sbjct: 1663 RQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPA 1722
Query: 2196 HALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAK 2255
L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T
Sbjct: 1723 QELQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNAPD 1781
Query: 2256 FIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKN 2315
F NI+ R+GVP++IITDNG V + CE+F I+ +S P NG VE AN
Sbjct: 1782 FF-INIVHRFGVPNRIITDNGRQFTGGVFKDCCEDFGIKICYASVAHPMSNGQVERANGM 1840
Query: 2316 I-----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEI 2370
I R+ ++ W E LP L RTT +TG +PF LVYG EA+LP EVE
Sbjct: 1841 ILQGIKARVFDRLKPYAGKWVEQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEF 1900
Query: 2371 PSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKV 2430
SLR E + + R D L+ +EE R A+ + Y ++ ++ VR R F V
Sbjct: 1901 ESLRF---RNFREERYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRAFLV 1957
Query: 2431 GELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKY 2490
G+LVL++ Q R K +P +EGP+++ G+ L DG + N N + ++++
Sbjct: 1958 GDLVLRK--IQTTRDRHKLSPLWEGPFIISAVTRPGSYRLKREDGTLVDNSWNIEHLRRF 2015
Query: 2491 FA 2492
+A
Sbjct: 2016 YA 2017
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 789 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 848
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 849 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 892
>gb|AAP53095.1| putative retroelement [Oryza sativa (japonica cultivar-group)]
gi|37533012|ref|NP_920808.1| putative retroelement [Oryza
sativa (japonica cultivar-group)]
gi|19881590|gb|AAM00991.1| Putative retroelement [Oryza
sativa]
Length = 2017
Score = 692 bits (1786), Expect = 0.0
Identities = 400/1085 (36%), Positives = 589/1085 (53%), Gaps = 58/1085 (5%)
Query: 1422 DIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLM 1481
DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 977 DIFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIK 1036
Query: 1482 TVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 1541
V +P+W+AN V V KK G+ RMCVD+ DLNK PKD F LP ID +VD+TA ++ SF+
Sbjct: 1037 EVLHPDWLANPVLVRKKTGQWRMCVDYTDLNKCCPKDPFGLPRIDQVVDSTAGCELLSFL 1096
Query: 1542 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEV 1601
D +SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + V
Sbjct: 1097 DCYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNV 1156
Query: 1602 EVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 1661
E YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1157 EAYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQ 1216
Query: 1662 VDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDE 1721
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1217 ANPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFHWGPE 1276
Query: 1722 CQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIY 1777
Q+AF+ KK L EPP+L P PL++Y++ + + VL +++E G K + IY
Sbjct: 1277 AQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEDGHVQKVQRPIY 1336
Query: 1778 YLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGK 1837
++S+ D +TRY ++K + ++L HY H +++ P+ I G+
Sbjct: 1337 FVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHLVTVVTSF-PLGDILHNREANGR 1395
Query: 1838 IARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEE 1897
IA+W + L DI FK + +IK LAD +A ++ + D P E++ +
Sbjct: 1396 IAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAEKVEH-------- 1442
Query: 1898 PLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEAC 1957
W + FDG+ G G G V++SP G + + I F ++N+AEYEA
Sbjct: 1443 ------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEAL 1490
Query: 1958 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 2017
+ G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H
Sbjct: 1491 LHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSH 1550
Query: 2018 IPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDY-- 2075
+ R N+ AD LA S V + + V+ L P+ D AG + V
Sbjct: 1551 VLRHNNKAADRLANFGSKREVAPSD----VFVEHLYAPT--VPHKDTTQVAGTHDVAMVE 1604
Query: 2076 KPWYYDIKQFLLSREYPPGASKQDKKTLRRLASR---FLLDGDILYKRNYDMVLLRCVDE 2132
W + +FL S+E P QDK R++ R +++ LYK++ +L RCV
Sbjct: 1605 ADWREPLIRFLTSQELP-----QDKDEAERISRRSRLYVMHEAELYKKSPSGILQRCVSL 1659
Query: 2133 HEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIH 2192
E QL+ D+H G G HA T+ K R G++W D + R C CQ +A +IH
Sbjct: 1660 EEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIH 1719
Query: 2193 VPPHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQV 2252
+P L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T
Sbjct: 1720 LPAQELQTIPLSWPFAVWGLDMVGPFK-KAFGGYTHLFVAIDKFSKWIEAKPVVTITADN 1778
Query: 2253 VAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAA 2312
F NI+ R+GVP++IITDNG V + CE+F I+ +S P NG VE A
Sbjct: 1779 ARDFF-INIVHRFGVPNRIITDNGRQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERA 1837
Query: 2313 NKNI-----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLE 2367
N I R+ ++ W E LP L RTT +TG +PF LVYG EA+LP E
Sbjct: 1838 NGMILQGIKARVFDRLKPYAGKWVEQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSE 1897
Query: 2368 VEIPSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPRE 2427
VE SLR E + + R D L+ +EE R A+ + Y ++ ++ VR R
Sbjct: 1898 VEFESLRF---RNFREERYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRA 1954
Query: 2428 FKVGELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIV 2487
F VG+LVL++ Q R K +P +EGP+++ + G+ L DG + N N + +
Sbjct: 1955 FLVGDLVLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLIDNSWNIEHL 2012
Query: 2488 KKYFA 2492
++++A
Sbjct: 2013 RRFYA 2017
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 789 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 848
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 849 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 892
>ref|XP_475035.1| OSJNBa0042F21.5 [Oryza sativa (japonica cultivar-group)]
gi|38347303|emb|CAE02298.2| OSJNBa0042F21.5 [Oryza sativa
(japonica cultivar-group)]
Length = 1950
Score = 691 bits (1784), Expect = 0.0
Identities = 398/1082 (36%), Positives = 589/1082 (53%), Gaps = 52/1082 (4%)
Query: 1422 DIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLM 1481
DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 910 DIFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIK 969
Query: 1482 TVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 1541
V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+
Sbjct: 970 EVLHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFL 1029
Query: 1542 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEV 1601
D +SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + V
Sbjct: 1030 DCYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNV 1089
Query: 1602 EVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 1661
E YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1090 EAYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQ 1149
Query: 1662 VDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDE 1721
+P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K W E
Sbjct: 1150 ANPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDNFQWGPE 1209
Query: 1722 CQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIY 1777
Q+AF+ KK L EPPIL P PL++Y++ + + VL +++E G K + IY
Sbjct: 1210 AQKAFEDFKKLLTEPPILASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIY 1269
Query: 1778 YLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGK 1837
++S+ D +TRY ++K + ++L HY H+ +++ P+ I G+
Sbjct: 1270 FVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGR 1328
Query: 1838 IARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEE 1897
IA+W + L DI FK + +IK LAD +A ++ + D P E + +
Sbjct: 1329 IAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAENMEH-------- 1375
Query: 1898 PLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEAC 1957
W + FDG+ G G G V++SP G + + I F ++N+AEYEA
Sbjct: 1376 ------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEAL 1423
Query: 1958 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 2017
+ G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H
Sbjct: 1424 LHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSH 1483
Query: 2018 IPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPS--HVFAIGDVIDQAGENVVDY 2075
+ R N+ AD LA S + + + V+ L P+ H D D +V+
Sbjct: 1484 VLRHNNEAADRLANFGSKREMAPSD----VFVEHLYTPTVPHKDTTQDA-DTHDVALVE- 1537
Query: 2076 KPWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEA 2135
W +FL S+E P K + + + R + + + LYK++ +L RCV E
Sbjct: 1538 ADWREPFIRFLTSQELP--QDKDEAERISRRSKLYAMHEAELYKKSPSGILQRCVSLEEG 1595
Query: 2136 EQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPP 2195
QL+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P
Sbjct: 1596 RQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPA 1655
Query: 2196 HALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAK 2255
L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T
Sbjct: 1656 QELQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARD 1714
Query: 2256 FIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKN 2315
F NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG VE AN
Sbjct: 1715 FF-INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGM 1773
Query: 2316 I-----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEI 2370
I R+ ++ W + LP L RTT +TG +PF LVYG EA+LP EVE
Sbjct: 1774 ILQGIKARVFDRLKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEF 1833
Query: 2371 PSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKV 2430
SLR E + + R D L+ +EE R A+ R Y ++ ++ VR R F V
Sbjct: 1834 ESLRF---RNFREERYEEDRVDDLHRLEEAREAALIRSARYLQGLRRYHNRNVRSRAFLV 1890
Query: 2431 GELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKY 2490
G+LVL++ Q R K +P +EGP+++ + G+ L DG + N N + ++++
Sbjct: 1891 GDLVLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRF 1948
Query: 2491 FA 2492
+A
Sbjct: 1949 YA 1950
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 722 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 781
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 782 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 825
>gb|AAP52499.1| putative gag-pol precursor [Oryza sativa (japonica cultivar-group)]
gi|37531820|ref|NP_920212.1| putative gag-pol precursor
[Oryza sativa (japonica cultivar-group)]
gi|22128689|gb|AAM92802.1| putative gag-pol precursor
[Oryza sativa (japonica cultivar-group)]
Length = 2017
Score = 691 bits (1783), Expect = 0.0
Identities = 408/1130 (36%), Positives = 605/1130 (53%), Gaps = 60/1130 (5%)
Query: 1382 QEEIELVNIGTEENK-REIKIGATLEEGVKRKIIQLLREYP-------DIFAWSYEDMPG 1433
+E+I L E + KI + E K K I L P DIFAW DMPG
Sbjct: 929 REDIRLAAATASEGEVPATKISKSGESEAKTKKIPLDPSDPTKTANNKDIFAWKPSDMPG 988
Query: 1434 LDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIV 1493
+ ++EH + K + P++Q+LRR D IK E+ K + AGF+ V +P+W+AN V
Sbjct: 989 IPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPV 1048
Query: 1494 PVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMS 1553
V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+TA ++ SF+D +SGY+QI++
Sbjct: 1049 LVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLK 1108
Query: 1554 PEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKST 1613
D KTSFITP+G +CY MPFGL NAGATYQR + F + VE YVDD++VK+
Sbjct: 1109 ESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTK 1168
Query: 1614 DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREM 1673
++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+ +P+KV AI M
Sbjct: 1169 QKDDLILDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNM 1228
Query: 1674 PAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYL 1733
P T+K V+ G + +SRF+S + P FKLL+K W E Q+AF+ KK L
Sbjct: 1229 KPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFQWGPEAQKAFEDFKKLL 1288
Query: 1734 LEPPILVPPVEGRPLIMYLAVFDESMGCVLG-QQDETG---KKEHAIYYLSKKFTDCETR 1789
EPP+L P PL++Y++ + + VL +++E G K + IY++S+ D +TR
Sbjct: 1289 TEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTR 1348
Query: 1790 YTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYD 1849
Y ++K + ++L HY H+ +++ P+ I G+IA+W + L D
Sbjct: 1349 YPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGRIAKWALELMSLD 1407
Query: 1850 IVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSK 1909
+ FK + +IK LAD +A + +C+E + +
Sbjct: 1408 LSFKPRISIKSQALADFVA----------------------EWTECQEDTTVKKME---H 1442
Query: 1910 WGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRI 1969
W + FDG+ G G G V++SP G + + I F ++N+AEYEA + G+ AI + I
Sbjct: 1443 WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGI 1502
Query: 1970 KHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADAL 2029
K L + GDS LV+NQ+ EW + YR R+L F +EL H+ R +N+ AD L
Sbjct: 1503 KRLIVRGDSQLVVNQVMKEWSCLDDNMKAYRQEVRKLEDKFDGLELSHVLRHDNEAADRL 1562
Query: 2030 ATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDY--KPWYYDIKQFLL 2087
A S V + + V+ L P+ D AG + V W + +FL
Sbjct: 1563 ANFGSKREVAPSD----VFVEHLYTPT--VPHKDTTQAAGIHDVAMVETDWREPLIRFLT 1616
Query: 2088 SREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTF 2147
S+E P K + + + R + +++ LYK++ +L RCV E QL+ D+H G
Sbjct: 1617 SQELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGIC 1674
Query: 2148 GTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPF 2207
G HA T+ K R G++W D + R C CQ +A +IH+P L + WPF
Sbjct: 1675 GNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPF 1734
Query: 2208 SMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGV 2267
++WG+DM+G + KA G+ + VAID F+KW+EA +T F NI+ R+GV
Sbjct: 1735 AVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF-INIVHRFGV 1792
Query: 2268 PSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-----KRIVQK 2322
P++IITDNGT V + CE+F I+ +S P NG VE AN I R+ +
Sbjct: 1793 PNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDR 1852
Query: 2323 MVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLS 2382
+ W + LP L RTT +TG +PF LVYG EA+LP EVE SLR
Sbjct: 1853 LKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESLRF---RNFR 1909
Query: 2383 EAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQ 2442
E + + R D L+ +EE R A+ R Y ++ ++ VR R F VG+LVL++ Q
Sbjct: 1910 EERYEEDRVDDLHRLEEVREAALIRSARYLQGLRRYHNRNVRSRAFLVGDLVLRK--IQT 1967
Query: 2443 PDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1968 TRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEYLRRFYA 2017
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 789 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 848
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 849 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 892
>ref|XP_475120.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
gi|46063437|gb|AAS79740.1| putative polyprotein [Oryza
sativa (japonica cultivar-group)]
Length = 1756
Score = 690 bits (1780), Expect = 0.0
Identities = 404/1128 (35%), Positives = 602/1128 (52%), Gaps = 56/1128 (4%)
Query: 1382 QEEIEL-VNIGTEENKREIKIGATLEEGVKRKIIQL-------LREYPDIFAWSYEDMPG 1433
+E+I L V +E KI + E K K I L DIFAW DMPG
Sbjct: 668 REDIRLAVATASEGEVPATKISKSGESEAKTKKIPLDPSDSTKTANNKDIFAWKPSDMPG 727
Query: 1434 LDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLMTVEYPEWVANIV 1493
+ ++EH + K + P++Q+LRR D IK E+ K + AGF+ V +P+W+AN V
Sbjct: 728 IPREVIEHSLHVKEDAKPIKQRLRRFVQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPV 787
Query: 1494 PVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFMDGFSGYNQIKMS 1553
V KK G+ RMCVD+ DLNK PKD F LP ID +VD+TA ++ SF+D +SGY+QI++
Sbjct: 788 LVRKKTGQWRMCVDYTDLNKCCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLK 847
Query: 1554 PEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEVEVYVDDMIVKST 1613
D KTSFITP G +CY MPFGL NAGATYQR + F + VE YVDD+++K+
Sbjct: 848 ESDCLKTSFITPIGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVIKTK 907
Query: 1614 DEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIEVDPDKVRAIREM 1673
++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+ +P+KV AI M
Sbjct: 908 QKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNM 967
Query: 1674 PAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDECQEAFDSIKKYL 1733
P T+K V+ G + +SRF+S + P FKLL+K W E Q+AF+ KK+L
Sbjct: 968 KPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFQWGPEAQKAFEDFKKFL 1027
Query: 1734 LEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIYYLSKKFTDCETR 1789
EPP+L P PL++Y++ + + VL +++E G K + IY++S+ D +TR
Sbjct: 1028 TEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTR 1087
Query: 1790 YTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGKIARWQMLLSEYD 1849
Y ++K + ++L HY H+ +++ P+ I G+IA+W + L D
Sbjct: 1088 YPQVQKLLYGILITTRKLSHYFQGHSVTVVTSF-PLGDILHNREANGRIAKWALELMSLD 1146
Query: 1850 IVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEEPLIEEGPDPNSK 1909
I FK + +IK LAD +A ++ + D P E++ +
Sbjct: 1147 ISFKPRISIKSQALADFVA-----EWTECQEDTPAEKMEH-------------------- 1181
Query: 1910 WGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEACIFGIEEAIDMRI 1969
W + FDG+ G G G V++SP G + + I F ++N+AEYEA + G+ AI + I
Sbjct: 1182 WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGI 1241
Query: 1970 KHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHHIPRDENQMADAL 2029
K L + GDS LV+NQ+ EW ++ YR R+L F +EL H+ R N+ AD L
Sbjct: 1242 KRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRL 1301
Query: 2030 ATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKPWYYDIKQFLLSR 2089
A S V + + V+ L P+ I + + W +FL S+
Sbjct: 1302 ANFGSKREVAPSD----VFVEHLYTPTVPHKDTTQIAGTHDVALVEADWREPFIRFLTSQ 1357
Query: 2090 EYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQLMHDVHDGTFGT 2149
E P K + + + R + +++ LYK++ +L RCV E QL+ D+H G G
Sbjct: 1358 ELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGN 1415
Query: 2150 HATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHALNVMSSPWPFSM 2209
HA T+ K R G++W D + R C CQ +A +IH+P L + WPF++
Sbjct: 1416 HAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAV 1475
Query: 2210 WGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFIKNNIICRYGVPS 2269
WG+DM+G + KA G+ + VAID F+KW+EA +T F NI+ R+GVP+
Sbjct: 1476 WGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF-INIVHRFGVPN 1533
Query: 2270 KIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI-----KRIVQKMV 2324
+IITDNG V + CE+F I+ +S P NG VE AN I R+ ++
Sbjct: 1534 RIITDNGRQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLK 1593
Query: 2325 TTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPSLRVIMEAKLSEA 2384
W + LP L RTT +TG +PF LVYG EA+LP EVE SLR E
Sbjct: 1594 PYAGKWVKQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFESLRF---RNFREE 1650
Query: 2385 EWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGELVLKRRISQQPD 2444
+ + R D L+ +EE R A+ + Y ++ ++ VR R F VG+LVL++ Q
Sbjct: 1651 RYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRAFLVGDLVLRK--IQTTR 1708
Query: 2445 PRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
R K +P +EGP+++ + G+ L DG + N N + +++++A
Sbjct: 1709 DRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 1756
Score = 48.9 bits (115), Expect = 0.002
Identities = 33/117 (28%), Positives = 57/117 (48%), Gaps = 5/117 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 528 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 587
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLKFVKNGKLVTIHGE 1198
ENF ++F+V D +Y +LGRP + AV + +K +++++ +
Sbjct: 588 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMKMPGPRRVLSLRSD 644
>ref|XP_474845.1| OSJNBa0035O13.3 [Oryza sativa (japonica cultivar-group)]
gi|21741410|emb|CAD40114.1| OSJNBa0035O13.3 [Oryza sativa
(japonica cultivar-group)]
Length = 2008
Score = 685 bits (1768), Expect = 0.0
Identities = 395/1089 (36%), Positives = 587/1089 (53%), Gaps = 62/1089 (5%)
Query: 1413 IIQLLREYPDIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQ 1472
+I L+ DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+
Sbjct: 973 LITFLQNNKDIFAWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELT 1032
Query: 1473 KQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNT 1532
K + AGF+ V +P+W+AN V V KK G+ RMCVD+ DLNK+ PKD F LP ID +VD+T
Sbjct: 1033 KLLAAGFIKEVLHPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDST 1092
Query: 1533 AQSKVFSFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTL 1592
A ++ SF+D +SGY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR +
Sbjct: 1093 AGCELLSFLDCYSGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRC 1152
Query: 1593 FHDMTHKEVEVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLG 1652
F + VE YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLG
Sbjct: 1153 FSTQIGRNVEAYVDDVVVKTKQKDDLISDLEETFVSIRAFRMKLNPEKCTFGVPSGKLLG 1212
Query: 1653 FIVSQKGIEVDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRK 1712
F+VS +GI+ +P+KV AI M P T+K V+ G + +SRF+S + P FKLL+K
Sbjct: 1213 FMVSHRGIQANPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKK 1272
Query: 1713 NQPVVWNDECQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVLG-QQDETG- 1770
W E Q+AF+ KK L EPP+L P PL++Y++ + + VL +++E G
Sbjct: 1273 TDNFQWGPEAQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLAVEREEEGH 1332
Query: 1771 --KKEHAIYYLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYI 1828
K + IY++S+ D +TRY ++K L H+ +++ P+ I
Sbjct: 1333 VQKVQRPIYFVSEVLADSKTRYPQVQK--------------LLYGHSVTVVTSF-PLGDI 1377
Query: 1829 FEKAAVTGKIARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIM 1888
G+IA+W + L DI FK + +IK LAD +A ++ + D P E +
Sbjct: 1378 LHNREANGRIAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAENME 1432
Query: 1889 YLKSKDCEEPLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECT 1948
+ W + FDG+ G G G V++SP G + + I F +
Sbjct: 1433 H--------------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSAS 1472
Query: 1949 NNMAEYEACIFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLT 2008
+N+AEYEA + G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L
Sbjct: 1473 HNVAEYEALLHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLED 1532
Query: 2009 YFTKVELHHIPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQA 2068
F +EL H+ R N+ AD LA S V + + V+ L P+ +
Sbjct: 1533 KFDGLELSHVLRHNNEAADRLANFGSKREVAPSD----VFVEHLYTPTVPHKDTTQVAGT 1588
Query: 2069 GENVVDYKPWYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLR 2128
+ V W + +FL S+E P K + + + R + +++ LYK++ +L R
Sbjct: 1589 HDVAVVETDWREPLIRFLTSQELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQR 1646
Query: 2129 CVDEHEAEQLMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYA 2188
CV E QL+ D+H G G HA T+ K R G++W D + R C CQ +A
Sbjct: 1647 CVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFA 1706
Query: 2189 DKIHVPPHALNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNV 2248
+IH+P L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +
Sbjct: 1707 RQIHLPAQELQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTI 1765
Query: 2249 TKQVVAKFIKNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGA 2308
T F NI+ R+GVP++IITDNGT V + CE+F I+ +S P NG
Sbjct: 1766 TADNARDFF-INIVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAHPMSNGQ 1824
Query: 2309 VEAANKNI-----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAV 2363
VE AN I R+ ++ W + LP L RTT +TG +PF LVYG EA+
Sbjct: 1825 VERANGMILQGIKARVFDRLKPYAGKWVQQLPSVLWSLRTTPSRATGQSPFFLVYGAEAM 1884
Query: 2364 LPLEVEIPSLRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKV 2423
LP EVE SLR E + + R D L+ +EE R A+ R Y ++ ++ V
Sbjct: 1885 LPSEVEFESLRF---RNFREERYEEDRVDDLHRLEEAREAALIRSARYLQGLRRYHNRNV 1941
Query: 2424 RPREFKVGELVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVN 2483
R R F VG+LVL++ Q R K +P +EGP+++ + G+ L DG + N N
Sbjct: 1942 RSRAFLVGDLVLRK--IQTTRDRHKLSPLWEGPFIISEVTRPGSYRLKREDGTLVDNSWN 1999
Query: 2484 ADIVKKYFA 2492
+ +++++A
Sbjct: 2000 IEHLRRFYA 2008
Score = 48.5 bits (114), Expect = 0.003
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 785 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 844
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 845 RENFRTENISFEVADFETAYHAILGRPALAKFMAVPHYTYMMMK 888
>gb|AAP54912.1| gag-pol precursor [Oryza sativa (japonica cultivar-group)]
gi|37536646|ref|NP_922625.1| gag-pol precursor [Oryza
sativa (japonica cultivar-group)]
gi|13876521|gb|AAK43497.1| gag-pol precursor [Oryza
sativa (japonica cultivar-group)]
Length = 2017
Score = 682 bits (1760), Expect = 0.0
Identities = 389/1080 (36%), Positives = 585/1080 (54%), Gaps = 48/1080 (4%)
Query: 1422 DIFAWSYEDMPGLDPMIVEHRIPTKPECPPVRQKLRRTHPDMALKIKSEVQKQIDAGFLM 1481
DIFAW DMPG+ ++EH + K + P++Q+LRR D IK E+ K + AGF+
Sbjct: 977 DIFAWKPSDMPGIPREVIEHSLYVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIK 1036
Query: 1482 TVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLPHIDVLVDNTAQSKVFSFM 1541
V +P+W+AN V V KK G+ RMCVD+ DLN + PKD F LP ID +VD+TA ++ SF+
Sbjct: 1037 EVLHPDWLANPVLVRKKTGQWRMCVDYTDLNISCPKDPFGLPRIDQVVDSTAGCELLSFL 1096
Query: 1542 DGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPFGLINAGATYQRGMTTLFHDMTHKEV 1601
D + GY+QI++ D KTSFITP+G +CY MPFGL NAGATYQR + F + V
Sbjct: 1097 DCYLGYHQIRLKESDCLKTSFITPFGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNV 1156
Query: 1602 EVYVDDMIVKSTDEEQHVEYLTKMFERLRKYKLRLNPNKCTFGVRSGKLLGFIVSQKGIE 1661
E YVDD++VK+ ++ + L + F +R ++++LNP KCTFGV SGKLLGF+VS +GI+
Sbjct: 1157 EAYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQ 1216
Query: 1662 VDPDKVRAIREMPAPRTEKQVRGFLGRLNYISRFISHMTATCGPIFKLLRKNQPVVWNDE 1721
+P+K+ AI M P T+K V+ G + +SRF+S + P FKLL+K W +
Sbjct: 1217 ANPEKITAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLLKKTDDFQWGPD 1276
Query: 1722 CQEAFDSIKKYLLEPPILVPPVEGRPLIMYLAVFDESMGCVL-GQQDETG---KKEHAIY 1777
Q+AF+ KK L EPP+L P PL++Y+A + + VL +++E G K + IY
Sbjct: 1277 AQKAFEDFKKLLTEPPVLASPHPQEPLLLYIAAASQVVSTVLVVEREEEGHVQKVQRPIY 1336
Query: 1778 YLSKKFTDCETRYTMLEKTCCALAWAAKRLRHYLVNHTTWLISRMDPIKYIFEKAAVTGK 1837
++S+ D +TRY ++K + ++L HY +H+ +++ + I G+
Sbjct: 1337 FVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQSHSVTVVTSFS-LGDILHNREANGR 1395
Query: 1838 IARWQMLLSEYDIVFKTQKAIKGSILADHLAYQPLDDYQPIEFDFPDEEIMYLKSKDCEE 1897
IA+W + L DI FK + +IK LAD +A ++ + D P E++ +
Sbjct: 1396 IAKWALELMSLDISFKPRISIKSQALADFVA-----EWTECQEDTPAEKMEH-------- 1442
Query: 1898 PLIEEGPDPNSKWGLVFDGAVNAYGKGIGAVIVSPQGHHIPFTARILFECTNNMAEYEAC 1957
W + FDG+ G G G V++SP G + + I F ++N+AEYEA
Sbjct: 1443 ------------WTMHFDGSKRLSGTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEAL 1490
Query: 1958 IFGIEEAIDMRIKHLDIYGDSALVINQIKGEWETHHAKLIPYRDYARRLLTYFTKVELHH 2017
+ G+ AI + IK L + GDS LV+NQ+ EW ++ YR R+L F +EL H
Sbjct: 1491 LHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSH 1550
Query: 2018 IPRDENQMADALATLSSMFRVNHWNDVPLIKVQRLERPSHVFAIGDVIDQAGENVVDYKP 2077
+ R N+ AD LA S V + + V+ L P+ I + +
Sbjct: 1551 VLRHNNEAADRLANFGSKREVAPSD----VFVEHLYTPTVPHKDTTQIAGTHDVALVEAD 1606
Query: 2078 WYYDIKQFLLSREYPPGASKQDKKTLRRLASRFLLDGDILYKRNYDMVLLRCVDEHEAEQ 2137
W + +FL S+E P K + + + R + +++ LYK++ +L RCV E Q
Sbjct: 1607 WREPLIRFLTSQELP--QDKDEAERISRRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQ 1664
Query: 2138 LMHDVHDGTFGTHATGHTMSRKLLRAGYYWMAMEHDCYQYARKCHKCQIYADKIHVPPHA 2197
L+ D+H G G HA T+ K R G++W D + R C CQ +A +IH+P
Sbjct: 1665 LLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCEGCQFFARQIHLPAQE 1724
Query: 2198 LNVMSSPWPFSMWGIDMIGRIEPKASNGHRFILVAIDYFTKWVEAASYTNVTKQVVAKFI 2257
L + WPF++WG+DM+G + KA G+ + VAID F+KW+EA +T F
Sbjct: 1725 LQTIPLSWPFAVWGLDMVGPFK-KAVGGYTHLFVAIDKFSKWIEAKPVVTITADNARDFF 1783
Query: 2258 KNNIICRYGVPSKIITDNGTNLNNNVVQALCEEFKIEHHNSSPYRPQMNGAVEAANKNI- 2316
NI+ R+GVP++IITDNG V + CE+F I+ +S P NG VE AN I
Sbjct: 1784 -INIVHRFGVPNRIITDNGRQFTGGVFKDFCEDFGIKICYASVAHPMSNGQVERANGMIL 1842
Query: 2317 ----KRIVQKMVTTYKDWHEMLPYALHGYRTTVRSSTGATPFSLVYGMEAVLPLEVEIPS 2372
R+ ++ W + LP L RTT +TG +PF LVYG EA+LP EVE S
Sbjct: 1843 QGIKARVFDRLKPYAGKWVKQLPSVLWSLRTTPSRATGQSPFFLVYGAEAMLPSEVEFES 1902
Query: 2373 LRVIMEAKLSEAEWCQSRYDQLNLIEEKRMDAMARGQSYQARMKTAFDKKVRPREFKVGE 2432
LR E + + R D L+ +EE R A+ + Y ++ ++ VR R F VG+
Sbjct: 1903 LRF---RNYREERYEEDRVDDLHRLEEVREAALIQSARYLQGLRRYHNRNVRSRAFLVGD 1959
Query: 2433 LVLKRRISQQPDPRGKWAPNYEGPYVVKKAFSGGALILTHMDGVELPNPVNADIVKKYFA 2492
LVL++ Q R K +P +EGP+++ G+ L DG + N N + +++++A
Sbjct: 1960 LVLRK--IQTTRDRHKLSPLWEGPFIISDVTRPGSYRLKREDGTLVDNSWNIEHLRRFYA 2017
Score = 47.8 bits (112), Expect = 0.005
Identities = 33/104 (31%), Positives = 51/104 (48%), Gaps = 5/104 (4%)
Query: 1087 ISNVLVDTGSSLNVMPKTTLDQLSYRGTPLRRSTFLVKA-FDGSRKNVLGEIDLPITIGP 1145
+ L+D GS+LN++ TLD + + L+ S G LG+I LP+T G
Sbjct: 789 LRRTLIDGGSALNILFAKTLDDMQIPRSELKPSNAPFHGVIPGLSATPLGQITLPVTFGT 848
Query: 1146 -ENFL---VTFQVMDINASYSCLLGRPWIHDAGAVTSTLHQKLK 1185
ENF ++F+V D +Y +LGRP + AV + +K
Sbjct: 849 RENFRTENISFEVDDFETAYHAILGRPALAKFMAVPHYTYMMMK 892
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,417,879,666
Number of Sequences: 2540612
Number of extensions: 200640539
Number of successful extensions: 779979
Number of sequences better than 10.0: 36450
Number of HSP's better than 10.0 without gapping: 4963
Number of HSP's successfully gapped in prelim test: 31613
Number of HSP's that attempted gapping in prelim test: 724496
Number of HSP's gapped (non-prelim): 54223
length of query: 2492
length of database: 863,360,394
effective HSP length: 145
effective length of query: 2347
effective length of database: 494,971,654
effective search space: 1161698471938
effective search space used: 1161698471938
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 84 (37.0 bits)
Medicago: description of AC146557.10


