

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146556.4 + phase: 0
(887 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
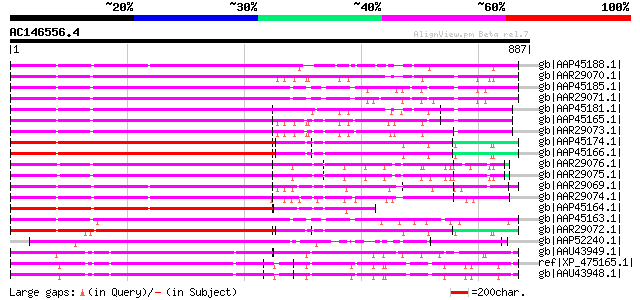
Score E
Sequences producing significant alignments: (bits) Value
gb|AAP45188.1| putative disease resistant protein rga2 [Solanum ... 493 e-137
gb|AAR29070.1| blight resistance protein RGA1 [Solanum bulbocast... 478 e-133
gb|AAP45185.1| putative disease resistant protein rga1 [Solanum ... 473 e-131
gb|AAR29071.1| blight resistance protein RGA3 [Solanum bulbocast... 464 e-129
gb|AAP45181.1| putative disease resistant protein rga3 [Solanum ... 464 e-129
gb|AAP45165.1| putative disease resistant protein RGA3 [Solanum ... 452 e-125
gb|AAR29073.1| blight resistance protein B149 [Solanum bulbocast... 449 e-124
gb|AAP45174.1| putative disease resistant protein rga4 [Solanum ... 433 e-119
gb|AAP45166.1| putative disease resistant protein RGA4 [Solanum ... 431 e-119
gb|AAR29076.1| blight resistance protein T118 [Solanum tarijense] 429 e-118
gb|AAR29075.1| blight resistance protein SH20 [Solanum tuberosum] 427 e-118
gb|AAR29069.1| blight resistance protein RPI [Solanum bulbocasta... 425 e-117
gb|AAR29074.1| blight resistance protein SH10 [Solanum tuberosum] 421 e-116
gb|AAP45164.1| putative disease resistant protein RGA2 [Solanum ... 420 e-115
gb|AAP45163.1| putative disease resistant protein RGA1 [Solanum ... 412 e-113
gb|AAR29072.1| blight resistance protein RGA4 [Solanum bulbocast... 406 e-111
gb|AAP52240.1| putative NBS-LRR type resistance protein [Oryza s... 326 2e-87
gb|AAU43949.1| putative NBS-LRR protein [Oryza sativa (japonica ... 313 2e-83
ref|XP_475165.1| putative NBS-LRR resistance protein [Oryza sati... 311 8e-83
gb|AAU43948.1| putative NBS-LRR resistance protein [Oryza sativa... 309 2e-82
>gb|AAP45188.1| putative disease resistant protein rga2 [Solanum bulbocastanum]
Length = 872
Score = 493 bits (1268), Expect = e-137
Identities = 331/910 (36%), Positives = 483/910 (52%), Gaps = 85/910 (9%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L+ NL SF++ ELA G + Q LS + I+AVL+DA++KQ+ N ++
Sbjct: 1 MAEAFIQVLLDNLTSFLKGELALLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNNKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T + R+HP I R +GKRM ++ K++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFSQSEYGRYHPKVIPFRHKVGKRMDQVMKKLKAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F LH ++ERQ RR+T SV+TE +VYGRDK+K+ IV+ L+ + D++ LS
Sbjct: 120 AEERKNFHLHEKIVERQAV---RRETGSVLTEPQVYGRDKEKDEIVKILINNVSDAQHLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+RV HF KIW+CVS D + +++++I+E+ G+ P
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRVTEHFHSKIWICVSEDFDEKRLIKAIVESIEGR-PL 235
Query: 241 LSSLE--SMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLD 298
L ++ +Q+K+QE+L RYLLVLDDVW ED++KW L+++L G GAS+L TTRL+
Sbjct: 236 LGEMDLAPLQKKLQELLNGKRYLLVLDDVWNEDQQKWANLRAVLKVGASGASVLTTTRLE 295
Query: 299 IVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAK 358
V SIMGT + L++LS +D W LF Q+AFG E LVAIGK++V+K G PLAAK
Sbjct: 296 KVGSIMGTLQPYELSNLSQEDCWLLFMQRAFGHQEEINPNLVAIGKEIVKKSGGVPLAAK 355
Query: 359 VLGSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPK 417
LG LC E W V +S WNLP+ + SI+ ALR+SY L L L+ CFA+CAVFPK
Sbjct: 356 TLGGILCFKREERAWEHVRDSPIWNLPQDESSILPALRLSYHQLPLDLKQCFAYCAVFPK 415
Query: 418 GFEMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARET 477
+M KE LI LWMA+G + S+GN+++E VGDEL + +G +H++ L + +
Sbjct: 416 DAKMEKEKLISLWMAHGFLLSKGNMELEDVGDELPS-SIGDLVHLRYLNLYGSGMRSLPK 474
Query: 478 NLISKKDLDRLYLSWGN------DTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGG-T 530
L ++L L L + S++GS L++ ++G T
Sbjct: 475 QLCKLQNLQTLDLQYCTKLCCLPKETSKLGS----------------LRNLLLDGSQSLT 518
Query: 531 IFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYIDDDLYEPE 590
P + + + LK L ++ K + L G L + +S + +K D D E
Sbjct: 519 CMPPRIGSLTCLKTLGQFVVGRKKGYQ-LGELGNLNLYGSIKISHLERVKN-DKDAKEAN 576
Query: 591 TEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASG 650
A +L LS+ + E + V++L L +N+ L + +
Sbjct: 577 L-SAKGNLHSLSMSWNNFGPHIYESEEVKVLEAL--KPHSNLTSLKIYGFRGIH--LPEW 631
Query: 651 GNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKELPVELGPLTALESLSIERCNEMES 710
N +LK N+ S+ IS F N LP G L LESL + +
Sbjct: 632 MNHSVLK--------------NIVSILISNFRNCSCLP-PFGDLPCLESLELHWGSADVE 676
Query: 711 FSEHL----------LKGLSSLRNMSVFSCSGFKSL--SDGMRHLTCLETLHIYYCPQLV 758
+ E + SLR + ++ K L +G LE + I+ CP L
Sbjct: 677 YVEEVDIDVHSGFPTRIRFPSLRKLDIWDFGSLKGLLKKEGEEQFPVLEEMIIHECPFLT 736
Query: 759 FPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVL 818
N+ +L SLR + + + +L+ L + ++K LP L ++ +L+ L
Sbjct: 737 LSSNLRALTSLRICYNKVATSFPEEMFKNLANLKYLTISRCNNLKELPTSLASLNALKSL 796
Query: 819 AICDFPE-------------------LSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIG 859
A+ PE L LP+ Q L L +L I GCP L KRC++GIG
Sbjct: 797 ALESLPEEGLEGLSSLTELFVEHCNMLKCLPEGLQHLTTLTSLKIRGCPQLIKRCEKGIG 856
Query: 860 EDWHKIAHIP 869
EDWHKI+HIP
Sbjct: 857 EDWHKISHIP 866
>gb|AAR29070.1| blight resistance protein RGA1 [Solanum bulbocastanum]
Length = 992
Score = 478 bits (1231), Expect = e-133
Identities = 352/1019 (34%), Positives = 506/1019 (49%), Gaps = 183/1019 (17%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L+ NL F+Q EL G + + LS ++I+AVL+DA++KQ+ A+K
Sbjct: 1 MAEAFLQVLLDNLTFFIQGELGLVFGFEKEFKKLSSMFSMIQAVLEDAQEKQLKYKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDILD+C T A + R+HP I +GKRMKE+ +++D I
Sbjct: 61 WLQKLNVAAYEVDDILDDCK-TEAARFKQAVLGRYHPRTITFCYKVGKRMKEMMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ RRQT V+TE KVYGR+K+++ IV+ L+ + SEE+
Sbjct: 120 AEERRNFHLDERIIERQA---ARRQTGFVLTEPKVYGREKEEDEIVKILINNVSYSEEVP 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+R+ HF+LKIWVCVS D + +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRITEHFNLKIWVCVSDDFDEKRLIKAIVESIEGKSLG 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+EKW+ L+++L G GASILITTRL+ +
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQEKWDNLRAVLKIGASGASILITTRLEKI 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LFKQ+AF E +L+ IGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQLYQLSNLSQEDCWLLFKQRAFCHQTETSPKLMEIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLP-EVDSIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +SE WNLP + +S++ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 357 GGLLRFKREESEWEHVRDSEIWNLPQDENSVLPALRLSYHHLPLDLRQCFAYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHN-LQL----------GGKLHIKCLENV 468
++ KE LI LWMA+ + S+GN+++E VG+E+ N L L GK + K + +
Sbjct: 417 KIEKEYLIALWMAHSFLLSKGNMELEDVGNEVWNELYLRSFFQEIEVKSGKTYFKMHDLI 476
Query: 469 -----------SNEEDARETNLISKKDL--------DRLYLSWGNDTNSQVGSVDAE--- 506
++ R+ N+ +D+ D + + + +S S+
Sbjct: 477 HDLATSMFSASASSRSIRQINVKDDEDMMFIVTNYKDMMSIGFSEVVSSYSPSLFKRFVS 536
Query: 507 -RVL-------EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRH 558
RVL E L L H G S K L+ L ++ LYNC++
Sbjct: 537 LRVLNLSNSEFEQLPSSVGDLVHLRYLDLSGNKICSLPKRLCKLQNLQTLDLYNCQSLSC 596
Query: 559 LPP------------------------FGKLPCLTIL--YLSGMR--------------- 577
LP G L CL L ++ G R
Sbjct: 597 LPKQTSKLCSLRNLVLDHCPLTSMPPRIGLLTCLKTLGYFVVGERKGYQLGELRNLNLRG 656
Query: 578 -----YIKYIDDDLYEPETE-KAFTSLKKLSLH-DLPNLERVLEVDGVEML---PQLLNL 627
+++ + +D+ E A +L LS+ D PN EV +E L P L L
Sbjct: 657 AISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEVKVLEALKPHPNLKYL 716
Query: 628 DITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL 687
+I + L + N +LK N+ S+ IS N L
Sbjct: 717 EIIDFCGFCLPDWM----------NHSVLK--------------NVVSILISGCENCSCL 752
Query: 688 PVELGPLTALESLSIE----RCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLS--DGMR 741
P G L LESL ++ +E + SLR + + K L G
Sbjct: 753 P-PFGELPCLESLELQDGSVEVEYVEDSGFLTRRRFPSLRKLHIGGFCNLKGLQRMKGAE 811
Query: 742 HLTCLETLHIYYCPQLVFPHNMNSLASLRQL-LLVECNESILDGIEGIPSLQKLRLFN-- 798
LE + I CP VFP +L+S+++L + E + L I + +L L++F+
Sbjct: 812 QFPVLEEMKISDCPMFVFP----TLSSVKKLEIWGEADAGGLSSISNLSTLTSLKIFSNH 867
Query: 799 -----------------------FPSIKSLPDWLGAMTSLQVL------AICDFPE---- 825
++K LP L ++ +L+ L A+ PE
Sbjct: 868 TVTSLLEEMFKNLENLIYLSVSFLENLKELPTSLASLNNLKCLDIRYCYALESLPEEGLE 927
Query: 826 ---------------LSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L LP+ Q L L +L I GCP L KRC++GIGEDWHKI+HIP
Sbjct: 928 GLSSLTELFVEHCNMLKCLPEGLQHLTTLTSLKIRGCPQLIKRCEKGIGEDWHKISHIP 986
>gb|AAP45185.1| putative disease resistant protein rga1 [Solanum bulbocastanum]
Length = 923
Score = 473 bits (1217), Expect = e-131
Identities = 335/952 (35%), Positives = 496/952 (51%), Gaps = 118/952 (12%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +++ NL SF++ EL G + Q LS + I+AVL+DA++KQ+ + ++
Sbjct: 1 MAEAFIQVVLDNLTSFLKGELVLLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNDKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T R+HP I R +GKRM ++ K+++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFLLSEYGRYHPKVIPFRHKVGKRMDQVMKKLNAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ R+T SV+TES+VYGRDK+K+ IV+ L A D+++LS
Sbjct: 120 AEERKNFHLQEKIIERQA---ATRETGSVLTESQVYGRDKEKDEIVKILTNTASDAQKLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTL+Q VFND+RV F KIW+CVS D N +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLSQMVFNDQRVTERFYPKIWICVSDDFNEKRLIKAIVESIEGKSLS 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+ KW L+++L G GA +L TTRL+ V
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQHKWANLRAVLKVGASGAFVLTTTRLEKV 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LF Q+AFG E LVAIGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQPYELSNLSPEDCWFLFMQRAFGHQEEINPNLVAIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +S WNLP+ + SI+ ALR+SY +L L LR CF +CAVFPK
Sbjct: 357 GGILRFKREEREWEHVRDSPIWNLPQDESSILPALRLSYHHLPLDLRQCFVYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNL 479
+M KENLI WMA+G + S+GNL++E VG+E+ N +L + + +E S + + +L
Sbjct: 417 KMAKENLIAFWMAHGFLLSKGNLELEDVGNEVWN-ELYLRSFFQEIEVESGKTYFKMHDL 475
Query: 480 ISKKDLDRLYLSWGNDTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGGTIFPSWMKNT 539
I DL S N ++S + ++A + + G + P
Sbjct: 476 I--HDLATSLFS-ANTSSSNIREINAN--------YDGYMMSIGFAEVVSSYSP------ 518
Query: 540 SILKGLVSIILYNCK--NCRHLP-PFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFT 596
S+L+ VS+ + N + N LP G L L L LSG I+ + P
Sbjct: 519 SLLQKFVSLRVLNLRNSNLNQLPSSIGDLVHLRYLDLSGNVRIRSL------PRRLCKLQ 572
Query: 597 SLKKLSLHDLPNL----------------ERVLEVDGVEMLPQLLNLDITNVPKLTLTSL 640
+L+ L LH +L E+ ++ ++ L ++ IT + ++ +
Sbjct: 573 NLQTLDLHYCDSLSCLPKQTISKLLCYWQEKGYQLGELKNLNLYGSISITKLDRVKKDTD 632
Query: 641 LSVESLSASGGNEELLKSF----FYNNCSEDVAG----NNLKSLSISKFANLKELPVELG 692
+LSA L S+ + SE + +NLK L I+ F + LP +
Sbjct: 633 AKEANLSAKANLHSLCLSWDLDGKHRYDSEVLEALKPHSNLKYLEINGFGGIL-LPDWMN 691
Query: 693 PLTALESLSI-----ERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCLE 747
+SI E C+ + F E L L SL + + + ++G + LE
Sbjct: 692 QSVLKNVVSIRIRGCENCSCLPPFGE--LPCLESLELHTGSAEVEYVEDNEGEKQFPVLE 749
Query: 748 TLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLFN--------- 798
+ Y+CP V P +L+S++ L ++ + ++L I + +L L + N
Sbjct: 750 EMTFYWCPMFVIP----TLSSVKTLKVIATDATVLRSISNLRALTSLDISNNVEATSLPE 805
Query: 799 ----------------FPSIKSLPDWLGA-------------------------MTSLQV 817
F ++K LP L + +TSL
Sbjct: 806 EMFKSLANLKYLNISFFRNLKELPTSLASLNALKSLKFEFCDALESLPEEGVKGLTSLTE 865
Query: 818 LAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L++ + L LP+ Q L L TLTI+ CPI+ KRC+RGIGEDWHKI+HIP
Sbjct: 866 LSVSNCMMLKCLPEGLQHLTALTTLTITQCPIVFKRCERGIGEDWHKISHIP 917
>gb|AAR29071.1| blight resistance protein RGA3 [Solanum bulbocastanum]
Length = 979
Score = 464 bits (1195), Expect = e-129
Identities = 343/1010 (33%), Positives = 506/1010 (49%), Gaps = 178/1010 (17%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +++ NL SF++ EL G + Q LS + I+AVL+DA++KQ+ + ++
Sbjct: 1 MAEAFIQVVLDNLTSFLKGELVLLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNDKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T R+HP I R +GKRM ++ K+++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFLQSEYGRYHPKVIPFRHKVGKRMDQVMKKLNAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ R+T SV+TE +VYGRDK+K+ IV+ L+ + D+++LS
Sbjct: 120 AEERKNFHLQEKIIERQA---ATRETGSVLTEPQVYGRDKEKDEIVKILINNVSDAQKLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTL+Q VFND+RV F KIW+CVS D + +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLSQMVFNDQRVTERFYPKIWICVSDDFDEKRLIKAIVESIEGKSLS 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+ KW L+++L G GA +L TTRL+ V
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQHKWANLRAVLKVGASGAFVLTTTRLEKV 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LF Q+AFG E LVAIGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQPYELSNLSPEDCWFLFMQRAFGHQEEINPNLVAIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +S WNLP+ + SI+ ALR+SY +L L LR CF +CAVFPK
Sbjct: 357 GGILRFKREEREWEHVRDSPIWNLPQDESSILPALRLSYHHLPLDLRQCFVYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNL 479
+M KENLI WMA+G + S+GNL++E VG+E+ N +L + + +E S + + +L
Sbjct: 417 KMAKENLIAFWMAHGFLLSKGNLELEDVGNEVWN-ELYLRSFFQEIEVESGKTYFKMHDL 475
Query: 480 ISKKDLDRLYLSWGNDTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGGTIFPSWMKNT 539
I DL S N ++S + ++A + + G + P
Sbjct: 476 I--HDLATSLFS-ANTSSSNIREINAN--------YDGYMMSIGFAEVVSSYSP------ 518
Query: 540 SILKGLVSIILYNCK--NCRHLP-PFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFT 596
S+L+ VS+ + N + N LP G L L L LSG I+ + P+
Sbjct: 519 SLLQKFVSLRVLNLRNSNLNQLPSSIGDLVHLRYLDLSGNFRIRNL------PKRLCRLQ 572
Query: 597 SLKKLSLHDLPNLE------------RVLEVDGVEM------------------------ 620
+L+ L LH +L R L +DG +
Sbjct: 573 NLQTLDLHYCDSLSCLPKQTSKLGSLRNLLLDGCSLTSTPPRIGLLTCLKSLSCFVIGKR 632
Query: 621 ----LPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNC-SEDVAG----- 670
L +L NL++ +++T L V+ S + K+ ++ C S D+ G
Sbjct: 633 KGYQLGELKNLNLYG--SISITKLDRVKKDSDAKEANLSAKANLHSLCLSWDLDGKHRYD 690
Query: 671 ----------NNLKSLSISKFANLKELPVELGPLTALESLSI-----ERCNEMESFSEHL 715
+NLK L I+ F ++ LP + +SI E C+ + F E
Sbjct: 691 SEVLEALKPHSNLKYLEINGFGGIR-LPDWMNQSVLKNVVSIRIRGCENCSCLPPFGE-- 747
Query: 716 LKGLSSL------------------------RNMSVFSCSGFKSL--SDGMRHLTCLETL 749
L L SL R + ++ S K L +G + LE +
Sbjct: 748 LPCLESLELHTGSADVEYVEDNVHPGRFPSLRKLVIWDFSNLKGLLKKEGEKQFPVLEEM 807
Query: 750 HIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLFN----------- 798
Y+CP V P +L+S++ L ++ + ++L I + +L L + N
Sbjct: 808 TFYWCPMFVIP----TLSSVKTLKVIATDATVLRSISNLRALTSLDISNNVEATSLPEEM 863
Query: 799 --------------FPSIKSLPDWLGA-------------------------MTSLQVLA 819
F ++K LP L + +TSL L+
Sbjct: 864 FKSLANLKYLNISFFRNLKELPTSLASLNALKSLKFEFCNALESLPEEGVKGLTSLTELS 923
Query: 820 ICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
+ + L LP+ Q L L TLTI+ CPI+ KRC+RGIGEDWHKIAHIP
Sbjct: 924 VSNCMMLKCLPEGLQHLTALTTLTITQCPIVFKRCERGIGEDWHKIAHIP 973
>gb|AAP45181.1| putative disease resistant protein rga3 [Solanum bulbocastanum]
Length = 908
Score = 464 bits (1195), Expect = e-129
Identities = 326/922 (35%), Positives = 487/922 (52%), Gaps = 99/922 (10%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L+ NL F+Q EL G + + LS ++I+AVL+DA++KQ+ A+K
Sbjct: 1 MAEAFLQVLLDNLTFFIQGELGLVFGFEKEFKKLSSMFSMIQAVLEDAQEKQLKYKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDILD+C T A + R+HP I +GKRMKE+ +++D I
Sbjct: 61 WLQKLNVAAYEVDDILDDCK-TEAARFKQAVLGRYHPRTITFCYKVGKRMKEMMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ RRQT V+TE KVYG++K+++ IV+ L+ + S+E+
Sbjct: 120 AEERRNFHLDERIIERQA---ARRQTGFVLTEPKVYGKEKEEDEIVKILINNVSYSKEVP 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+R+ HF+LKIWVCVS D + +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRITEHFNLKIWVCVSDDFDEKRLIKAIVESIEGKSLG 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+EKW+ L+++L G GASILITTRL+ +
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQEKWDNLRAVLKIGASGASILITTRLEKI 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LFKQ+AF E +L+ IGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQLYQLSNLSQEDCWLLFKQRAFCHQTETSPKLMEIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLP-EVDSIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +SE WNLP + +S++ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 357 GGLLRFKREESEWEHVRDSEIWNLPQDENSVLPALRLSYHHLPLDLRQCFAYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNL 479
++ KE LI LWMA+ + S+GN+++E VG+E+ N +L + + +E S + + +L
Sbjct: 417 KIEKEYLIALWMAHSFLLSKGNMELEDVGNEVWN-ELYLRSFFQEIEVKSGKTYFKMHDL 475
Query: 480 ISKKDLDRLYLSWGNDTNSQVGSVDAERVLEALEPH----SSGLKHFGVNGYGGTIFPSW 535
I S + + Q+ D E ++ + + S G V+ Y ++F S
Sbjct: 476 IHDLATSMFSASASSRSIRQINVKDDEDMMFIVTNYKDMMSIGFSEV-VSSYSPSLFKSL 534
Query: 536 MKNTSILKGLVSIILYNCKNCRHLPP------------------------FGKLPCLTIL 571
K L+ L ++ LYNC++ LP G L CL L
Sbjct: 535 PKRLCKLQNLQTLDLYNCQSLSCLPKQTSKLCSLRNLVLDHCPLTSMPPRIGLLTCLKTL 594
Query: 572 --YLSGMR--------------------YIKYIDDDLYEPETE-KAFTSLKKLSLH-DLP 607
++ G R +++ + +D+ E A +L LS+ D P
Sbjct: 595 GYFVVGERKGYQLGELRNLNLRGAISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRP 654
Query: 608 NLERVLEVDGVEML---PQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNC 664
N EV +E L P L L+I + L + N +LK
Sbjct: 655 NRYESEEVKVLEALKPHPNLKYLEIIDFCGFCLPDWM----------NHSVLK------- 697
Query: 665 SEDVAGNNLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEH----LLKGLS 720
N+ S+ IS N LP G L LESL ++ + F E +
Sbjct: 698 -------NVVSILISGCENCSCLP-PFGELPCLESLELQDGSVEVEFVEDSGFPTRRRFP 749
Query: 721 SLRNMSVFSCSGFKSLS--DGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQL-LLVEC 777
SLR + + K L +G LE + I CP VFP +L+S+++L + E
Sbjct: 750 SLRKLHIGGFCNLKGLQRMEGEEQFPVLEEMKISDCPMFVFP----TLSSVKKLEIWGEA 805
Query: 778 NESILDGIEGIPSLQKLRLFNFPSIKS-LPDWLGAMTSLQVLAICDFPELSSLPDNFQQL 836
+ L I + +L L++F+ ++ S L + ++ +L+ L++ L LP + L
Sbjct: 806 DARGLSSISNLSTLTSLKIFSNHTVTSLLEEMFKSLENLKYLSVSYLENLKELPTSLASL 865
Query: 837 QNLQTLTISGCPILEKRCKRGI 858
NL+ L I C LE + G+
Sbjct: 866 NNLKCLDIRYCYALESLPEEGL 887
Score = 149 bits (377), Expect = 3e-34
Identities = 108/305 (35%), Positives = 155/305 (50%), Gaps = 29/305 (9%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ +A+E NL +K +L L +SW + V +VL
Sbjct: 609 ELRNLNLRGAISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEV---KVL 665
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + + G P WM N S+LK +VSI++ C+NC LPPFG+LPCL
Sbjct: 666 EALKPHPN-LKYLEIIDFCGFCLPDWM-NHSVLKNVVSILISGCENCSCLPPFGELPCLE 723
Query: 570 ILYL-SGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLD 628
L L G +++++D + T + F SL+KL + NL+ + ++G E P L +
Sbjct: 724 SLELQDGSVEVEFVEDSGF--PTRRRFPSLRKLHIGGFCNLKGLQRMEGEEQFPVLEEMK 781
Query: 629 ITNVPKLTLTSLLSVESLSASG----------GNEELLKS--FFYNNCSEDV------AG 670
I++ P +L SV+ L G N L S F N+ + +
Sbjct: 782 ISDCPMFVFPTLSSVKKLEIWGEADARGLSSISNLSTLTSLKIFSNHTVTSLLEEMFKSL 841
Query: 671 NNLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSC 730
NLK LS+S NLKELP L L L+ L I C +ES E +GL + N F
Sbjct: 842 ENLKYLSVSYLENLKELPTSLASLNNLKCLDIRYCYALESLPE---EGLEAPNNPHKFKN 898
Query: 731 SGFKS 735
SG +
Sbjct: 899 SGMST 903
>gb|AAP45165.1| putative disease resistant protein RGA3 [Solanum bulbocastanum]
gi|46576966|sp|Q7XA40|RGA3_SOLBU Putative disease
resistance protein RGA3 (RGA1-blb) (Blight resistance
protein B149)
Length = 947
Score = 452 bits (1163), Expect = e-125
Identities = 332/959 (34%), Positives = 485/959 (49%), Gaps = 134/959 (13%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L+ NL F+Q EL G + + LS ++I+AVL+DA++KQ+ A+K
Sbjct: 1 MAEAFLQVLLDNLTFFIQGELGLVFGFEKEFKKLSSMFSMIQAVLEDAQEKQLKYKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDILD+C T A + R+HP I +GKRMKE+ +++D I
Sbjct: 61 WLQKLNVAAYEVDDILDDCK-TEAARFKQAVLGRYHPRTITFCYKVGKRMKEMMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ RRQT V+TE KVYGR+K+++ IV+ L+ + SEE+
Sbjct: 120 AEERRNFHLDERIIERQA---ARRQTGFVLTEPKVYGREKEEDEIVKILINNVSYSEEVP 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+R+ HF+LKIWVCVS D + +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRITEHFNLKIWVCVSDDFDEKRLIKAIVESIEGKSLG 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+EKW+ L+++L G GASILITTRL+ +
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQEKWDNLRAVLKIGASGASILITTRLEKI 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LFKQ+AF E +L+ IGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQLYQLSNLSQEDCWLLFKQRAFCHQTETSPKLMEIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLP-EVDSIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +SE WNLP + +S++ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 357 GGLLRFKREESEWEHVRDSEIWNLPQDENSVLPALRLSYHHLPLDLRQCFAYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHN-LQL----------GGKLHIKCLENV 468
++ KE LI LWMA+ + S+GN+++E VG+E+ N L L GK + K + +
Sbjct: 417 KIEKEYLIALWMAHSFLLSKGNMELEDVGNEVWNELYLRSFFQEIEVKSGKTYFKMHDLI 476
Query: 469 -----------SNEEDARETNLISKKDL--------DRLYLSWGNDTNSQVGSVDAE--- 506
++ R+ N+ +D+ D + + + +S S+
Sbjct: 477 HDLATSMFSASASSRSIRQINVKDDEDMMFIVTNYKDMMSIGFSEVVSSYSPSLFKRFVS 536
Query: 507 -RVL-------EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRH 558
RVL E L L H G S K L+ L ++ LYNC++
Sbjct: 537 LRVLNLSNSEFEQLPSSVGDLVHLRYLDLSGNKICSLPKRLCKLQNLQTLDLYNCQSLSC 596
Query: 559 LPP------------------------FGKLPCLTIL--YLSGMR--------------- 577
LP G L CL L ++ G R
Sbjct: 597 LPKQTSKLCSLRNLVLDHCPLTSMPPRIGLLTCLKTLGYFVVGERKGYQLGELRNLNLRG 656
Query: 578 -----YIKYIDDDLYEPETE-KAFTSLKKLSLH-DLPNLERVLEVDGVEML---PQLLNL 627
+++ + +D+ E A +L LS+ D PN EV +E L P L L
Sbjct: 657 AISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEVKVLEALKPHPNLKYL 716
Query: 628 DITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL 687
+I + L + N +LK N+ S+ IS N L
Sbjct: 717 EIIDFCGFCLPDWM----------NHSVLK--------------NVVSILISGCENCSCL 752
Query: 688 PVELGPLTALESLSIE----RCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLS--DGMR 741
P G L LESL ++ +E + SLR + + K L G
Sbjct: 753 P-PFGELPCLESLELQDGSVEVEYVEDSGFLTRRRFPSLRKLHIGGFCNLKGLQRMKGAE 811
Query: 742 HLTCLETLHIYYCPQLVFPHNMNSLASLRQL-LLVECNESILDGIEGIPSLQKLRLFNFP 800
LE + I CP VFP +L+S+++L + E + L I + +L L++F+
Sbjct: 812 QFPVLEEMKISDCPMFVFP----TLSSVKKLEIWGEADAGGLSSISNLSTLTSLKIFSNH 867
Query: 801 SIKS-LPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGI 858
++ S L + + +L L++ L LP + L NL+ L I C LE + G+
Sbjct: 868 TVTSLLEEMFKNLENLIYLSVSFLENLKELPTSLASLNNLKCLDIRYCYALESLPEEGL 926
Score = 148 bits (373), Expect = 9e-34
Identities = 109/305 (35%), Positives = 158/305 (51%), Gaps = 29/305 (9%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ +A+E NL +K +L L +SW + V +VL
Sbjct: 648 ELRNLNLRGAISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEV---KVL 704
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + + G P WM N S+LK +VSI++ C+NC LPPFG+LPCL
Sbjct: 705 EALKPHPN-LKYLEIIDFCGFCLPDWM-NHSVLKNVVSILISGCENCSCLPPFGELPCLE 762
Query: 570 ILYL-SGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLD 628
L L G ++Y++D + T + F SL+KL + NL+ + + G E P L +
Sbjct: 763 SLELQDGSVEVEYVEDSGF--LTRRRFPSLRKLHIGGFCNLKGLQRMKGAEQFPVLEEMK 820
Query: 629 ITNVPKLTLTSLLSVESL-----SASGGNEEL--------LKSFFYNNCS---EDVAGN- 671
I++ P +L SV+ L + +GG + LK F + + E++ N
Sbjct: 821 ISDCPMFVFPTLSSVKKLEIWGEADAGGLSSISNLSTLTSLKIFSNHTVTSLLEEMFKNL 880
Query: 672 -NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSC 730
NL LS+S NLKELP L L L+ L I C +ES E +GL + N F
Sbjct: 881 ENLIYLSVSFLENLKELPTSLASLNNLKCLDIRYCYALESLPE---EGLEAPNNPHKFKN 937
Query: 731 SGFKS 735
SG +
Sbjct: 938 SGMST 942
>gb|AAR29073.1| blight resistance protein B149 [Solanum bulbocastanum]
Length = 971
Score = 449 bits (1155), Expect = e-124
Identities = 330/959 (34%), Positives = 484/959 (50%), Gaps = 134/959 (13%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L+ NL F+Q EL G + + LS ++I+AVL+DA++KQ+ A+K
Sbjct: 1 MAEAFIQVLLDNLTFFIQGELGLVFGFEKEFKKLSSMFSMIQAVLEDAQEKQLKYKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDILD+C T A + R+HP I +GKRMKE+ +++D I
Sbjct: 61 WLQKLNVAAYEVDDILDDCK-TEAARFKQAVLGRYHPRTITFCYKVGKRMKEMMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F L +IERQ RRQT V+TE KVYGR+K+++ IV+ L+ + SEE+
Sbjct: 120 AEERRNFHLDERIIERQA---ARRQTGFVLTEPKVYGREKEEDEIVKILINNVSYSEEVP 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+R+ HF+LKIWVCVS D + +++++I+E+ GK+
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRITEHFNLKIWVCVSDDFDEKRLIKAIVESIEGKSLG 236
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
L +Q+K+QE+L RY LVLDDVW ED+EKW+ L+++L G GASILITTRL+ +
Sbjct: 237 DMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQEKWDNLRAVLKIGASGASILITTRLEKI 296
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS +D W LFKQ+AF E +L+ IGK++V+KC G PLAAK L
Sbjct: 297 GSIMGTLQLYQLSNLSQEDCWLLFKQRAFCHQTETSPKLMEIGKEIVKKCGGVPLAAKTL 356
Query: 361 GSSLCCTSNEHQWISVLESEFWNLP-EVDSIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V +SE W LP + +S++ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 357 GGLLRFKREESEWEHVRDSEIWXLPQDENSVLPALRLSYHHLPLDLRQCFAYCAVFPKDT 416
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHN-LQL----------GGKLHIKCLENV 468
++ KE LI LWMA+ + S+GN+++E VG+E+ N L L GK + K + +
Sbjct: 417 KIEKEYLIALWMAHSFLLSKGNMELEDVGNEVWNELYLRSFFQGIEVKSGKTYFKMHDLI 476
Query: 469 -----------SNEEDARETNLISKKDL--------DRLYLSWGNDTNSQVGSVDAE--- 506
++ R+ N+ +D+ D + + + +S S+
Sbjct: 477 HDLATSMFSASASSRSIRQINVKDDEDMMFIVTNYKDMMSIGFSEVVSSYSPSLFKRFVS 536
Query: 507 -RVL-------EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRH 558
RVL E L L H G S K L+ L ++ LYNC++
Sbjct: 537 LRVLNLSNSEFEQLPSSVGDLVHLRYLDLSGNKICSLPKRLCKLRNLQTLDLYNCQSLSC 596
Query: 559 LPP------------------------FGKLPCLTIL--YLSGMR--------------- 577
LP G L CL L ++ G R
Sbjct: 597 LPKQTSKLCSLRNLVLDHCPLTSMPPRIGLLTCLKTLGYFVVGERKGYQLGELRNLNLRG 656
Query: 578 -----YIKYIDDDLYEPETE-KAFTSLKKLSLH-DLPNLERVLEVDGVEML---PQLLNL 627
+++ + +D+ E A +L LS+ D PN EV +E L P L L
Sbjct: 657 AISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEVKVLEALKPHPNLKYL 716
Query: 628 DITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL 687
+I + L + N +LK N+ S+ IS N L
Sbjct: 717 EIIDFCGFCLPDWM----------NHSVLK--------------NVVSILISGCENCSCL 752
Query: 688 PVELGPLTALESLSIE----RCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLS--DGMR 741
P G L LESL ++ +E + SLR + + K L G
Sbjct: 753 P-PFGELPCLESLELQDGSVEVEYVEDSGFLTRRRFPSLRKLHIGGFCNLKGLQRMKGAE 811
Query: 742 HLTCLETLHIYYCPQLVFPHNMNSLASLRQL-LLVECNESILDGIEGIPSLQKLRLFNFP 800
LE + I CP VFP +L+S+++L + E + L I + +L L++F+
Sbjct: 812 QFPVLEEMKISDCPMFVFP----TLSSVKKLEIWGEADAGGLSSISNLSTLTSLKIFSNH 867
Query: 801 SIKS-LPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGI 858
++ S L + + +L L++ L LP + L NL+ L I C LE + G+
Sbjct: 868 TVTSLLEEMFKNLENLIYLSVSFLENLKELPTSLASLNNLKCLDIRYCYALESLPEEGL 926
Score = 181 bits (460), Expect = 7e-44
Identities = 124/328 (37%), Positives = 177/328 (53%), Gaps = 26/328 (7%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ +A+E NL +K +L L +SW + V +VL
Sbjct: 648 ELRNLNLRGAISITHLERVKNDMEAKEANLSAKANLHSLSMSWDRPNRYESEEV---KVL 704
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + + G P WM N S+LK +VSI++ C+NC LPPFG+LPCL
Sbjct: 705 EALKPHPN-LKYLEIIDFCGFCLPDWM-NHSVLKNVVSILISGCENCSCLPPFGELPCLE 762
Query: 570 ILYLS-GMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLD 628
L L G ++Y++D + T + F SL+KL + NL+ + + G E P L +
Sbjct: 763 SLELQDGSVEVEYVEDSGFL--TRRRFPSLRKLHIGGFCNLKGLQRMKGAEQFPVLEEMK 820
Query: 629 ITNVPKLTLTSLLSVESLSA-----SGGNEEL--------LKSFFYNNCS---EDVAGN- 671
I++ P +L SV+ L +GG + LK F + + E++ N
Sbjct: 821 ISDCPMFVFPTLSSVKKLEIWGEADAGGLSSISNLSTLTSLKIFSNHTVTSLLEEMFKNL 880
Query: 672 -NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSC 730
NL LS+S NLKELP L L L+ L I C +ES E L+GLSSL + V C
Sbjct: 881 ENLIYLSVSFLENLKELPTSLASLNNLKCLDIRYCYALESLPEEGLEGLSSLTELFVEHC 940
Query: 731 SGFKSLSDGMRHLTCLETLHIYYCPQLV 758
+ K L +G++HLT L +L I CPQL+
Sbjct: 941 NMLKCLPEGLQHLTTLTSLKIRGCPQLI 968
>gb|AAP45174.1| putative disease resistant protein rga4 [Solanum bulbocastanum]
Length = 988
Score = 433 bits (1114), Expect = e-119
Identities = 226/454 (49%), Positives = 312/454 (67%), Gaps = 3/454 (0%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L++NL SF+ ++L G + + LS + I+AV++DA++KQ+ + A++
Sbjct: 1 MAEAFLQVLLENLTSFIGDKLVLIFGFEKECEKLSSVFSTIQAVVQDAQEKQLKDKAIEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDIL EC R+ +HP I R IG+RMKEI +++D I
Sbjct: 61 WLQKLNSAAYEVDDILGECK-NEAIRFEQSRLGFYHPGIINFRHKIGRRMKEIMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER KF + ERQ R+T V+TE KVYGRDK+++ IV+ L+ + +EEL
Sbjct: 120 AEERRKFHFLEKITERQAA-AATRETGFVLTEPKVYGRDKEEDEIVKILINNVNVAEELP 178
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V+ I+G GG GKTTLAQ +FNDERV HF+ KIWVCVS D + +++++II N +PH
Sbjct: 179 VFPIIGMGGLGKTTLAQMIFNDERVTKHFNPKIWVCVSDDFDEKRLIKTIIGNIERSSPH 238
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
+ L S Q+K+QE+L RYLLVLDDVW +D EKW KL+++L G +GASIL TTRL+ V
Sbjct: 239 VEDLASFQKKLQELLNGKRYLLVLDDVWNDDLEKWAKLRAVLTVGARGASILATTRLEKV 298
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGTS +HL++LS D LF Q+AFG+ +E LVAIGK++V+KC G PLAAK L
Sbjct: 299 GSIMGTSQPYHLSNLSPHDSLLLFMQRAFGQQKEANPNLVAIGKEIVKKCGGVPLAAKTL 358
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V ++E W+LP+ + SI+ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 359 GGLLRFKREESEWEHVRDNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDT 418
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHN 453
+M+KENLI LWMA+G + S+GNL++E VG+E+ N
Sbjct: 419 KMIKENLITLWMAHGFLLSKGNLELEDVGNEVWN 452
Score = 185 bits (469), Expect = 7e-45
Identities = 123/329 (37%), Positives = 179/329 (54%), Gaps = 25/329 (7%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ DA E NL +K +L L +SW ND ++ S + +VL
Sbjct: 640 ELKNLNLCGSISITHLERVKNDTDA-EANLSAKANLQSLSMSWDNDGPNRYESEEV-KVL 697
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + +GG FPSW+ N S+L+ ++S+ + +CKNC LPPFG+LPCL
Sbjct: 698 EALKPHPN-LKYLEIIAFGGFRFPSWI-NHSVLEKVISVRIKSCKNCLCLPPFGELPCLE 755
Query: 570 ILYL-SGMRYIKYIDDDLYEPE--TEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLN 626
L L +G ++Y+++D T ++F SLKKL + +L+ +++ +G E P L
Sbjct: 756 NLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKFPMLEE 815
Query: 627 LDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNC---SEDVAGN------------ 671
+ I P +L SV+ L G S N S + N
Sbjct: 816 MAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLPEEMFT 875
Query: 672 ---NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVF 728
NL+ LS F NLK+LP L L AL+ L IE C+ +ESF E L+GL+SL + V
Sbjct: 876 SLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLTQLFVK 935
Query: 729 SCSGFKSLSDGMRHLTCLETLHIYYCPQL 757
C K L +G++HLT L L + CP++
Sbjct: 936 YCKMLKCLPEGLQHLTALTNLGVSGCPEV 964
Score = 89.0 bits (219), Expect = 6e-16
Identities = 108/413 (26%), Positives = 163/413 (39%), Gaps = 86/413 (20%)
Query: 517 SGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGM 576
S L+H V+G T P + + LK L I+ + K + G+L L + +
Sbjct: 597 SSLRHLVVDGCPLTSTPPRIGLLTCLKTLGFFIVGSKKGYQ----LGELKNLNLCGSISI 652
Query: 577 RYIKYIDDDLYEPETEKAFTSLKKLSL---HDLPNLERVLEVDGVEMLPQLLNLDITNVP 633
+++ + +D A +L+ LS+ +D PN EV +E L NL +
Sbjct: 653 THLERVKNDTDAEANLSAKANLQSLSMSWDNDGPNRYESEEVKVLEALKPHPNLKYLEI- 711
Query: 634 KLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKELPVELGP 693
A GG S+ ++ E V +KS N LP G
Sbjct: 712 -------------IAFGGFR--FPSWINHSVLEKVISVRIKSCK-----NCLCLP-PFGE 750
Query: 694 LTALESLSIERCNEMESFSEH--------LLKGLSSLRNMSVFSCSGFKSL--SDGMRHL 743
L LE+L ++ + + E + SL+ + ++ K L +G
Sbjct: 751 LPCLENLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKF 810
Query: 744 TCLETLHIYYCPQLVFP----------------------HNMNSLASLRQLLLVECNESI 781
LE + I YCP VFP N+++L SLR
Sbjct: 811 PMLEEMAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLP 870
Query: 782 LDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAI--CD----FPE---------- 825
+ + +L+ L F+F ++K LP L ++ +L+ L I CD FPE
Sbjct: 871 EEMFTSLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLT 930
Query: 826 ---------LSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L LP+ Q L L L +SGCP +EKRC + IGEDWHKIAHIP
Sbjct: 931 QLFVKYCKMLKCLPEGLQHLTALTNLGVSGCPEVEKRCDKEIGEDWHKIAHIP 983
Score = 39.3 bits (90), Expect = 0.58
Identities = 76/314 (24%), Positives = 128/314 (40%), Gaps = 60/314 (19%)
Query: 583 DDDLYE-PETEKAFTSLKKLSLHDLPNLER-------VLEVDGVEMLPQLLNLDITNVPK 634
D++++ P+ E + +LS H LP R V D + L+ L + +
Sbjct: 376 DNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDTKMIKENLITLWMAHGFL 435
Query: 635 LTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNN-------LKSLSISKFA----- 682
L+ +L +E + NE L+SFF E +GN + L+ S F+
Sbjct: 436 LSKGNL-ELEDVGNEVWNELYLRSFFQE--IEAKSGNTYFKIHDLIHDLATSLFSASASC 492
Query: 683 -NLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSV-------------- 727
N++E+ V+ ++SI + S+S LLK SLR +++
Sbjct: 493 GNIREINVK----DYKHTVSIGFSAVVSSYSPSLLKKFVSLRVLNLSYSKLEQLPSSIGD 548
Query: 728 --------FSCSGFKSLSDGMRHLTCLETLHIYYCPQL-VFPHNMNSLASLRQLLLVECN 778
SC+ F+SL + + L L+TL ++ C L P + L+SLR L++ C
Sbjct: 549 LLHLRYLDLSCNNFRSLPERLCKLQNLQTLDVHNCYSLNCLPKQTSKLSSLRHLVVDGCP 608
Query: 779 -ESILDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPD------ 831
S I + L+ L F S K LG + +L + L + +
Sbjct: 609 LTSTPPRIGLLTCLKTLGFFIVGSKKGYQ--LGELKNLNLCGSISITHLERVKNDTDAEA 666
Query: 832 NFQQLQNLQTLTIS 845
N NLQ+L++S
Sbjct: 667 NLSAKANLQSLSMS 680
>gb|AAP45166.1| putative disease resistant protein RGA4 [Solanum bulbocastanum]
gi|46576965|sp|Q7XA39|RGA4_SOLBU Putative disease
resistance protein RGA4 (RGA4-blb)
Length = 988
Score = 431 bits (1108), Expect = e-119
Identities = 225/454 (49%), Positives = 311/454 (67%), Gaps = 3/454 (0%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L++NL SF+ ++L G + + LS + I+AVL+DA++KQ+ + A++
Sbjct: 1 MAEAFLQVLLENLTSFIGDKLVLIFGFEKECEKLSSVFSTIQAVLQDAQEKQLKDKAIEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDIL EC R+ +HP I R IG+RMKEI +++D I
Sbjct: 61 WLQKLNSAAYEVDDILGECK-NEAIRFEQSRLGFYHPGIINFRHKIGRRMKEIMEKLDAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
+EER KF + ERQ R+T V+TE KVYGRDK+++ IV+ L+ + +EEL
Sbjct: 120 SEERRKFHFLEKITERQAA-AATRETGFVLTEPKVYGRDKEEDEIVKILINNVNVAEELP 178
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V+ I+G GG GKTTLAQ +FNDERV HF+ KIWVCVS D + +++++II N +PH
Sbjct: 179 VFPIIGMGGLGKTTLAQMIFNDERVTKHFNPKIWVCVSDDFDEKRLIKTIIGNIERSSPH 238
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
+ L S Q+K+QE+L RYLLVLDDVW +D EKW KL+++L G +GASIL TTRL+ V
Sbjct: 239 VEDLASFQKKLQELLNGKRYLLVLDDVWNDDLEKWAKLRAVLTVGARGASILATTRLEKV 298
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT +HL++LS D LF Q+AFG+ +E LVAIGK++V+KC G PLAAK L
Sbjct: 299 GSIMGTLQPYHLSNLSPHDSLLLFMQRAFGQQKEANPNLVAIGKEIVKKCGGVPLAAKTL 358
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L E +W V ++E W+LP+ + SI+ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 359 GGLLRFKREESEWEHVRDNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDT 418
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHN 453
+M+KENLI LWMA+G + S+GNL++E VG+E+ N
Sbjct: 419 KMIKENLITLWMAHGFLLSKGNLELEDVGNEVWN 452
Score = 185 bits (469), Expect = 7e-45
Identities = 123/329 (37%), Positives = 179/329 (54%), Gaps = 25/329 (7%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ DA E NL +K +L L +SW ND ++ S + +VL
Sbjct: 640 ELKNLNLCGSISITHLERVKNDTDA-EANLSAKANLQSLSMSWDNDGPNRYESKEV-KVL 697
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + +GG FPSW+ N S+L+ ++S+ + +CKNC LPPFG+LPCL
Sbjct: 698 EALKPHPN-LKYLEIIAFGGFRFPSWI-NHSVLEKVISVRIKSCKNCLCLPPFGELPCLE 755
Query: 570 ILYL-SGMRYIKYIDDDLYEPE--TEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLN 626
L L +G ++Y+++D T ++F SLKKL + +L+ +++ +G E P L
Sbjct: 756 NLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKFPMLEE 815
Query: 627 LDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNC---SEDVAGN------------ 671
+ I P +L SV+ L G S N S + N
Sbjct: 816 MAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLPEEMFT 875
Query: 672 ---NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVF 728
NL+ LS F NLK+LP L L AL+ L IE C+ +ESF E L+GL+SL + V
Sbjct: 876 SLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLTQLFVK 935
Query: 729 SCSGFKSLSDGMRHLTCLETLHIYYCPQL 757
C K L +G++HLT L L + CP++
Sbjct: 936 YCKMLKCLPEGLQHLTALTNLGVSGCPEV 964
Score = 89.4 bits (220), Expect = 5e-16
Identities = 108/413 (26%), Positives = 163/413 (39%), Gaps = 86/413 (20%)
Query: 517 SGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGM 576
S L+H V+G T P + + LK L I+ + K + G+L L + +
Sbjct: 597 SSLRHLVVDGCPLTSTPPRIGLLTCLKTLGFFIVGSKKGYQ----LGELKNLNLCGSISI 652
Query: 577 RYIKYIDDDLYEPETEKAFTSLKKLSL---HDLPNLERVLEVDGVEMLPQLLNLDITNVP 633
+++ + +D A +L+ LS+ +D PN EV +E L NL +
Sbjct: 653 THLERVKNDTDAEANLSAKANLQSLSMSWDNDGPNRYESKEVKVLEALKPHPNLKYLEI- 711
Query: 634 KLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKELPVELGP 693
A GG S+ ++ E V +KS N LP G
Sbjct: 712 -------------IAFGGFR--FPSWINHSVLEKVISVRIKSCK-----NCLCLP-PFGE 750
Query: 694 LTALESLSIERCNEMESFSEH--------LLKGLSSLRNMSVFSCSGFKSL--SDGMRHL 743
L LE+L ++ + + E + SL+ + ++ K L +G
Sbjct: 751 LPCLENLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKF 810
Query: 744 TCLETLHIYYCPQLVFP----------------------HNMNSLASLRQLLLVECNESI 781
LE + I YCP VFP N+++L SLR
Sbjct: 811 PMLEEMAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLP 870
Query: 782 LDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAI--CD----FPE---------- 825
+ + +L+ L F+F ++K LP L ++ +L+ L I CD FPE
Sbjct: 871 EEMFTSLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLT 930
Query: 826 ---------LSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L LP+ Q L L L +SGCP +EKRC + IGEDWHKIAHIP
Sbjct: 931 QLFVKYCKMLKCLPEGLQHLTALTNLGVSGCPEVEKRCDKEIGEDWHKIAHIP 983
Score = 39.7 bits (91), Expect = 0.44
Identities = 76/314 (24%), Positives = 128/314 (40%), Gaps = 60/314 (19%)
Query: 583 DDDLYE-PETEKAFTSLKKLSLHDLPNLER-------VLEVDGVEMLPQLLNLDITNVPK 634
D++++ P+ E + +LS H LP R V D + L+ L + +
Sbjct: 376 DNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDTKMIKENLITLWMAHGFL 435
Query: 635 LTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNN-------LKSLSISKFA----- 682
L+ +L +E + NE L+SFF E +GN + L+ S F+
Sbjct: 436 LSKGNL-ELEDVGNEVWNELYLRSFFQE--IEAKSGNTYFKIHDLIHDLATSLFSASASC 492
Query: 683 -NLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSV-------------- 727
N++E+ V+ ++SI + S+S LLK SLR +++
Sbjct: 493 GNIREINVK----DYKHTVSIGFAAVVSSYSPSLLKKFVSLRVLNLSYSKLEQLPSSIGD 548
Query: 728 --------FSCSGFKSLSDGMRHLTCLETLHIYYCPQL-VFPHNMNSLASLRQLLLVECN 778
SC+ F+SL + + L L+TL ++ C L P + L+SLR L++ C
Sbjct: 549 LLHLRYLDLSCNNFRSLPERLCKLQNLQTLDVHNCYSLNCLPKQTSKLSSLRHLVVDGCP 608
Query: 779 -ESILDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPD------ 831
S I + L+ L F S K LG + +L + L + +
Sbjct: 609 LTSTPPRIGLLTCLKTLGFFIVGSKKGYQ--LGELKNLNLCGSISITHLERVKNDTDAEA 666
Query: 832 NFQQLQNLQTLTIS 845
N NLQ+L++S
Sbjct: 667 NLSAKANLQSLSMS 680
>gb|AAR29076.1| blight resistance protein T118 [Solanum tarijense]
Length = 948
Score = 429 bits (1104), Expect = e-118
Identities = 324/909 (35%), Positives = 479/909 (52%), Gaps = 107/909 (11%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L++N+ SF+Q EL LG +++S + + I+AVL+DA++KQ+ + A+K
Sbjct: 1 MAEAFIQVLLENITSFIQGELGLLLGFENEFENISSRFSTIQAVLEDAQEKQLKDKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DD+LDEC A R+ R HP I+ R IGKR+KE+ +++D I
Sbjct: 61 WLQKLNAAAYKVDDLLDECKA---ARLEQSRLGRHHPKAIVFRHKIGKRIKEMMEKLDAI 117
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
A+ER F LH +IERQ R +T V+TE +VYGRDK+++ IV+ L+ + ++ ELS
Sbjct: 118 AKERTDFHLHEKIIERQV---ARPETGPVLTEPQVYGRDKEEDEIVKILINNVSNALELS 174
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+RV HF KIW+CVS D + +++E+II N +
Sbjct: 175 VLPILGMGGLGKTTLAQMVFNDQRVTEHFYPKIWICVSDDFDEKRLIETIIGNIERSSLD 234
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
+ L S Q+K+Q++L RYLLVLDDVW ED++KW+ L+++L G GAS+L TTRL+ V
Sbjct: 235 VKDLASFQKKLQQLLNGKRYLLVLDDVWNEDQQKWDNLRAVLKVGASGASVLTTTRLEKV 294
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
SIMGT + L++LS DD W LF Q+A+ E LVAIGK++V+K G PLAAK L
Sbjct: 295 GSIMGTLQPYQLSNLSQDDCWLLFIQRAYRHQEEISPNLVAIGKEIVKKSGGVPLAAKTL 354
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L + +W V + E WNLP+ + SI+ LR+SY +L L LR CFA+CAVFPK
Sbjct: 355 GGLLRFKREKREWEHVRDREIWNLPQDEMSILPVLRLSYHHLPLDLRQCFAYCAVFPKDT 414
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNL 479
+M K+ +I LWMA+G + SR NL++E VG+E+ N +L+ L + E + R N
Sbjct: 415 KMEKKKVISLWMAHGFLLSRRNLELEDVGNEVWN-----ELY---LRSFFQEIEVRYGNT 466
Query: 480 ISK-----KDLDRLYLSWGNDTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGGTIFPS 534
K DL S N ++S + ++ E + S G V+ Y
Sbjct: 467 YFKMHDLIHDLATSLFS-ANTSSSNIREINVESYTHMM--MSIGFSEV-VSSY------- 515
Query: 535 WMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKA 594
+ S+L+ VS+ + N + F +LP +I L +RY+ + +++ K
Sbjct: 516 ---SPSLLQKFVSLRVLNLSYSK----FEELPS-SIGDLVHLRYMD-LSNNIEIRSLPKQ 566
Query: 595 FTSLKKLSLHDLPNLERVLEV-DGVEMLPQLLNLDITNVPKLTLT-----SLLSVESLSA 648
L+ L DL R+ + L L NL + +LT T SL +++L
Sbjct: 567 LCKLQNLQTLDLQYCTRLCCLPKQTSKLGSLRNLLLHGCHRLTRTPPRIGSLTCLKTLGQ 626
Query: 649 SGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL-PVELGPLTALESLSI----- 702
+ K + ++K + + N KE L L SLS+
Sbjct: 627 FVVKRK--KGYQLGELGSLNLYGSIKISHLERVKNDKEAKEANLSAKENLHSLSMKWDDD 684
Query: 703 ERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKS--LSDGMRH------------------ 742
ER + ES +L+ L N++ + SGF+ L D M H
Sbjct: 685 ERPHRYESEEVEVLEALKPHSNLTCLTISGFRGIRLPDWMNHSVLKNIVLIEISGCKNCS 744
Query: 743 -------LTCLETLHIY-----YCPQL-------VFPHNMNSLASLRQLLLVECNESILD 783
L CLE+L +Y Y ++ FP + SLR+L + C L
Sbjct: 745 CLPPFGDLPCLESLQLYRGSAEYVEEVDIDVEDSGFPTRIR-FPSLRKLCI--CKFDNLK 801
Query: 784 GI------EGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDN-FQQL 836
G+ E P L+++ + P I +L L A+TSL I D E +S P+ F+ L
Sbjct: 802 GLVKKEGGEQFPVLEEMEIRYCP-IPTLSSNLKALTSLN---ISDNKEATSFPEEMFKSL 857
Query: 837 QNLQTLTIS 845
NL+ L IS
Sbjct: 858 ANLKYLNIS 866
Score = 192 bits (489), Expect = 3e-47
Identities = 124/314 (39%), Positives = 178/314 (56%), Gaps = 12/314 (3%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL +L L G + I LE V N+++A+E NL +K++L L + W +D + VL
Sbjct: 639 ELGSLNLYGSIKISHLERVKNDKEAKEANLSAKENLHSLSMKWDDDERPHRYESEEVEVL 698
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PHS+ L ++G+ G P WM N S+LK +V I + CKNC LPPFG LPCL
Sbjct: 699 EALKPHSN-LTCLTISGFRGIRLPDWM-NHSVLKNIVLIEISGCKNCSCLPPFGDLPCLE 756
Query: 570 IL--YLSGMRYIKYIDDDLYEP--ETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLL 625
L Y Y++ +D D+ + T F SL+KL + NL+ +++ +G E P L
Sbjct: 757 SLQLYRGSAEYVEEVDIDVEDSGFPTRIRFPSLRKLCICKFDNLKGLVKKEGGEQFPVLE 816
Query: 626 NLDITNVPKLTLTS-LLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANL 684
++I P TL+S L ++ SL+ S E + F + +A NLK L+IS F NL
Sbjct: 817 EMEIRYCPIPTLSSNLKALTSLNISDNKEA---TSFPEEMFKSLA--NLKYLNISHFKNL 871
Query: 685 KELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLT 744
KELP L L AL+SL I+ C +ES E +KGL+SL + V C K L +G++HLT
Sbjct: 872 KELPTSLASLNALKSLKIQWCCALESIPEEGVKGLTSLTELIVKFCKMLKCLPEGLQHLT 931
Query: 745 CLETLHIYYCPQLV 758
L + I+ CPQL+
Sbjct: 932 ALTRVKIWGCPQLI 945
Score = 58.5 bits (140), Expect = 9e-07
Identities = 93/383 (24%), Positives = 140/383 (36%), Gaps = 88/383 (22%)
Query: 537 KNTSILKGLVSIILYNCKNCRHLPP-FGKLPCLTIL-------------------YLSGM 576
K TS L L +++L+ C PP G L CL L L G
Sbjct: 589 KQTSKLGSLRNLLLHGCHRLTRTPPRIGSLTCLKTLGQFVVKRKKGYQLGELGSLNLYGS 648
Query: 577 RYIKYIDDDLYEPETEKAFTSLKKLSLHDL---------PNLERVLEVDGVEMLPQLLNL 627
I +++ + E ++A S K+ +LH L P+ EV+ +E L NL
Sbjct: 649 IKISHLERVKNDKEAKEANLSAKE-NLHSLSMKWDDDERPHRYESEEVEVLEALKPHSNL 707
Query: 628 DITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL 687
LT++ + N +LK N+ + IS N L
Sbjct: 708 TC-----LTISGFRGIRL--PDWMNHSVLK--------------NIVLIEISGCKNCSCL 746
Query: 688 PVELGPLTALESLSIERCN---------EMESFSEHLLKGLSSLRNMSVFSCSGFKSL-- 736
P G L LESL + R + ++E SLR + + K L
Sbjct: 747 P-PFGDLPCLESLQLYRGSAEYVEEVDIDVEDSGFPTRIRFPSLRKLCICKFDNLKGLVK 805
Query: 737 SDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRL 796
+G LE + I YCP N+ +L SL E + + + +L+ L +
Sbjct: 806 KEGGEQFPVLEEMEIRYCPIPTLSSNLKALTSLNISDNKEATSFPEEMFKSLANLKYLNI 865
Query: 797 FNFPSIKSLPDWLGAMTSLQVL------AICDFPE-------------------LSSLPD 831
+F ++K LP L ++ +L+ L A+ PE L LP+
Sbjct: 866 SHFKNLKELPTSLASLNALKSLKIQWCCALESIPEEGVKGLTSLTELIVKFCKMLKCLPE 925
Query: 832 NFQQLQNLQTLTISGCPILEKRC 854
Q L L + I GCP L KRC
Sbjct: 926 GLQHLTALTRVKIWGCPQLIKRC 948
Score = 39.7 bits (91), Expect = 0.44
Identities = 49/196 (25%), Positives = 85/196 (43%), Gaps = 23/196 (11%)
Query: 672 NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCS 731
+L+ + +S ++ LP +L L L++L ++ C + + K L SLRN+ + C
Sbjct: 548 HLRYMDLSNNIEIRSLPKQLCKLQNLQTLDLQYCTRLCCLPKQTSK-LGSLRNLLLHGCH 606
Query: 732 GFKSLSDGMRHLTCLETLHIYYCP-----QLVFPHNMNSLASLRQLLL------VECNES 780
+ LTCL+TL + QL ++N S++ L E E+
Sbjct: 607 RLTRTPPRIGSLTCLKTLGQFVVKRKKGYQLGELGSLNLYGSIKISHLERVKNDKEAKEA 666
Query: 781 ILDGIEGIPSL-------QKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPD-- 831
L E + SL ++ + ++ L + L ++L L I F + LPD
Sbjct: 667 NLSAKENLHSLSMKWDDDERPHRYESEEVEVL-EALKPHSNLTCLTISGFRGI-RLPDWM 724
Query: 832 NFQQLQNLQTLTISGC 847
N L+N+ + ISGC
Sbjct: 725 NHSVLKNIVLIEISGC 740
Score = 35.4 bits (80), Expect = 8.4
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query: 790 SLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGC 847
SL+ L L ++ + LP +G + L+ + + + E+ SLP +LQNLQTL + C
Sbjct: 525 SLRVLNL-SYSKFEELPSSIGDLVHLRYMDLSNNIEIRSLPKQLCKLQNLQTLDLQYC 581
>gb|AAR29075.1| blight resistance protein SH20 [Solanum tuberosum]
Length = 947
Score = 427 bits (1099), Expect = e-118
Identities = 324/906 (35%), Positives = 482/906 (52%), Gaps = 102/906 (11%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L++N+ SF+Q EL LG +++S + + I+AVL+DA++KQ+ + A+K
Sbjct: 1 MAEAFIQVLLENITSFIQGELGLLLGFENDFENISSRFSTIQAVLEDAQEKQLKDKAIKN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DD+LDEC A R+ HP I+ R IGKR+KE+ +++D I
Sbjct: 61 WLQKLNAAVYKVDDLLDECKA---ARLEQSRLGCHHPKAIVFRHKIGKRIKEMMEKLDAI 117
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
A+ER F LH +IERQ R +T V+TE +VYGRDK+++ IV+ L+ + +++ELS
Sbjct: 118 AKERTDFHLHEKIIERQV---ARPETGFVLTEPQVYGRDKEEDEIVKILINNVSNAQELS 174
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+RV HF KIW+CVS D + +++E+II N +
Sbjct: 175 VLPILGMGGLGKTTLAQMVFNDQRVTEHFYPKIWICVSDDFDEKRLIENIIGNIERSSLD 234
Query: 241 LSSLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIV 300
+ L S Q+K+Q++L RYLLVLDDVW ED++KW+ L+ +L G GAS+L TTRL+ V
Sbjct: 235 VKDLASFQKKLQQLLNGKRYLLVLDDVWNEDQQKWDNLRVVLKVGASGASVLTTTRLEKV 294
Query: 301 ASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVL 360
S+MGT + L++LS DD W LF Q+AF E LVAIGK++V+K G PLAAK L
Sbjct: 295 GSVMGTLQPYQLSNLSQDDCWLLFIQRAFRHQEEISPNLVAIGKEIVKKSGGVPLAAKTL 354
Query: 361 GSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGF 419
G L + +W V +SE WNLP+ + SI+ ALR+SY +L L+LR CFA+CAVFPK
Sbjct: 355 GGLLRFKREKREWEHVRDSEIWNLPQDEMSILPALRLSYHHLPLALRQCFAYCAVFPKDT 414
Query: 420 EMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNL 479
+M K+ +I LWMA+G + SR NL++E V +E N +L+ L + E + R N
Sbjct: 415 KMEKKKVISLWMAHGFLLSRRNLELEDVRNEGWN-----ELY---LRSFFQEIEVRYGNT 466
Query: 480 ISK-----KDLDRLYLSWGNDTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGGTIFPS 534
K DL LS N ++S + ++ E + S G V+ Y
Sbjct: 467 YFKMXDLIHDLAXSLLS-ANTSSSNIREINVESYTHMM--MSIGFSEV-VSSY------- 515
Query: 535 WMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKA 594
+ S+L+ VS+ + N + F +LP +I L +RY+ + +++ K
Sbjct: 516 ---SPSLLQKFVSLRVLNLSYSK----FEELPS-SIGDLVHLRYMD-LSNNIEIRSLPKQ 566
Query: 595 FTSLKKLSLHDLPNLERVLEV-DGVEMLPQLLNLDITNVPKLTLT-----SLLSVESLSA 648
L+ L DL R+ + L L NL + +LT T SL +++L
Sbjct: 567 LCKLQNLQTLDLQYCTRLCCLPKQTSKLGSLRNLLLHGCHRLTRTPPRIGSLTCLKTLGQ 626
Query: 649 SGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL-PVELGPLTALESLSI----- 702
S + K + ++K + + N KE L L SLS+
Sbjct: 627 SVVKRK--KGYQLGELGSLNLYGSIKISHLERVKNDKEAKEANLSAKENLHSLSMKWDDD 684
Query: 703 ERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKS--LSDGMRH------------------ 742
E + ES +L+ L N++ SGF+ L D M H
Sbjct: 685 EHPHRYESEEVEVLEALKPHSNLTCLKISGFRGIRLPDWMNHSVLKNIVLIEISGCKNCS 744
Query: 743 -------LTCLETLHIY-----YCPQL------VFPHNMNSLASLRQLLLVECN--ESIL 782
L CLE+L +Y Y ++ FP + L SLR+L + + + + +L
Sbjct: 745 CLPPFGDLPCLESLELYRGSAEYVEEVDIDVDSGFPTRIR-LPSLRKLCICKFDNLKGLL 803
Query: 783 --DGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDN-FQQLQNL 839
+G E P L+++ + P I +L L A+TSL I D E +S P+ F+ L NL
Sbjct: 804 KKEGGEQFPVLEEMEIRYCP-IPTLSPNLKALTSLN---ISDNKEATSFPEEMFKSLANL 859
Query: 840 QTLTIS 845
+ L IS
Sbjct: 860 KYLNIS 865
Score = 184 bits (466), Expect = 1e-44
Identities = 121/313 (38%), Positives = 175/313 (55%), Gaps = 11/313 (3%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL +L L G + I LE V N+++A+E NL +K++L L + W +D + + VL
Sbjct: 639 ELGSLNLYGSIKISHLERVKNDKEAKEANLSAKENLHSLSMKWDDDEHPHRYESEEVEVL 698
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PHS+ L ++G+ G P WM N S+LK +V I + CKNC LPPFG LPCL
Sbjct: 699 EALKPHSN-LTCLKISGFRGIRLPDWM-NHSVLKNIVLIEISGCKNCSCLPPFGDLPCLE 756
Query: 570 IL--YLSGMRYIKYIDDDLYEP-ETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLN 626
L Y Y++ +D D+ T SL+KL + NL+ +L+ +G E P L
Sbjct: 757 SLELYRGSAEYVEEVDIDVDSGFPTRIRLPSLRKLCICKFDNLKGLLKKEGGEQFPVLEE 816
Query: 627 LDITNVPKLTLT-SLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLK 685
++I P TL+ +L ++ SL+ S E + F + +A NLK L+IS F NLK
Sbjct: 817 MEIRYCPIPTLSPNLKALTSLNISDNKEA---TSFPEEMFKSLA--NLKYLNISHFKNLK 871
Query: 686 ELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTC 745
ELP L L AL+SL I+ C +E+ + +KGL+SL + V K L +G+ HLT
Sbjct: 872 ELPTSLASLNALKSLKIQWCCALENIPKEGVKGLTSLTELIVKFSKVLKCLPEGLHHLTA 931
Query: 746 LETLHIYYCPQLV 758
L L I+ CPQL+
Sbjct: 932 LTRLKIWGCPQLI 944
Score = 56.6 bits (135), Expect = 4e-06
Identities = 93/382 (24%), Positives = 138/382 (35%), Gaps = 87/382 (22%)
Query: 537 KNTSILKGLVSIILYNCKNCRHLPP-FGKLPCLTIL-------------------YLSGM 576
K TS L L +++L+ C PP G L CL L L G
Sbjct: 589 KQTSKLGSLRNLLLHGCHRLTRTPPRIGSLTCLKTLGQSVVKRKKGYQLGELGSLNLYGS 648
Query: 577 RYIKYIDDDLYEPETEKAFTSLKKLSLHDL---------PNLERVLEVDGVEMLPQLLNL 627
I +++ + E ++A S K+ +LH L P+ EV+ +E L NL
Sbjct: 649 IKISHLERVKNDKEAKEANLSAKE-NLHSLSMKWDDDEHPHRYESEEVEVLEALKPHSNL 707
Query: 628 DITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKEL 687
+ L N +LK N+ + IS N L
Sbjct: 708 TCLKISGFRGIRL-------PDWMNHSVLK--------------NIVLIEISGCKNCSCL 746
Query: 688 PVELGPLTALESLSIER--CNEMESFSEHLLKG------LSSLRNMSVFSCSGFKSL--S 737
P G L LESL + R +E + G L SLR + + K L
Sbjct: 747 P-PFGDLPCLESLELYRGSAEYVEEVDIDVDSGFPTRIRLPSLRKLCICKFDNLKGLLKK 805
Query: 738 DGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIPSLQKLRLF 797
+G LE + I YCP N+ +L SL E + + + +L+ L +
Sbjct: 806 EGGEQFPVLEEMEIRYCPIPTLSPNLKALTSLNISDNKEATSFPEEMFKSLANLKYLNIS 865
Query: 798 NFPSIKSLPDWLGAMTSLQVL------AICDFPE-------------------LSSLPDN 832
+F ++K LP L ++ +L+ L A+ + P+ L LP+
Sbjct: 866 HFKNLKELPTSLASLNALKSLKIQWCCALENIPKEGVKGLTSLTELIVKFSKVLKCLPEG 925
Query: 833 FQQLQNLQTLTISGCPILEKRC 854
L L L I GCP L KRC
Sbjct: 926 LHHLTALTRLKIWGCPQLIKRC 947
Score = 35.4 bits (80), Expect = 8.4
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 1/58 (1%)
Query: 790 SLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGC 847
SL+ L L ++ + LP +G + L+ + + + E+ SLP +LQNLQTL + C
Sbjct: 525 SLRVLNL-SYSKFEELPSSIGDLVHLRYMDLSNNIEIRSLPKQLCKLQNLQTLDLQYC 581
>gb|AAR29069.1| blight resistance protein RPI [Solanum bulbocastanum]
gi|32693281|gb|AAP86601.1| putative disease resistant
protein RGA2 [Solanum bulbocastanum]
gi|46576968|sp|Q7XBQ9|RGA2_SOLBU Disease resistance
protein RGA2 (RGA2-blb) (Blight resistance protein RPI)
Length = 970
Score = 425 bits (1092), Expect = e-117
Identities = 319/920 (34%), Positives = 481/920 (51%), Gaps = 114/920 (12%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L+ NL SF++ EL G + Q LS + I+AVL+DA++KQ+ N ++
Sbjct: 1 MAEAFIQVLLDNLTSFLKGELVLLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNNKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T + R+HP I R +GKRM ++ K++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFSQSEYGRYHPKVIPFRHKVGKRMDQVMKKLKAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F LH ++ERQ RR+T SV+TE +VYGRDK+K+ IV+ L+ + D++ LS
Sbjct: 120 AEERKNFHLHEKIVERQAV---RRETGSVLTEPQVYGRDKEKDEIVKILINNVSDAQHLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+RV HF KIW+CVS D + +++++I+E+ G+ P
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRVTEHFHSKIWICVSEDFDEKRLIKAIVESIEGR-PL 235
Query: 241 LSSLE--SMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLD 298
L ++ +Q+K+QE+L RYLLVLDDVW ED++KW L+++L G GAS+L TTRL+
Sbjct: 236 LGEMDLAPLQKKLQELLNGKRYLLVLDDVWNEDQQKWANLRAVLKVGASGASVLTTTRLE 295
Query: 299 IVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAK 358
V SIMGT + L++LS +D W LF Q+AFG E LVAIGK++V+K G PLAAK
Sbjct: 296 KVGSIMGTLQPYELSNLSQEDCWLLFMQRAFGHQEEINPNLVAIGKEIVKKSGGVPLAAK 355
Query: 359 VLGSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPK 417
LG LC E W V +S WNLP+ + SI+ ALR+SY L L L+ CFA+CAVFPK
Sbjct: 356 TLGGILCFKREERAWEHVRDSPIWNLPQDESSILPALRLSYHQLPLDLKQCFAYCAVFPK 415
Query: 418 GFEMVKENLIHLWMANGLVTSRGNLQMEHVGDEL-HNLQL----------GGKLHIKCLE 466
+M KE LI LWMA+G + S+GN+++E VGDE+ L L GK + K +
Sbjct: 416 DAKMEKEKLISLWMAHGFLLSKGNMELEDVGDEVWKELYLRSFFQEIEVKDGKTYFKMHD 475
Query: 467 NV-----------SNEEDARETNLIS---------------------KKDLDRLYLSWGN 494
+ ++ + RE N S +K + L+ G+
Sbjct: 476 LIHDLATSLFSANTSSSNIREINKHSYTHMMSIGFAEVVFFYTLPPLEKFISLRVLNLGD 535
Query: 495 DTNSQVGSVDAERV-LEALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNC 553
T +++ S + V L L + SG++ S K L+ L ++ L C
Sbjct: 536 STFNKLPSSIGDLVHLRYLNLYGSGMR-------------SLPKQLCKLQNLQTLDLQYC 582
Query: 554 KNCRHLP-PFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERV 612
LP KL L L L G + + + P + T LK L + ++
Sbjct: 583 TKLCCLPKETSKLGSLRNLLLDGSQSLTCM------PPRIGSLTCLKTLG-QFVVGRKKG 635
Query: 613 LEVDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDV-AGN 671
++ + L ++ I+++ ++ +LSA G L S +NN +
Sbjct: 636 YQLGELGNLNLYGSIKISHLERVKNDKDAKEANLSAKGNLHSL--SMSWNNFGPHIYESE 693
Query: 672 NLKSL-SISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSC 730
+K L ++ +NL T+L+ + E + +LK + S+ + +C
Sbjct: 694 EVKVLEALKPHSNL----------TSLKIYGFRGIHLPEWMNHSVLKNIVSILISNFRNC 743
Query: 731 SGFKSLSDGMRHLTCLETLHIYYCPQLV-------------FPHNMNSLASLRQLLLVEC 777
S D L CLE+L +++ V FP + SLR+L + +
Sbjct: 744 SCLPPFGD----LPCLESLELHWGSADVEYVEEVDIDVHSGFPTRIR-FPSLRKLDIWDF 798
Query: 778 N--ESIL--DGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDN- 832
+ +L +G E P L+++ + P + +L L A+TSL+ IC +S P+
Sbjct: 799 GSLKGLLKKEGEEQFPVLEEMIIHECPFL-TLSSNLRALTSLR---ICYNKVATSFPEEM 854
Query: 833 FQQLQNLQTLTISGCPILEK 852
F+ L NL+ LTIS C L++
Sbjct: 855 FKNLANLKYLTISRCNNLKE 874
Score = 195 bits (496), Expect = 5e-48
Identities = 130/321 (40%), Positives = 184/321 (56%), Gaps = 26/321 (8%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N++DA+E NL +K +L L +SW N+ + + +VL
Sbjct: 640 ELGNLNLYGSIKISHLERVKNDKDAKEANLSAKGNLHSLSMSW-NNFGPHIYESEEVKVL 698
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PHS+ L + G+ G P WM N S+LK +VSI++ N +NC LPPFG LPCL
Sbjct: 699 EALKPHSN-LTSLKIYGFRGIHLPEWM-NHSVLKNIVSILISNFRNCSCLPPFGDLPCLE 756
Query: 570 ILYL----SGMRYIKYIDDDLYEP-ETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQL 624
L L + + Y++ +D D++ T F SL+KL + D +L+ +L+ +G E P L
Sbjct: 757 SLELHWGSADVEYVEEVDIDVHSGFPTRIRFPSLRKLDIWDFGSLKGLLKKEGEEQFPVL 816
Query: 625 LNLDITNVPKLTLTS-LLSVESLS------ASGGNEELLKSFFYNNCSEDVAGNNLKSLS 677
+ I P LTL+S L ++ SL A+ EE+ K+ NLK L+
Sbjct: 817 EEMIIHECPFLTLSSNLRALTSLRICYNKVATSFPEEMFKNLA-----------NLKYLT 865
Query: 678 ISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLS 737
IS+ NLKELP L L AL+SL I+ C +ES E L+GLSSL + V C+ K L
Sbjct: 866 ISRCNNLKELPTSLASLNALKSLKIQLCCALESLPEEGLEGLSSLTELFVEHCNMLKCLP 925
Query: 738 DGMRHLTCLETLHIYYCPQLV 758
+G++HLT L +L I CPQL+
Sbjct: 926 EGLQHLTTLTSLKIRGCPQLI 946
Score = 89.7 bits (221), Expect = 4e-16
Identities = 71/235 (30%), Positives = 107/235 (45%), Gaps = 38/235 (16%)
Query: 672 NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSS---------- 721
N+ S+ IS F N LP G L LESL + + + E + + S
Sbjct: 731 NIVSILISNFRNCSCLP-PFGDLPCLESLELHWGSADVEYVEEVDIDVHSGFPTRIRFPS 789
Query: 722 LRNMSVFSCSGFKSL--SDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLR--------- 770
LR + ++ K L +G LE + I+ CP L N+ +L SLR
Sbjct: 790 LRKLDIWDFGSLKGLLKKEGEEQFPVLEEMIIHECPFLTLSSNLRALTSLRICYNKVATS 849
Query: 771 -------------QLLLVECN--ESILDGIEGIPSLQKLRLFNFPSIKSLPD-WLGAMTS 814
L + CN + + + + +L+ L++ +++SLP+ L ++S
Sbjct: 850 FPEEMFKNLANLKYLTISRCNNLKELPTSLASLNALKSLKIQLCCALESLPEEGLEGLSS 909
Query: 815 LQVLAICDFPELSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L L + L LP+ Q L L +L I GCP L KRC++GIGEDWHKI+HIP
Sbjct: 910 LTELFVEHCNMLKCLPEGLQHLTTLTSLKIRGCPQLIKRCEKGIGEDWHKISHIP 964
>gb|AAR29074.1| blight resistance protein SH10 [Solanum tuberosum]
Length = 948
Score = 421 bits (1083), Expect = e-116
Identities = 320/966 (33%), Positives = 475/966 (49%), Gaps = 130/966 (13%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +LI NL SF++ EL G Q LS + I+AVL+DA++KQ+ + ++
Sbjct: 1 MAEAFIQVLIDNLTSFLKGELVLLFGFQNEFQRLSSIFSTIQAVLEDAQEKQLNDKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T + R+HP I R +GKRM ++ K+++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFSQSAYGRYHPKVIPFRHKVGKRMDQVMKKLNAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F LH +IERQ RR+T SV+TE +VYGRDK+++ IV+ L+ + D++ LS
Sbjct: 120 AEERKNFHLHEKIIERQAV---RRETGSVLTEPQVYGRDKEEDEIVKILINNVSDAQHLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+R+ HF KIW+CVS D + ++L++IIE+ G+ P
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRITEHFHSKIWICVSEDFDEKRLLKAIIESIEGR-PL 235
Query: 241 LSSLE--SMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLD 298
L ++ +Q+K+QE+L RY LVLDDVW ED++KW L+++L G GA +L TTRL+
Sbjct: 236 LGEMDLAPLQKKLQELLNGKRYFLVLDDVWNEDQQKWANLRAVLKVGASGAFVLATTRLE 295
Query: 299 IVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAK 358
V SIMGT + L++LS +D W LF Q AFG E LVAIGK++V+K G PLAAK
Sbjct: 296 KVGSIMGTLQPYELSNLSQEDCWLLFIQCAFGHQEEINPNLVAIGKEIVKKSGGVPLAAK 355
Query: 359 VLGSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPK 417
LG L E +W V +SE WNLP+ + SI+ ALR+SY +L L LR CFA+CAVFPK
Sbjct: 356 TLGGILRFKREEREWEHVRDSEIWNLPQEERSILPALRLSYHHLPLDLRQCFAYCAVFPK 415
Query: 418 GFEMVKENLIHLWMANGLVTSRGNLQME----HVGDELHNLQLGGKLHIKCLENV----- 468
+M KE LI LWMA+G + G LQ E V EL ++ KC +
Sbjct: 416 DTKMEKEKLISLWMAHGFLLLEGKLQPEDVGNEVSKELCLRSFFQEIEAKCGKTYFKMHD 475
Query: 469 -------------SNEEDARETNL----------------------ISKKDLDRLYLSWG 493
++ + RE N+ +S+K + L+
Sbjct: 476 LHHDLATSLFSASTSSSNIREINVKGYPHKMMSIGFTEVVSSYSPSLSQKFVSLRVLNLS 535
Query: 494 N----DTNSQVGSVDAERVLEALEPHSSGLKHFGV--------------NGYGGTIFPSW 535
N + +S +G + R L+ E +SG++ N Y + P
Sbjct: 536 NLHFEELSSSIGDLVHMRCLDLSE--NSGIRSLPKQLCKLQNLQTLDLHNCYSLSCLP-- 591
Query: 536 MKNTSILKGLVSIILYNCKNCRHLPP-FGKLPCLTIL-------YLSGMRYIKYIDDDLY 587
K S L L ++ + C +PP G L L L G + K D +LY
Sbjct: 592 -KEPSKLGSLRNLFFHGCDELNSMPPRIGSLTFLKTLKWICCGIQKKGYQLGKLRDVNLY 650
Query: 588 EP------ETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLDITNVPKLTLTSLL 641
E K K+ +L NL ++ ++ P + + V + L
Sbjct: 651 GSIEITHLERVKNVMDAKEANLSAKGNLHSLI-MNWSRKGPHIYESEEVRVIE-ALKPHP 708
Query: 642 SVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKELPVELGPLTALESLS 701
++ L+ SG + F + N+ S+ IS N LP G L L+ L
Sbjct: 709 NLTCLTISG-----FRGFRFPEWMNHSVLKNVVSIEISGCKNCSCLP-PFGELPCLKRLE 762
Query: 702 IERCNEMESFSEH---LLKGLSSLRNMSVFSCSGFKSL--SDGMRHLTCLETLHIYYCPQ 756
+++ + + + + SLR + + K L +G LE + I+YC
Sbjct: 763 LQKGSAEVEYVDSGFPTRRRFPSLRKLFIGEFPNLKGLLKKEGEEKFPVLERMTIFYCHM 822
Query: 757 LVFPHNMNSLASLRQLLLVECNE--------------------SILDGIEGIPS------ 790
V+ ++ +L L + NE S+ ++ +PS
Sbjct: 823 FVYTTLSSNFRALTSLHISHNNEATSLPEEIFKSFANLKYLKISLFYNLKELPSSLACLN 882
Query: 791 -LQKLRLFNFPSIKSLPD-WLGAMTSLQVLAICDFPELSSLPDNFQQLQNLQTLTISGCP 848
L+ L + + +++SLP+ + +TSL L + D L LP+ Q L L +L + CP
Sbjct: 883 ALKTLEIHSCSALESLPEEGVKGLTSLTELFVYDCEMLKFLPEGLQHLTALTSLKLRRCP 942
Query: 849 ILEKRC 854
L KRC
Sbjct: 943 QLIKRC 948
Score = 191 bits (485), Expect = 9e-47
Identities = 125/320 (39%), Positives = 175/320 (54%), Gaps = 28/320 (8%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
+L ++ L G + I LE V N DA+E NL +K +L L ++W + + RV+
Sbjct: 643 KLRDVNLYGSIEITHLERVKNVMDAKEANLSAKGNLHSLIMNWSRK-GPHIYESEEVRVI 701
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + L ++G+ G FP WM N S+LK +VSI + CKNC LPPFG+LPCL
Sbjct: 702 EALKPHPN-LTCLTISGFRGFRFPEWM-NHSVLKNVVSIEISGCKNCSCLPPFGELPCLK 759
Query: 570 ILYLS-GMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLNLD 628
L L G ++Y+D T + F SL+KL + + PNL+ +L+ +G E P L +
Sbjct: 760 RLELQKGSAEVEYVDSGF---PTRRRFPSLRKLFIGEFPNLKGLLKKEGEEKFPVLERMT 816
Query: 629 ITNVPKLTLTSLLS----VESLSASGGNE------ELLKSFFYNNCSEDVAGNNLKSLSI 678
I T+L S + SL S NE E+ KSF NLK L I
Sbjct: 817 IFYCHMFVYTTLSSNFRALTSLHISHNNEATSLPEEIFKSFA-----------NLKYLKI 865
Query: 679 SKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSD 738
S F NLKELP L L AL++L I C+ +ES E +KGL+SL + V+ C K L +
Sbjct: 866 SLFYNLKELPSSLACLNALKTLEIHSCSALESLPEEGVKGLTSLTELFVYDCEMLKFLPE 925
Query: 739 GMRHLTCLETLHIYYCPQLV 758
G++HLT L +L + CPQL+
Sbjct: 926 GLQHLTALTSLKLRRCPQLI 945
>gb|AAP45164.1| putative disease resistant protein RGA2 [Solanum bulbocastanum]
Length = 819
Score = 420 bits (1079), Expect = e-115
Identities = 220/454 (48%), Positives = 304/454 (66%), Gaps = 8/454 (1%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +L+ NL SF++ EL G + Q LS + I+AVL+DA++KQ+ N ++
Sbjct: 1 MAEAFIQVLLDNLTSFLKGELVLLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNNKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T + R+HP I R +GKRM ++ K++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFSQSEYGRYHPKVIPFRHKVGKRMDQVMKKLKAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELS 180
AEER F LH ++ERQ RR+T SV+TE +VYGRDK+K+ IV+ L+ + D++ LS
Sbjct: 120 AEERKNFHLHEKIVERQAV---RRETGSVLTEPQVYGRDKEKDEIVKILINNVSDAQHLS 176
Query: 181 VYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPH 240
V I+G GG GKTTLAQ VFND+RV HF KIW+CVS D + +++++I+E+ G+ P
Sbjct: 177 VLPILGMGGLGKTTLAQMVFNDQRVTEHFHSKIWICVSEDFDEKRLIKAIVESIEGR-PL 235
Query: 241 LSSLE--SMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLD 298
L ++ +Q+K+QE+L RYLLVLDDVW ED++KW L+++L G GAS+L TTRL+
Sbjct: 236 LGEMDLAPLQKKLQELLNGKRYLLVLDDVWNEDQQKWANLRAVLKVGASGASVLTTTRLE 295
Query: 299 IVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAK 358
V SIMGT + L++LS +D W LF Q+AFG E LVAIGK++V+K G PLAAK
Sbjct: 296 KVGSIMGTLQPYELSNLSQEDCWLLFMQRAFGHQEEINPNLVAIGKEIVKKSGGVPLAAK 355
Query: 359 VLGSSLCCTSNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPK 417
LG LC E W V +S WNLP+ + SI+ ALR+SY L L L+ CFA+CAVFPK
Sbjct: 356 TLGGILCFKREERAWEHVRDSPIWNLPQDESSILPALRLSYHQLPLDLKQCFAYCAVFPK 415
Query: 418 GFEMVKENLIHLWMANGLVTSRGNLQMEHVGDEL 451
+M KE LI LWMA+G + S+GN+++E VGDE+
Sbjct: 416 DAKMEKEKLISLWMAHGFLLSKGNMELEDVGDEV 449
Score = 120 bits (302), Expect = 2e-25
Identities = 73/180 (40%), Positives = 107/180 (58%), Gaps = 8/180 (4%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N++DA+E NL +K +L L +SW N+ + + +VL
Sbjct: 640 ELGNLNLYGSIKISHLERVKNDKDAKEANLSAKGNLHSLSMSW-NNFGPHIYESEEVKVL 698
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PHS+ L + G+ G P WM N S+LK +VSI++ N +NC LPPFG LPCL
Sbjct: 699 EALKPHSN-LTSLKIYGFRGIHLPEWM-NHSVLKNIVSILISNFRNCSCLPPFGDLPCLE 756
Query: 570 ILYL----SGMRYIKYIDDDLYEP-ETEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQL 624
L L + + Y++ +D D++ T F SL+KL + D +L+ +L+ +G E P L
Sbjct: 757 SLELHWGSADVEYVEEVDIDVHSGFPTRIRFPSLRKLDIWDFGSLKGLLKKEGEEQFPVL 816
>gb|AAP45163.1| putative disease resistant protein RGA1 [Solanum bulbocastanum]
gi|46576967|sp|Q7XA42|RGA1_SOLBU Putative disease
resistance protein RGA1 (RGA3-blb)
Length = 1025
Score = 412 bits (1058), Expect = e-113
Identities = 331/1051 (31%), Positives = 490/1051 (46%), Gaps = 214/1051 (20%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A + +++ NL SF++ EL G + Q LS + I+AVL+DA++KQ+ + ++
Sbjct: 1 MAEAFIQVVLDNLTSFLKGELVLLFGFQDEFQRLSSMFSTIQAVLEDAQEKQLNDKPLEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L A Y +DDILDE T R+HP I R +GKRM ++ K+++ I
Sbjct: 61 WLQKLNAATYEVDDILDEYK-TKATRFLQSEYGRYHPKVIPFRHKVGKRMDQVMKKLNAI 119
Query: 121 AEERMKFGLHVGVIERQPEDEGRRQTTS-------------------------------- 148
AEER KF L +IERQ R+T S
Sbjct: 120 AEERKKFHLQEKIIERQA---ATRETVSMFSTCSSSHPYLFIVQNPSLFNYFLPTPKELE 176
Query: 149 --------------VITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSIVGHGGYGKTT 194
V+TE +VYGRDK+K+ IV+ L+ A D+++LSV I+G GG GKTT
Sbjct: 177 CNNILIWTLLAPGSVLTEPQVYGRDKEKDEIVKILINTASDAQKLSVLPILGMGGLGKTT 236
Query: 195 LAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPHLSSLESMQQKVQEI 254
L+Q VFND+RV F KIW+C+S D N +++++I+E+ GK+ L +Q+K+QE+
Sbjct: 237 LSQMVFNDQRVTERFYPKIWICISDDFNEKRLIKAIVESIEGKSLSDMDLAPLQKKLQEL 296
Query: 255 LQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIVASIMGTSDAHHLAS 314
L RY LVLDDVW ED+ KW L+++L G GA +L TTRL+ V SIMGT + L++
Sbjct: 297 LNGKRYFLVLDDVWNEDQHKWANLRAVLKVGASGAFVLTTTRLEKVGSIMGTLQPYELSN 356
Query: 315 LSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVLGSSLCCTSNEHQWI 374
LS +D W LF Q+AFG E L+AIGK++V+KC G PLAAK LG L E +W
Sbjct: 357 LSPEDCWFLFMQRAFGHQEEINPNLMAIGKEIVKKCGGVPLAAKTLGGILRFKREEREWE 416
Query: 375 SVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGFEMVKENLIHLWMAN 433
V +S WNLP+ + SI+ ALR+SY +L L LR CF +CAVFPK +M KENLI WMA+
Sbjct: 417 HVRDSPIWNLPQDESSILPALRLSYHHLPLDLRQCFVYCAVFPKDTKMAKENLIAFWMAH 476
Query: 434 GLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWG 493
G + S+GNL++E VG+E+ N +L + + +E S + + +LI DL S
Sbjct: 477 GFLLSKGNLELEDVGNEVWN-ELYLRSFFQEIEVESGKTYFKMHDLI--HDLATSLFS-A 532
Query: 494 NDTNSQVGSVDAERVLEALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNC 553
N ++S + ++A + + G + P S+L+ VS+ + N
Sbjct: 533 NTSSSNIREINAN--------YDGYMMSIGFAEVVSSYSP------SLLQKFVSLRVLNL 578
Query: 554 K--NCRHLP-PFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLE 610
+ N LP G L L L LSG I+ + P+ +L+ L LH +L
Sbjct: 579 RNSNLNQLPSSIGDLVHLRYLDLSGNFRIRNL------PKRLCKLQNLQTLDLHYCDSLS 632
Query: 611 RV-LEVDGVEMLPQLL--NLDITNVPK----LTLTSLLSVESLSASGGNE--ELLKSFFY 661
+ + + L LL +T+ P LT LS + G++ EL Y
Sbjct: 633 CLPKQTSKLGSLRNLLLDGCSLTSTPPRIGLLTCLKSLSCFVIGKRKGHQLGELKNLNLY 692
Query: 662 NNCS-----EDVAGNNLKSLSISKFANLKELPV----------ELGPLTALESLSIERCN 706
+ S + K ++S ANL L + + L AL+ S +
Sbjct: 693 GSISITKLDRVKKDTDAKEANLSAKANLHSLCLSWDLDGKHRYDSEVLEALKPHSNLKYL 752
Query: 707 EMESF---------SEHLLKGLSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIY----- 752
E+ F ++ +LK + S+R +CS + L CLE+L ++
Sbjct: 753 EINGFGGIRLPDWMNQSVLKNVVSIRIRGCENCSCLPPFGE----LPCLESLELHTGSAD 808
Query: 753 --YCPQLVFPHNMNSL------------------------------------------AS 768
Y V P SL +S
Sbjct: 809 VEYVEDNVHPGRFPSLRKLVIWDFSNLKGLLKMEGEKQFPVLEEMTFYWCPMFVIPTLSS 868
Query: 769 LRQLLLVECNESILDGIEGIPSLQKLRLFNFPSIKSLP-DWLGAMTSLQVLAICDFPELS 827
++ L ++ + ++L I + +L L + + SLP + ++ +L+ L I F L
Sbjct: 869 VKTLKVIVTDATVLRSISNLRALTSLDISDNVEATSLPEEMFKSLANLKYLKISFFRNLK 928
Query: 828 SLPDNFQQLQNLQTLTISGCPILE------------------------------------ 851
LP + L L++L C LE
Sbjct: 929 ELPTSLASLNALKSLKFEFCDALESLPEEGVKGLTSLTELSVSNCMMLKCLPEGLQHLTA 988
Query: 852 -------------KRCKRGIGEDWHKIAHIP 869
KRC+RGIGEDWHKIAHIP
Sbjct: 989 LTTLTITQCPIVFKRCERGIGEDWHKIAHIP 1019
>gb|AAR29072.1| blight resistance protein RGA4 [Solanum bulbocastanum]
Length = 1040
Score = 406 bits (1044), Expect = e-111
Identities = 227/507 (44%), Positives = 314/507 (61%), Gaps = 57/507 (11%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
MA+A L +L++NL SF+ ++L G + + LS + I+AVL+DA++KQ+ + A++
Sbjct: 1 MAEAFLQVLLENLTSFIGDKLVLIFGFEKECEKLSSVFSTIQAVLQDAQEKQLKDKAIEN 60
Query: 61 WLQQLRDAAYVLDDILDECSITLKAHGNNKRITRFHPMKILVRRNIGKRMKEIAKEIDDI 120
WLQ+L AAY +DDIL EC R+ +HP I R IG+RMKEI +++D I
Sbjct: 61 WLQKLNSAAYEVDDILGECK-NEAIRFEQSRLGFYHPGIINFRHKIGRRMKEIMEKLDAI 119
Query: 121 AEERMKFG-------------------------------LHVGVIER------------- 136
+EER KF L +GV+ R
Sbjct: 120 SEERRKFHFLEKITERQAAAATRETVGWQWGWARLEYKRLLLGVLMRIMSLRMHVSTCST 179
Query: 137 ---------QPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSIVGH 187
P+ RR V+TE KVYGRDK+++ IV+ L+ + +EEL V+ I+G
Sbjct: 180 LYEFKFYLCTPKVGARR--CFVLTEPKVYGRDKEEDEIVKILINNVNVAEELPVFPIIGM 237
Query: 188 GGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPHLSSLESM 247
GG GKTTLAQ +FNDERV HF+ KIWVCVS D + +++++II N +PH+ L S
Sbjct: 238 GGLGKTTLAQMIFNDERVTKHFNPKIWVCVSDDFDEKRLIKTIIGNIERSSPHVEDLASF 297
Query: 248 QQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIVASIMGTS 307
Q+K+QE+L RYLLVLDDVW +D EKW KL+++L G +GASIL TTRL+ V SIMGT
Sbjct: 298 QKKLQELLNGKRYLLVLDDVWNDDLEKWAKLRAVLTVGARGASILATTRLEKVGSIMGTL 357
Query: 308 DAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVRKCVGSPLAAKVLGSSLCCT 367
+HL++LS D LF Q+AFG+ +E LVAIGK++V+KC G PLAAK LG L
Sbjct: 358 QPYHLSNLSPHDSLLLFMQRAFGQQKEANPNLVAIGKEIVKKCGGVPLAAKTLGGLLRFK 417
Query: 368 SNEHQWISVLESEFWNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGFEMVKENL 426
E +W V ++E W+LP+ + SI+ ALR+SY +L L LR CFA+CAVFPK +M+KENL
Sbjct: 418 REESEWEHVRDNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDTKMIKENL 477
Query: 427 IHLWMANGLVTSRGNLQMEHVGDELHN 453
I LWMA+G + S+GNL++E VG+E+ N
Sbjct: 478 ITLWMAHGFLLSKGNLELEDVGNEVWN 504
Score = 185 bits (469), Expect = 7e-45
Identities = 123/329 (37%), Positives = 179/329 (54%), Gaps = 25/329 (7%)
Query: 450 ELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDAERVL 509
EL NL L G + I LE V N+ DA E NL +K +L L +SW ND ++ S + +VL
Sbjct: 692 ELKNLNLCGSISITHLERVKNDTDA-EANLSAKANLQSLSMSWDNDGPNRYESKEV-KVL 749
Query: 510 EALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLT 569
EAL+PH + LK+ + +GG FPSW+ N S+L+ ++S+ + +CKNC LPPFG+LPCL
Sbjct: 750 EALKPHPN-LKYLEIIAFGGFRFPSWI-NHSVLEKVISVRIKSCKNCLCLPPFGELPCLE 807
Query: 570 ILYL-SGMRYIKYIDDDLYEPE--TEKAFTSLKKLSLHDLPNLERVLEVDGVEMLPQLLN 626
L L +G ++Y+++D T ++F SLKKL + +L+ +++ +G E P L
Sbjct: 808 NLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKFPMLEE 867
Query: 627 LDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNC---SEDVAGN------------ 671
+ I P +L SV+ L G S N S + N
Sbjct: 868 MAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLPEEMFT 927
Query: 672 ---NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVF 728
NL+ LS F NLK+LP L L AL+ L IE C+ +ESF E L+GL+SL + V
Sbjct: 928 SLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLTQLFVK 987
Query: 729 SCSGFKSLSDGMRHLTCLETLHIYYCPQL 757
C K L +G++HLT L L + CP++
Sbjct: 988 YCKMLKCLPEGLQHLTALTNLGVSGCPEV 1016
Score = 89.4 bits (220), Expect = 5e-16
Identities = 108/413 (26%), Positives = 163/413 (39%), Gaps = 86/413 (20%)
Query: 517 SGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGM 576
S L+H V+G T P + + LK L I+ + K + G+L L + +
Sbjct: 649 SSLRHLVVDGCPLTSTPPRIGLLTCLKTLGFFIVGSKKGYQ----LGELKNLNLCGSISI 704
Query: 577 RYIKYIDDDLYEPETEKAFTSLKKLSL---HDLPNLERVLEVDGVEMLPQLLNLDITNVP 633
+++ + +D A +L+ LS+ +D PN EV +E L NL +
Sbjct: 705 THLERVKNDTDAEANLSAKANLQSLSMSWDNDGPNRYESKEVKVLEALKPHPNLKYLEI- 763
Query: 634 KLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANLKELPVELGP 693
A GG S+ ++ E V +KS N LP G
Sbjct: 764 -------------IAFGGFR--FPSWINHSVLEKVISVRIKSCK-----NCLCLP-PFGE 802
Query: 694 LTALESLSIERCNEMESFSEH--------LLKGLSSLRNMSVFSCSGFKSL--SDGMRHL 743
L LE+L ++ + + E + SL+ + ++ K L +G
Sbjct: 803 LPCLENLELQNGSAEVEYVEEDDVHSRFSTRRSFPSLKKLRIWFFRSLKGLMKEEGEEKF 862
Query: 744 TCLETLHIYYCPQLVFP----------------------HNMNSLASLRQLLLVECNESI 781
LE + I YCP VFP N+++L SLR
Sbjct: 863 PMLEEMAILYCPLFVFPTLSSVKKLEVHGNTNTRGLSSISNLSTLTSLRIGANYRATSLP 922
Query: 782 LDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAI--CD----FPE---------- 825
+ + +L+ L F+F ++K LP L ++ +L+ L I CD FPE
Sbjct: 923 EEMFTSLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGLTSLT 982
Query: 826 ---------LSSLPDNFQQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
L LP+ Q L L L +SGCP +EKRC + IGEDWHKIAHIP
Sbjct: 983 QLFVKYCKMLKCLPEGLQHLTALTNLGVSGCPEVEKRCDKEIGEDWHKIAHIP 1035
Score = 39.7 bits (91), Expect = 0.44
Identities = 76/314 (24%), Positives = 128/314 (40%), Gaps = 60/314 (19%)
Query: 583 DDDLYE-PETEKAFTSLKKLSLHDLPNLER-------VLEVDGVEMLPQLLNLDITNVPK 634
D++++ P+ E + +LS H LP R V D + L+ L + +
Sbjct: 428 DNEIWSLPQDESSILPALRLSYHHLPLDLRQCFAYCAVFPKDTKMIKENLITLWMAHGFL 487
Query: 635 LTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGNN-------LKSLSISKFA----- 682
L+ +L +E + NE L+SFF E +GN + L+ S F+
Sbjct: 488 LSKGNL-ELEDVGNEVWNELYLRSFFQE--IEAKSGNTYFKIHDLIHDLATSLFSASASC 544
Query: 683 -NLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSV-------------- 727
N++E+ V+ ++SI + S+S LLK SLR +++
Sbjct: 545 GNIREINVK----DYKHTVSIGFAAVVSSYSPSLLKKFVSLRVLNLSYSKLEQLPSSIGD 600
Query: 728 --------FSCSGFKSLSDGMRHLTCLETLHIYYCPQL-VFPHNMNSLASLRQLLLVECN 778
SC+ F+SL + + L L+TL ++ C L P + L+SLR L++ C
Sbjct: 601 LLHLRYLDLSCNNFRSLPERLCKLQNLQTLDVHNCYSLNCLPKQTSKLSSLRHLVVDGCP 660
Query: 779 -ESILDGIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPD------ 831
S I + L+ L F S K LG + +L + L + +
Sbjct: 661 LTSTPPRIGLLTCLKTLGFFIVGSKKGYQ--LGELKNLNLCGSISITHLERVKNDTDAEA 718
Query: 832 NFQQLQNLQTLTIS 845
N NLQ+L++S
Sbjct: 719 NLSAKANLQSLSMS 732
>gb|AAP52240.1| putative NBS-LRR type resistance protein [Oryza sativa (japonica
cultivar-group)] gi|37531302|ref|NP_919953.1| putative
NBS-LRR type resistance protein [Oryza sativa (japonica
cultivar-group)] gi|22655799|gb|AAN04216.1| Putative
NBS-LRR type resistance protein [Oryza sativa (japonica
cultivar-group)] gi|18652501|gb|AAL77135.1| Putative
NBS-LRR type resistance protein [Oryza sativa]
Length = 804
Score = 326 bits (836), Expect = 2e-87
Identities = 257/832 (30%), Positives = 409/832 (48%), Gaps = 100/832 (12%)
Query: 34 LSRKLTLIRAVLKDAEKKQITNDAVKEWLQQLRDAAYVLDDILDECSI-TLKAHGNNKRI 92
L L I VL DAE+KQ T+ A+KEWL++L+D Y +DDILD+ S TLK + +
Sbjct: 42 LQNSLQAISGVLLDAERKQSTSSALKEWLRKLKDVMYDIDDILDDASTETLKRRVSKDVV 101
Query: 93 TR---FHPMKILVRRNIGKRMK-------EIAKEIDDIAEERMKFGLHVGVIERQPEDEG 142
T+ H ++ +RR + KR K E+ +++++IA + FGL + Q +E
Sbjct: 102 TQTNCVHISRLKLRRKLLKRKKKWSSRIREVHEKLNEIAASKKDFGLTDWTVGGQCSEEP 161
Query: 143 RRQTTSVITESKVYGRDKDKEHIVEFLLRHAGDSEELSVYSIVGHGGYGKTTLAQTVFND 202
R++ S + + + GRD ++ IV +LR A + ++ V ++G GG GKT LA V++D
Sbjct: 162 ERESYSFVYQPDIIGRDDARDEIVSKILR-AAEHHDIFVLPLLGLGGIGKTELANMVYHD 220
Query: 203 ERVKTHFDLKIWVCVSGDINAMKVLESIIENTIGKNPHLSSLESMQQKVQEILQKNRYLL 262
++++ F +W CVS N +L+ IIE+ G++ +LE +Q K++ ILQ Y L
Sbjct: 221 QQIRERFSKMMWACVSNKFNLKNILQDIIESASGESCKHLNLEHLQNKLRGILQNGNYFL 280
Query: 263 VLDDVWTEDKEKWNKLKSLLLNGKKGASILITTRLDIVASIMGTSDAHHLASLSDDDIWS 322
VLDD+WT D +W +L++LL +G +G+ I++TTR ++VAS++GTSD + + +L +
Sbjct: 281 VLDDLWTRDVNEWRELRNLLSSGARGSVIIVTTRENVVASMVGTSDPYKVGALPFHECMQ 340
Query: 323 LFKQQAFGENREERAE-LVAIGKKLVRKCVGSPLAAKVLGSSLCCTSNEHQWISVLESEF 381
+F + AF + E + L+ IG+ +V+KC G PLA K LGS L +E QW+ V E
Sbjct: 341 IFTRVAFRQGEENKYPWLLKIGESIVKKCAGVPLAIKSLGSLLFTMRDETQWLRVKEDHL 400
Query: 382 WNLPEVD-SIMSALRISYFNLKLSLRPCFAFCAVFPKGFEMVKENLIHLWMANGLVTSRG 440
+ + D IM L++SY L +L+PC ++ ++FPK FE + +I WMA+GL+ S
Sbjct: 401 CKIVQGDRDIMPVLKLSYNALPAALKPCLSYLSIFPKDFEYYRRCIIMFWMAHGLLNSNK 460
Query: 441 NLQMEHVGDELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQV 500
+ VG++ +G L L + + I DL R L DT+ V
Sbjct: 461 LSEEIDVGNQYIIELIGSSLFQDSLITFDGSMPHCKLHDI-VHDLGRYVL----DTDLAV 515
Query: 501 GSVDAERVLEALEPHSSGLKHFG---------VNGYGGTIFPSWMKNTSILKGLVSIILY 551
+ + ++V E + LK F + F S + S+ K + ++L
Sbjct: 516 VNCEGQQVSETVRHLVWDLKDFTHEQEFPRHLIKARKARTFISSCNHGSLSKKFLEVLL- 574
Query: 552 NCKNCRHLPPFGKLPCLTILYLSGMRYIKYIDDDLYEPETEKAFTSLKKLSLHDLPNLER 611
KL L +L +SG+R ID E + +LK L DL
Sbjct: 575 -----------SKLLLLRVLIISGVR----ID------ELPDSIGNLKHLRYLDL----- 608
Query: 612 VLEVDGVEMLPQLLNLDITNVPKLTLTSLLSVESLSASGGNEELLKSFFYNNCSEDVAGN 671
N I +P +L L+++++L +L +S DV N
Sbjct: 609 ------------TWNKTIKYLPN-SLCKLINLQTL-------DLYRSDQLIELPRDV--N 646
Query: 672 NLKSLS-ISKFANLKELP-VELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFS 729
L SL +S + K+LP L +L SL + C+E+ S SE + L++L+ + +
Sbjct: 647 KLISLRYLSLTSKQKQLPEAGLRGWASLTSLQLHSCSELTSLSEG-IGSLTALQMLWISD 705
Query: 730 CSGFKSLSDGMRHLTCLETLHIYYCPQLVFPHNMNSLASLRQLLLVECNESILDGIEGIP 789
C SL M HL+ L L I CP+L H + ++G+
Sbjct: 706 CPKLPSLPASMTHLSSLRELFIDNCPELDLMHPE-------------------EAMDGLW 746
Query: 790 SLQKLRLFNFPSIKSLPDWL-GAMTSLQVLAICDFPELSSLPDNFQQLQNLQ 840
SL+ L++ P ++ LPD L A SL+ L I P L LP Q L N Q
Sbjct: 747 SLRSLQIIGLPKLERLPDTLCSASGSLRYLLIEQCPNLRELPSFMQDLTNHQ 798
Score = 71.6 bits (174), Expect = 1e-10
Identities = 47/135 (34%), Positives = 74/135 (54%), Gaps = 2/135 (1%)
Query: 719 LSSLRNMSVFSCSGFKSLSDGMRHLTCLETLHIYYCPQLV-FPHNMNSLASLRQLLLVEC 777
L LR + + K L + + L L+TL +Y QL+ P ++N L SLR L L
Sbjct: 600 LKHLRYLDLTWNKTIKYLPNSLCKLINLQTLDLYRSDQLIELPRDVNKLISLRYLSLTSK 659
Query: 778 NESILD-GIEGIPSLQKLRLFNFPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNFQQL 836
+ + + G+ G SL L+L + + SL + +G++T+LQ+L I D P+L SLP + L
Sbjct: 660 QKQLPEAGLRGWASLTSLQLHSCSELTSLSEGIGSLTALQMLWISDCPKLPSLPASMTHL 719
Query: 837 QNLQTLTISGCPILE 851
+L+ L I CP L+
Sbjct: 720 SSLRELFIDNCPELD 734
>gb|AAU43949.1| putative NBS-LRR protein [Oryza sativa (japonica cultivar-group)]
gi|47777415|gb|AAT38049.1| putative NBS-LRR resistance
protein [Oryza sativa (japonica cultivar-group)]
Length = 1222
Score = 313 bits (802), Expect = 2e-83
Identities = 180/463 (38%), Positives = 269/463 (57%), Gaps = 20/463 (4%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
+ ALL L++ G + E + G+ L L + V+ DAE + AVK
Sbjct: 4 LLSALLPALLKKAGESLGTEFSFIGGIERRRSELYTLLLAVNQVINDAEDQASKKPAVKS 63
Query: 61 WLQQLRDAAYVLDDILDE-------CSITLKAHGNNKRITRF---HPMKILVRRNIGKRM 110
W+ +L+ AA DD LDE C + H N + F H +L + IGKR+
Sbjct: 64 WIAKLKLAACDADDALDELHYEELRCEALRRGHKINTGVRAFFSSHYNPLLFKYRIGKRL 123
Query: 111 KEIAKEIDDIAEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLL 170
++I + ID + + +FG + P DE R QT S + E +V GRDK+++ IV LL
Sbjct: 124 QQIVERIDQLVSQMNRFGF---LNCSMPVDE-RMQTYSYVDEQEVIGRDKERDEIVHMLL 179
Query: 171 RHAGDSEELSVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESI 230
+ +++EL + IVG GG GKTTLAQ VFND +VK HF +WVCVS + + +++ I
Sbjct: 180 --SAETDELLILPIVGIGGLGKTTLAQLVFNDVKVKAHFQKHMWVCVSENFSVPVIVKGI 237
Query: 231 IENTIGKNPHLS--SLESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKG 288
I+ IG + L +LE +QQ+++E L + RYLLVLDDVW EDK+KW L++LL + G
Sbjct: 238 IDTAIGNDCGLKFDNLELLQQRLREELGQKRYLLVLDDVWNEDKQKWGALRTLLGSCGMG 297
Query: 289 ASILITTRLDIVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVR 348
+++++TTR VASIM + L +L+ +D W +F ++AFG E ELV +GK++V
Sbjct: 298 SAVVVTTRNVKVASIMESISPLCLENLNPEDSWIVFSRRAFGTGVVETPELVEVGKRIVE 357
Query: 349 KCVGSPLAAKVLGSSLCCTSNEHQWISVLESEFWNLPEVDSIMSALRISYFNLKLSLRPC 408
KC G PLA K +G+ + W+S+LES W+ E I+ AL + Y NL ++ C
Sbjct: 358 KCCGLPLAIKSMGALMSTKQETRDWLSILESNTWD--EESQILPALSLGYKNLPSHMKQC 415
Query: 409 FAFCAVFPKGFEMVKENLIHLWMANGLVTSRGNLQMEHVGDEL 451
FAFCAVFPK +E+ K++LIHLW++NG + S+ +E G+ +
Sbjct: 416 FAFCAVFPKDYEIDKDDLIHLWVSNGFIPSKKMSDIEENGNHV 458
Score = 149 bits (375), Expect = 5e-34
Identities = 155/544 (28%), Positives = 239/544 (43%), Gaps = 134/544 (24%)
Query: 449 DELHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLDRLYLSWGNDTNSQVGSVDA--- 505
+EL++L+LGGKL I L V+N +A+E NL K +L +L L WG ++++ + D
Sbjct: 683 NELNDLKLGGKLQIFNLIKVTNPIEAKEANLECKTNLQQLALCWGTSKSAELQAEDLHLY 742
Query: 506 --ERVLEALEPHSSGLKHFGVNGYGGTIFPSWMKNTSILKGLVSIILYNCKNCRHLPPFG 563
E VL+AL+P +GL + Y GT FP WM+N L+ +V + + + NC LP
Sbjct: 743 RHEEVLDALKP-PNGLTVLKLRQYMGTTFPIWMENGITLRNIVKLKVTDSINCMKLPSVW 801
Query: 564 KLPCLTILYLSGMRYIKYIDDDL-YEPETEK---AFTSLKKLSLHDLPNLERVLEVDGVE 619
KLP L +L L M+ +KY+ + + E + AF LK LSL + +LE E D +
Sbjct: 802 KLPFLEVLRLKDMKKLKYLCNGFCSDKECDHQLVAFPKLKLLSLERMESLENWQEYDVEQ 861
Query: 620 M----LPQLLNLDITNVPKLTL------------------------TSLLSVESLSASGG 651
+ P L ++I + PKLT S LS L AS G
Sbjct: 862 VTPANFPVLDAMEIIDCPKLTAMPNAPVLKSLSVIGNKILIGLSSSVSNLSYLYLGASQG 921
Query: 652 NEELLKSFFYNNCSEDVAG----------------NNLKSLSISKFANLKELPVE--LGP 693
+ E K+ Y + E++ G +L L + F+ L ++ G
Sbjct: 922 SLERKKTLIY-HYKENLEGTTDSKDHVLAHHFSSWGSLTKLHLQGFSALAPEDIQNISGH 980
Query: 694 LTALESLSI------------------------------ERCNEMESFSEHLLKGLSSLR 723
+ ++++L + E CN + + + L+SL+
Sbjct: 981 VMSVQNLDLISCDCFIQYDTLQSPLWFWKSFACLQHLTIEYCNSLTFWPGEEFQSLTSLK 1040
Query: 724 NMSVFSCSGF----------KSLSDGMRHLTCLETLHIYYCPQLV-FPHNMNSLASLRQL 772
+ + C+ F KS D H LE + I +C LV FP SL L
Sbjct: 1041 RLDIRYCNNFTGMPPAQVSVKSFEDEGMH--NLERIEIEFCYNLVAFP------TSLSYL 1092
Query: 773 LLVECN--ESILDGIEGIPSLQKLRLFNFPSIKSLPD-----------WLGA-------- 811
+ CN E + +G+ + +L+ L + P +KSLP +LG
Sbjct: 1093 RICSCNVLEDLPEGLGCLGALRSLSIDYNPRLKSLPPSIQRLSNLTRLYLGTNDSLTTLP 1152
Query: 812 -----MTSLQVLAICDFPELSSLPDNFQQ-LQNLQTLTISGCPILEKRCKRGIGEDWHKI 865
+T+L LAI + P L +LP+ QQ L +L+ L I CP L +RCKRG G+ W K+
Sbjct: 1153 EGMHNLTALNDLAIWNCPSLKALPEGLQQRLHSLEKLFIRQCPTLVRRCKRG-GDYWSKV 1211
Query: 866 AHIP 869
IP
Sbjct: 1212 KDIP 1215
>ref|XP_475165.1| putative NBS-LRR resistance protein [Oryza sativa (japonica
cultivar-group)] gi|47777417|gb|AAT38051.1| putative
NBS-LRR resistance protein [Oryza sativa (japonica
cultivar-group)]
Length = 1259
Score = 311 bits (796), Expect = 8e-83
Identities = 184/497 (37%), Positives = 282/497 (56%), Gaps = 25/497 (5%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
+ ALL L++ G + E A G+ L L I V+ DAE++ AVK
Sbjct: 4 LLSALLPALLKKAGESISTEFAFIGGIERKCSELKTSLLAINQVIYDAEEQASKKPAVKS 63
Query: 61 WLQQLRDAAYVLDDILDECSITL-------KAHGNNKRITRF---HPMKILVRRNIGKRM 110
W+ +L+ AA DD LDE + H N + F H +L + IGKR+
Sbjct: 64 WIAKLKMAACEADDALDELHYEALRSEALRRGHKINSGVRAFFTSHYNPLLFKYRIGKRL 123
Query: 111 KEIAKEIDDIAEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLL 170
++I ++ID + + +FG + P DE R QT S + E +V GR K+++ I+ LL
Sbjct: 124 QQIVEKIDKLVLQMNRFGF---LNCPMPVDE-RMQTYSYVDEQEVIGRQKERDEIIHMLL 179
Query: 171 RHAGDSEELSVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESI 230
+ S++L + IVG GG GKTTLAQ VFND +VK HF +WVCVS + + +++ I
Sbjct: 180 --SAKSDKLLILPIVGIGGLGKTTLAQLVFNDVKVKAHFQKHMWVCVSENFSVPDIVKGI 237
Query: 231 IENTIGKNPHLSS--LESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKG 288
I+ IG + L S LE +QQ+++E L + RYLLVLDDVW ED++KW L++LL + K G
Sbjct: 238 IDTAIGNDCGLKSDNLELLQQRLREELSQKRYLLVLDDVWNEDEQKWEALRTLLCSCKMG 297
Query: 289 ASILITTRLDIVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVR 348
+++++TTR VAS+MGT L LS +D W+LF ++AF + E V IG K+V+
Sbjct: 298 SAVVVTTRNSNVASVMGTVPPLALEQLSQEDSWTLFCERAFRTGVAKSCEFVEIGTKIVQ 357
Query: 349 KCVGSPLAAKVLGSSLCCTSNEHQWISVLESEFWNLPEVDSIMSALRISYFNLKLSLRPC 408
KC G PLA +G L + W+++L++ W E ++I++ L +SY +L ++ C
Sbjct: 358 KCSGVPLAINSMGGLLSRKHSVRDWLAILQNNTW---EENNILTVLSLSYKHLPSFMKQC 414
Query: 409 FAFCAVFPKGFEMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENV 468
FAFCAVFPK +E+ K++LIHLW++NG + S+ +E G+++ L+L L +N
Sbjct: 415 FAFCAVFPKDYEIDKDDLIHLWISNGFIPSKETSDIEETGNKVF-LEL---LWRSFFQNA 470
Query: 469 SNEEDARETNLISKKDL 485
+E + KD+
Sbjct: 471 KQTRSRKEEYIYGYKDV 487
Score = 140 bits (354), Expect = 1e-31
Identities = 157/576 (27%), Positives = 239/576 (41%), Gaps = 153/576 (26%)
Query: 434 GLVTSRGNLQMEHVGDE-------LHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLD 486
G ++S L M VG+E L +L+LGGKL I L V+N A+E NL +KK+L
Sbjct: 658 GQLSSLRTLTMYMVGNESDRRLHELKDLELGGKLQIHNLLKVTNPLQAKEANLENKKNLQ 717
Query: 487 RLYLSWG--NDTNSQVGSVDA-------ERVLEALEPHSSGLKHFGVNGYGGTIFPSWMK 537
+L L W N T S S D E VL+AL+P +GLK + Y G+ FP WM+
Sbjct: 718 QLALCWDSRNFTCSHSHSADEYLQLCCPEEVLDALKP-PNGLKVLKLRQYMGSDFPMWME 776
Query: 538 NTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYI-----DDDLYEPETE 592
+ L+ +V + L C LPP +LP L +L L M +KY+ D+ Y +
Sbjct: 777 DGVTLQNIVKLSLRGSVMCVKLPPVWQLPFLEVLRLKRMERLKYLCYRYPTDEEYGNQL- 835
Query: 593 KAFTSLKKLSLHDLPNLERVLEVDGVEM----LPQLLNLDITNVPKLT-------LTSL- 640
F LK LSL + +LE E D ++ P+L ++I + PKLT L SL
Sbjct: 836 VVFQKLKLLSLEWMESLENWHEYDTQQVTSVTFPKLDAMEIIDCPKLTALPNVPILKSLS 895
Query: 641 ----------------LSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANL 684
LS L AS G+ +++ +Y E + K I L
Sbjct: 896 LTGNKVLLGLVSGISNLSYLYLGASQGSSRRVRTLYYIYNGEREGSTDTKDEHI-----L 950
Query: 685 KELPVELGPLTA--LESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMR- 741
+ + G LT L+ + ++S S H++ S++++ + SC F +G++
Sbjct: 951 PDHLLSWGSLTKLHLQGFNTPAPENVKSISGHMM----SVQDLVLSSCDCFIQ-HEGLQS 1005
Query: 742 ------HLTCLETLHIYYCPQLVF--PHNMNSLASLRQLLLVEC---------------- 777
CL+ L I+YC L F SL SL +L +V+C
Sbjct: 1006 PLWFWISFGCLQQLEIWYCDSLTFWPEEEFRSLTSLEKLFIVDCKNFTGVPPDRLSARPS 1065
Query: 778 -----------------------------------NESILDGIEG----IPSLQKLRLFN 798
+ ++L+G+ G +L L +
Sbjct: 1066 TDGGPCNLEYLQIDRCPNLVVFPTNFICLRILVITHSNVLEGLPGGFGCQDTLTTLVILG 1125
Query: 799 FPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNF------------------------- 833
PS SLP + +++L+ L + L+SLP+
Sbjct: 1126 CPSFSSLPASIRCLSNLKSLELASNNSLTSLPEGMQNLTALKTLHFIKCPGITALPEGLQ 1185
Query: 834 QQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
Q+L LQT T+ CP L +RC+RG G+ W K+ IP
Sbjct: 1186 QRLHGLQTFTVEDCPALARRCRRG-GDYWEKVKDIP 1220
Score = 37.0 bits (84), Expect = 2.9
Identities = 23/81 (28%), Positives = 44/81 (53%), Gaps = 2/81 (2%)
Query: 672 NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCS 731
+L+ L +S +++K LP + L L+ L + RC + + + K + SLR++ + CS
Sbjct: 591 HLRYLDLSS-SDIKTLPEAVSALYNLQILMLNRCRGLTHLPDGM-KFMISLRHVYLDGCS 648
Query: 732 GFKSLSDGMRHLTCLETLHIY 752
+ + G+ L+ L TL +Y
Sbjct: 649 SLQRMPPGLGQLSSLRTLTMY 669
>gb|AAU43948.1| putative NBS-LRR resistance protein [Oryza sativa (japonica
cultivar-group)]
Length = 1259
Score = 309 bits (792), Expect = 2e-82
Identities = 183/497 (36%), Positives = 283/497 (56%), Gaps = 25/497 (5%)
Query: 1 MADALLGILIQNLGSFVQEELATYLGVGELTQSLSRKLTLIRAVLKDAEKKQITNDAVKE 60
+ ALL L++ G + E + G+ L L I V+ AE++ AVK
Sbjct: 4 LLSALLPALLKKAGESLSTEFSFIGGIEHRRSELYTLLLAINQVIYGAEEQASKKPAVKS 63
Query: 61 WLQQLRDAAYVLDDILDECSITL-------KAHGNNKRITRF---HPMKILVRRNIGKRM 110
W+ +L+ AA DD LDE + H N + F H +L + IGK++
Sbjct: 64 WITKLKLAACDADDALDELHYEALRSEALRRGHKINSGVRAFFSSHYNPLLFKYRIGKKL 123
Query: 111 KEIAKEIDDIAEERMKFGLHVGVIERQPEDEGRRQTTSVITESKVYGRDKDKEHIVEFLL 170
++I ++ID + + +FG + PEDE R QT S + E +V GRDK+++ I+ LL
Sbjct: 124 QQIVEQIDQLVSQMNQFGF---LNCPMPEDE-RMQTYSYVDEQEVIGRDKERDEIIHMLL 179
Query: 171 RHAGDSEELSVYSIVGHGGYGKTTLAQTVFNDERVKTHFDLKIWVCVSGDINAMKVLESI 230
+ S++L + IVG GG GKTTLAQ VFND +VK HF +WVCVS + + +++ I
Sbjct: 180 --SAKSDKLLILPIVGIGGLGKTTLAQLVFNDVKVKAHFQKHMWVCVSENFSVPDIVKGI 237
Query: 231 IENTIGKNPHLSS--LESMQQKVQEILQKNRYLLVLDDVWTEDKEKWNKLKSLLLNGKKG 288
I+ IG + L S LE +QQ+++E L + RYLLVLDDVW ED++KW L++LL + K G
Sbjct: 238 IDTAIGNDCGLKSDNLELLQQRLREELSQKRYLLVLDDVWNEDEQKWEALRTLLCSCKMG 297
Query: 289 ASILITTRLDIVASIMGTSDAHHLASLSDDDIWSLFKQQAFGENREERAELVAIGKKLVR 348
+++++TTR VAS+MGT L LS +D W+LF ++AF + E V IG K+V+
Sbjct: 298 SAVVVTTRNSNVASVMGTVPPLALEQLSQEDSWTLFCERAFRTGVAKSCEFVEIGTKIVQ 357
Query: 349 KCVGSPLAAKVLGSSLCCTSNEHQWISVLESEFWNLPEVDSIMSALRISYFNLKLSLRPC 408
KC G PLA +G L + W+++L++ W E ++I++ L +SY +L ++ C
Sbjct: 358 KCSGVPLAINSMGGLLSRKHSVRDWLAILQNNTW---EENNILTVLSLSYKHLPSFMKQC 414
Query: 409 FAFCAVFPKGFEMVKENLIHLWMANGLVTSRGNLQMEHVGDELHNLQLGGKLHIKCLENV 468
FAFCAVFPK +E+ K++LIHLW++NG + S+ +E G+++ L+L L +N
Sbjct: 415 FAFCAVFPKDYEIDKDDLIHLWISNGFIPSKETSDIEETGNKVF-LEL---LWRSFFQNA 470
Query: 469 SNEEDARETNLISKKDL 485
+E + KD+
Sbjct: 471 KQTRSRKEEYIYGYKDV 487
Score = 140 bits (353), Expect = 2e-31
Identities = 157/576 (27%), Positives = 239/576 (41%), Gaps = 153/576 (26%)
Query: 434 GLVTSRGNLQMEHVGDE-------LHNLQLGGKLHIKCLENVSNEEDARETNLISKKDLD 486
G ++S L M VG+E L +L+LGGKL I L V+N A+E NL +KK+L
Sbjct: 658 GQLSSLRTLTMYMVGNESDCRLHELKDLELGGKLQIHNLLKVTNPLQAKEANLENKKNLQ 717
Query: 487 RLYLSWG--NDTNSQVGSVDA-------ERVLEALEPHSSGLKHFGVNGYGGTIFPSWMK 537
+L L W N T S S D E VL+AL+P +GLK + Y G+ FP WM+
Sbjct: 718 QLALCWDSRNFTCSHCHSADEYLQLCRPEEVLDALKP-PNGLKVLKLRQYMGSNFPMWME 776
Query: 538 NTSILKGLVSIILYNCKNCRHLPPFGKLPCLTILYLSGMRYIKYI-----DDDLYEPETE 592
+ L+ +V + L C LPP +LP L +L L M +KY+ D+ Y +
Sbjct: 777 DGVTLQNIVKLSLRGSVMCVKLPPVWQLPFLEVLRLKRMERLKYLCYRYPTDEEYGNQL- 835
Query: 593 KAFTSLKKLSLHDLPNLERVLEVDGVEM----LPQLLNLDITNVPKLT-------LTSL- 640
F LK LSL + +LE E D ++ P+L ++I + PKLT L SL
Sbjct: 836 VVFQKLKLLSLEWMESLENWHEYDTQQVTSVTFPKLDAMEIIDCPKLTALPNVPILKSLS 895
Query: 641 ----------------LSVESLSASGGNEELLKSFFYNNCSEDVAGNNLKSLSISKFANL 684
LS L AS G+ +++ +Y E + K I L
Sbjct: 896 LTGNKVLLGLVSGISNLSYLYLGASQGSSRRVRTLYYIYNGEREGSTDTKDEHI-----L 950
Query: 685 KELPVELGPLTA--LESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCSGFKSLSDGMR- 741
+ + G LT L+ + ++S S H++ S++++ + SC F +G++
Sbjct: 951 PDHLLSWGSLTKLHLQGFNTPAPENVKSISGHMM----SVQDLVLSSCDCFIQ-HEGLQS 1005
Query: 742 ------HLTCLETLHIYYCPQLVF--PHNMNSLASLRQLLLVEC---------------- 777
CL+ L I+YC L F SL SL +L +V+C
Sbjct: 1006 PLWFWISFGCLQQLEIWYCDSLTFWPEEEFRSLTSLEKLFIVDCKNFTGVPPDRLSARPS 1065
Query: 778 -----------------------------------NESILDGIEG----IPSLQKLRLFN 798
+ ++L+G+ G +L L +
Sbjct: 1066 TDGGPCNLEYLQIDRCPNLVVFPTNFICLRILVITDSNVLEGLPGGFGCQGTLTTLVILG 1125
Query: 799 FPSIKSLPDWLGAMTSLQVLAICDFPELSSLPDNF------------------------- 833
PS SLP + +++L+ L + L+SLP+
Sbjct: 1126 CPSFSSLPASIRCLSNLKSLELTSNNSLTSLPEGMQNLTALKTLHFIKCPGITALPEGLQ 1185
Query: 834 QQLQNLQTLTISGCPILEKRCKRGIGEDWHKIAHIP 869
Q+L LQT T+ CP L +RC+RG G+ W K+ IP
Sbjct: 1186 QRLHGLQTFTVEDCPALARRCRRG-GDYWEKVKDIP 1220
Score = 37.0 bits (84), Expect = 2.9
Identities = 23/81 (28%), Positives = 44/81 (53%), Gaps = 2/81 (2%)
Query: 672 NLKSLSISKFANLKELPVELGPLTALESLSIERCNEMESFSEHLLKGLSSLRNMSVFSCS 731
+L+ L +S +++K LP + L L+ L + RC + + + K + SLR++ + CS
Sbjct: 591 HLRYLDLSS-SDIKTLPEAVSALYNLQILMLNRCRGLTHLPDGM-KFMISLRHVYLDGCS 648
Query: 732 GFKSLSDGMRHLTCLETLHIY 752
+ + G+ L+ L TL +Y
Sbjct: 649 SLQRMPPGLGQLSSLRTLTMY 669
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,466,143,262
Number of Sequences: 2540612
Number of extensions: 61370337
Number of successful extensions: 201179
Number of sequences better than 10.0: 3186
Number of HSP's better than 10.0 without gapping: 1393
Number of HSP's successfully gapped in prelim test: 1818
Number of HSP's that attempted gapping in prelim test: 186711
Number of HSP's gapped (non-prelim): 7716
length of query: 887
length of database: 863,360,394
effective HSP length: 137
effective length of query: 750
effective length of database: 515,296,550
effective search space: 386472412500
effective search space used: 386472412500
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146556.4


