

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146552.9 - phase: 0 /pseudo
(454 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
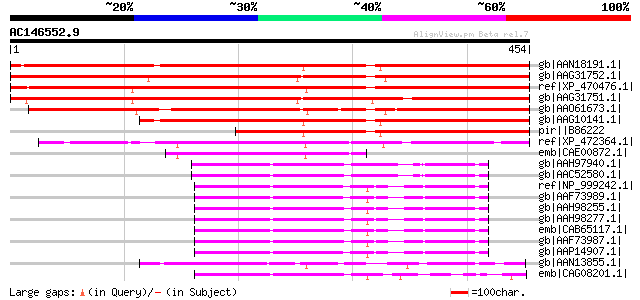
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN18191.1| At1g09020/F7G19_11 [Arabidopsis thaliana] gi|1839... 513 e-144
gb|AAG31752.1| protein kinase AKINbetagamma-2 [Zea mays] 513 e-144
ref|XP_470476.1| putative protein kinase AKINbetagamma-2 [Oryza ... 509 e-143
gb|AAG31751.1| protein kinase AKINbetagamma-1 [Zea mays] 496 e-139
gb|AAO61673.1| AKIN betagamma [Medicago truncatula] 476 e-133
gb|AAG10141.1| putative activator subunit of SNF1-related protei... 363 6e-99
pir||B86222 hypothetical protein [imported] - Arabidopsis thalia... 275 2e-72
ref|XP_472364.1| OSJNBb0012E08.3 [Oryza sativa (japonica cultiva... 253 9e-66
emb|CAE00872.1| NF protein [Oryza sativa (japonica cultivar-group)] 139 2e-31
gb|AAH97940.1| Unknown (protein for MGC:116060) [Rattus norvegic... 97 1e-18
gb|AAC52580.1| 5'-AMP-activated protein kinase, gamma-1 subunit 97 1e-18
ref|NP_999242.1| AMP-activated protein kinase gamma subunit [Sus... 97 1e-18
gb|AAF73989.1| AMPK gamma subunit [Sus scrofa] gi|37956600|gb|AA... 97 1e-18
gb|AAH98255.1| Unknown (protein for MGC:119343) [Homo sapiens] g... 96 2e-18
gb|AAH98277.1| Unknown (protein for MGC:119344) [Homo sapiens] g... 96 2e-18
emb|CAB65117.1| AMP-activated protein kinase gamma 3 subunit [Ho... 96 2e-18
gb|AAF73987.1| AMP-activated protein kinase gamma subunit [Homo ... 96 2e-18
gb|AAP14907.1| AMP-activated protein kinase gamma subunit [Sus s... 96 3e-18
gb|AAN13855.1| CG17299-PH, isoform H [Drosophila melanogaster] g... 95 4e-18
emb|CAG08201.1| unnamed protein product [Tetraodon nigroviridis] 82 3e-14
>gb|AAN18191.1| At1g09020/F7G19_11 [Arabidopsis thaliana]
gi|18390971|ref|NP_563834.1| protein kinase, putative
[Arabidopsis thaliana] gi|16612255|gb|AAL27498.1|
At1g09020/F7G19_11 [Arabidopsis thaliana]
Length = 487
Score = 513 bits (1320), Expect = e-144
Identities = 280/470 (59%), Positives = 333/470 (70%), Gaps = 30/470 (6%)
Query: 1 MFSPSMDSARDVGGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTV 60
MF ++DS+R G A+G +L P RFVWPYGGR V+LSGSFTRW+E + MSP+EGCPTV
Sbjct: 1 MFGSTLDSSR--GNSAASGQLLTPTRFVWPYGGRRVFLSGSFTRWTEHVPMSPLEGCPTV 58
Query: 61 FQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATDPFVPV-LPPDIVSG 119
FQVI NL PGYHQYKFFVDGEWRHDEH P ++G+ G+VNT+ + VP P+ +
Sbjct: 59 FQVICNLTPGYHQYKFFVDGEWRHDEHQPFVSGNGGVVNTIFITGPDMVPAGFSPETLGR 118
Query: 120 SNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGKVV 179
SNMDVD+ V T E +PR+S VD++ SR RIS LS RTAYELLPESGKV+
Sbjct: 119 SNMDVDD-----VFLRTADPSQEAVPRMSGVDLELSRHRISVLLSTRTAYELLPESGKVI 173
Query: 180 TLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEE 239
LDV+LPVKQAFHIL+EQGIP+APLWDF KGQFVGVL LDFILILRELG HGSNLTEEE
Sbjct: 174 ALDVNLPVKQAFHILYEQGIPLAPLWDFGKGQFVGVLGPLDFILILRELGTHGSNLTEEE 233
Query: 240 LETHTISAWKEGKWTL----------FSRRFIHAGPSDNLKDVALKILQNGISTVPIIHS 289
LETHTI+AWKEGK + + R + GP DNLKDVALKILQN ++ VP+I+S
Sbjct: 234 LETHTIAAWKEGKAHISRQYDGSGRPYPRPLVQVGPYDNLKDVALKILQNKVAAVPVIYS 293
Query: 290 SSADGSFPQLLHLASLSGILRCILGVVLVHCPYF--NFQFVQFLWAHGCP-KLGRQIAGL 346
S DGS+PQLLHLASLSGIL+CI C YF + + L C LG + +
Sbjct: 294 SLQDGSYPQLLHLASLSGILKCI-------CRYFRHSSSSLPILQQPICSIPLGTWVPRI 346
Query: 347 *QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYT 405
+ + L L+P + A+VSSIP+VD++DSL+DIY RSDITALAKD+AY
Sbjct: 347 GESSSKPLATLRPHASLGSALALLVQAEVSSIPVVDDNDSLIDIYSRSDITALAKDKAYA 406
Query: 406 HINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
I+LD+MTVHQALQL QDA P QRC MCLR+DSL KVMERLANP
Sbjct: 407 QIHLDDMTVHQALQLGQDASPPYGIFNGQRCHMCLRSDSLVKVMERLANP 456
>gb|AAG31752.1| protein kinase AKINbetagamma-2 [Zea mays]
Length = 496
Score = 513 bits (1320), Expect = e-144
Identities = 279/473 (58%), Positives = 338/473 (70%), Gaps = 27/473 (5%)
Query: 1 MFSPSMDSARDVGGV-VAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPT 59
MFS DSA D G V V++G +P RFVWPYGG+ V++SGSFTRWSE L MSP+EGCPT
Sbjct: 1 MFSHGADSAHDAGAVGVSSGGATVPTRFVWPYGGKRVFVSGSFTRWSEHLPMSPIEGCPT 60
Query: 60 VFQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATD-PFVPVLPPDIVS 118
VFQ I +L+PG H+YKFFVDGEWRHDE P I+G++GIVNT+ L + + L
Sbjct: 61 VFQAICSLSPGIHEYKFFVDGEWRHDERQPTISGEFGIVNTLYLTREYNQINTLSSPSTP 120
Query: 119 GS--NMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESG 176
GS NMDVDNE FQR V L+DGT+SE R+S+ +Q SR R+S YLS+ T Y+LLP+SG
Sbjct: 121 GSRMNMDVDNENFQRTVTLSDGTVSEGTLRVSEAAIQISRCRVSEYLSLHTCYDLLPDSG 180
Query: 177 KVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLT 236
KV+ LD++LPVKQ+FHILHEQGIP+APLWD +GQFVG+LS LDFILILREL HGSNLT
Sbjct: 181 KVIALDINLPVKQSFHILHEQGIPVAPLWDSFRGQFVGLLSPLDFILILRELETHGSNLT 240
Query: 237 EEELETHTISAWKE----------GKWTLFSRRFIHAGPSDNLKDVALKILQNGISTVPI 286
EE+LETHTISAWKE G+W + +HA P ++L+D+A+K+LQNGISTVP+
Sbjct: 241 EEQLETHTISAWKEAKRQTNGRNDGQWRP-QQHLVHATPYESLRDIAVKLLQNGISTVPV 299
Query: 287 IHSSSADGSFPQLLHLASLSGILRCILGVVLVHCPYFNFQF--VQFLWAHGCP-KLGRQI 343
I+SSS+DGSFPQLLHLASLSGIL+CI C YF + L C LG +
Sbjct: 300 IYSSSSDGSFPQLLHLASLSGILKCI-------CRYFKNSTGNLPILNQPVCSIPLGSWV 352
Query: 344 AGL*QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDR 402
+ + L +L+P N+ A VSSIPIVD++DSLLD Y RSDITALAK +
Sbjct: 353 PKIGDLNSRPLAMLRPNASLSSALNMLVQAGVSSIPIVDDNDSLLDTYSRSDITALAKAK 412
Query: 403 AYTHINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
YTH+ LDEMT+HQALQL QDA P QRCQMCLR+D L KVMERLANP
Sbjct: 413 VYTHVRLDEMTIHQALQLGQDANTPFGFFNGQRCQMCLRSDPLLKVMERLANP 465
>ref|XP_470476.1| putative protein kinase AKINbetagamma-2 [Oryza sativa (japonica
cultivar-group)] gi|30102976|gb|AAP21389.1| putative
protein kinase AKINbetagamma [Oryza sativa (japonica
cultivar-group)] gi|41393259|gb|AAS01982.1| putative
protein kinase AKINbetagamma-2 [Oryza sativa (japonica
cultivar-group)]
Length = 493
Score = 509 bits (1312), Expect = e-143
Identities = 280/471 (59%), Positives = 336/471 (70%), Gaps = 25/471 (5%)
Query: 1 MFSPSMDSARDVGGVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTV 60
MFS DSA D G V + G +P RFVWPYGG+ VYL+GSFTRW+E L MSPVEGCPTV
Sbjct: 1 MFSHGADSAHDAGAV-STGASGVPTRFVWPYGGKRVYLTGSFTRWTEHLPMSPVEGCPTV 59
Query: 61 FQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATD--PFVPVLPPDIV- 117
FQ I +L+PG HQYKF VDGEWRHDE P ITGDYG+VNT+ L D +L P
Sbjct: 60 FQAICSLSPGIHQYKFCVDGEWRHDERQPTITGDYGVVNTLCLTRDFDQINTILSPSTPG 119
Query: 118 SGSNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGK 177
S NMDVDN+ FQR V L+DG + E RIS+ +Q SR R++ +L+ +T Y+LLP+SGK
Sbjct: 120 SRMNMDVDNDNFQRTVSLSDGIIQEGPQRISEAAIQISRCRVADFLNGQTGYDLLPDSGK 179
Query: 178 VVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTE 237
V+ LDV+LPVKQ+FHILHEQGIP+APLWD +GQFVG+LS LDFILILREL HGSNLTE
Sbjct: 180 VIALDVNLPVKQSFHILHEQGIPVAPLWDSFRGQFVGLLSPLDFILILRELETHGSNLTE 239
Query: 238 EELETHTISAWKEGKWTLFSR---------RFIHAGPSDNLKDVALKILQNGISTVPIIH 288
E+LETHTISAWKE K ++R +HA P ++L+++A+KILQNG+STVPI+
Sbjct: 240 EQLETHTISAWKEAKRQTYARNEGSWRANHHLVHATPYESLREIAMKILQNGVSTVPIMF 299
Query: 289 SSSADGSFPQLLHLASLSGILRCILGVVLVHCPYF-NFQ-FVQFLWAHGCP-KLGRQIAG 345
SSS DGS+PQLLHLASLSGIL+CI C YF N Q + L C LG +
Sbjct: 300 SSSPDGSYPQLLHLASLSGILKCI-------CRYFKNSQGNLPILSQPVCTIPLGTWVPK 352
Query: 346 L*QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAY 404
+ + L +L+P N+ A VSSIPIVD++DSLLD Y RSDITALAKD+ Y
Sbjct: 353 IGDPNGRPLAMLRPNTSLSAALNLLVQAGVSSIPIVDDNDSLLDTYSRSDITALAKDKVY 412
Query: 405 THINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
THI LDEMT+HQALQL QDA +P QRCQMCLR+D+L KVMERLANP
Sbjct: 413 THIRLDEMTIHQALQLGQDANSPFGFFNGQRCQMCLRSDTLLKVMERLANP 463
>gb|AAG31751.1| protein kinase AKINbetagamma-1 [Zea mays]
Length = 497
Score = 496 bits (1278), Expect = e-139
Identities = 270/474 (56%), Positives = 329/474 (68%), Gaps = 28/474 (5%)
Query: 1 MFSPSMDSARDVG--GVVAAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCP 58
MFS DSA D G GV + +P RFVWPYGG+ V++SGSFTRWSE L MSPVEGCP
Sbjct: 1 MFSHGADSAHDAGTVGVSSGVGATVPARFVWPYGGKRVFVSGSFTRWSEHLPMSPVEGCP 60
Query: 59 TVFQVIHNLAPGYHQYKFFVDGEWRHDEHTPHITGDYGIVNTVLLATD--PFVPVLPPDI 116
TVFQ I +L+PG H+YKF+VDGEWRHDE P I+G++GIVNT+ L + +L P
Sbjct: 61 TVFQAICSLSPGIHEYKFYVDGEWRHDERQPTISGEFGIVNTLYLTREFNQINALLNPST 120
Query: 117 V-SGSNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPES 175
S NMDVDNE FQ V L+DGT+ E R+S+ +Q SR R+S YL++ T Y+LLP+S
Sbjct: 121 PGSRMNMDVDNENFQHTVTLSDGTIPEGTVRVSEAAIQISRCRVSEYLNLHTCYDLLPDS 180
Query: 176 GKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNL 235
GKV+ LD++LPVKQ+FHILHEQGIP+APLWD +GQFVG+LS LDFILILREL HGSNL
Sbjct: 181 GKVIALDINLPVKQSFHILHEQGIPVAPLWDSFRGQFVGLLSPLDFILILRELETHGSNL 240
Query: 236 TEEELETHTISAWKE----------GKWTLFSRRFIHAGPSDNLKDVALKILQNGISTVP 285
TE++LETHTISAWKE G+W + +HA P ++L+D+A+K+L N ISTVP
Sbjct: 241 TEDQLETHTISAWKEAKRQTCGRNDGQWRA-HQHLVHATPYESLRDIAVKLLLNDISTVP 299
Query: 286 IIHSSSADGSFPQLLHLASLSGILRCILGVV---LVHCPYFNFQFVQFLWAHGCPKLGRQ 342
+I+SSS+DGSFPQLLHLASLSGIL+CI + P N PK+G
Sbjct: 300 VIYSSSSDGSFPQLLHLASLSGILKCIFRYFKNSTGNLPILNQPVCSIPLGSWVPKIGDP 359
Query: 343 IAGL*QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKD 401
+ + L +L+P N+ A VSSIPIVDE+DSLLD Y RSDITALAK
Sbjct: 360 NS-------RPLAMLRPNASLSSALNMLVQAGVSSIPIVDENDSLLDTYSRSDITALAKA 412
Query: 402 RAYTHINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
+ YTH+ LDEM +HQALQL QDA P QRCQMCLR+D L KVMERLANP
Sbjct: 413 KVYTHVRLDEMAIHQALQLGQDANTPFGFFNGQRCQMCLRSDPLLKVMERLANP 466
>gb|AAO61673.1| AKIN betagamma [Medicago truncatula]
Length = 485
Score = 476 bits (1224), Expect = e-133
Identities = 263/458 (57%), Positives = 321/458 (69%), Gaps = 41/458 (8%)
Query: 17 AAGTVLIPVRFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKF 76
++G +LIP RFVWPYGG VYL GSFTRWSE + MSP+EGCP+VFQVI +L PGYHQ+KF
Sbjct: 19 SSGPILIPKRFVWPYGGTRVYLIGSFTRWSEHIPMSPMEGCPSVFQVICSLMPGYHQFKF 78
Query: 77 FVDGEWRHDEHTPHITGDYGIVNTVLLATDPFV--PVLPPDIVSGSNMDVDNETFQRVVR 134
VDG+WR+DE P + G+YGIVNT+ L +P + +L + S S+M+VDN+ F
Sbjct: 79 NVDGQWRYDEQQPFVNGNYGIVNTIYLVREPDILPAILSAETSSRSHMEVDNDVFGH--- 135
Query: 135 LTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVDLPVKQAFHIL 194
+E PR+S D++ SR+RIS +LS TAY+LLPESGKV+ LDV+LPVKQAFH+L
Sbjct: 136 ------AEANPRMSPSDLEVSRRRISKFLSEHTAYDLLPESGKVIALDVNLPVKQAFHVL 189
Query: 195 HEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEGKWT 254
+EQ + MAPLWDFCK QFVGVLS +DFILIL+ELG HGS+LTEE+LETHTI+AWKEGK
Sbjct: 190 YEQDVSMAPLWDFCKSQFVGVLSAMDFILILKELGTHGSHLTEEQLETHTIAAWKEGKSK 249
Query: 255 LFSRR------------FIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHL 302
RR F+HAGP + LKDVALK+LQN +STVPII S DGSFPQLLHL
Sbjct: 250 --QRRALDNNEGSNPHCFVHAGPKECLKDVALKVLQNKVSTVPII--SLEDGSFPQLLHL 305
Query: 303 ASLSGILRCILGVVLVHCPYFNFQF----VQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQ 358
ASLSGIL+CI C +F + L P LG + + Q L+ L+
Sbjct: 306 ASLSGILKCI-------CRHFEHSAGSLPILQLPIASIP-LGTWVPNVGDPNGQPLIRLR 357
Query: 359 P*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTVHQA 417
P ++F A+VSSIPIVDE+DSLLDIY RSDITALAKD+AY I+LDE +HQA
Sbjct: 358 PNASLGDALSMFVQAKVSSIPIVDENDSLLDIYSRSDITALAKDKAYARISLDETNIHQA 417
Query: 418 LQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
L L QDA +P + RC MCLR+DSLHKVMERLA P
Sbjct: 418 LILGQDANSPYGLNNGHRCHMCLRSDSLHKVMERLAKP 455
>gb|AAG10141.1| putative activator subunit of SNF1-related protein kinase SNF4
[Arabidopsis thaliana]
Length = 382
Score = 363 bits (932), Expect = 6e-99
Identities = 209/356 (58%), Positives = 247/356 (68%), Gaps = 27/356 (7%)
Query: 114 PDIVSGSNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRISTYLSMRTAYELLP 173
P+ + SNMDVD+ V T E +PR+S VD++ SR RIS LS RTAYELLP
Sbjct: 8 PETLGRSNMDVDD-----VFLRTADPSQEAVPRMSGVDLELSRHRISVLLSTRTAYELLP 62
Query: 174 ESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGS 233
ESGKV+ LDV+LPVKQAFHIL+EQGIP+APLWDF KGQFVGVL LDFILILRELG HGS
Sbjct: 63 ESGKVIALDVNLPVKQAFHILYEQGIPLAPLWDFGKGQFVGVLGPLDFILILRELGTHGS 122
Query: 234 NLTEEELETHTISAWKEGKWTL----------FSRRFIHAGPSDNLKDVALKILQNGIST 283
NLTEEELETHTI+AWKEGK + + R + GP DNLKDVALKILQN ++
Sbjct: 123 NLTEEELETHTIAAWKEGKAHISRQYDGSGRPYPRPLVQVGPYDNLKDVALKILQNKVAA 182
Query: 284 VPIIHSSSADGSFPQLLHLASLSGILRCILGVVLVHCPYF--NFQFVQFLWAHGCP-KLG 340
VP+I+SS DGS+PQLLHLASLSGIL+CI C YF + + L C LG
Sbjct: 183 VPVIYSSLQDGSYPQLLHLASLSGILKCI-------CRYFRHSSSSLPILQQPICSIPLG 235
Query: 341 RQIAGL*QH*DQMLLLLQP*I-Y*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALA 399
+ + + + L L+P + A+VSSIP+VD++DSL+DIY RSDITALA
Sbjct: 236 TWVPRIGESSSKPLATLRPHASLGSALALLVQAEVSSIPVVDDNDSLIDIYSRSDITALA 295
Query: 400 KDRAYTHINLDEMTVHQALQLSQDAFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
KD+AY I+LD+MTVHQALQL QDA P QRC MCLR+DSL KVMERLANP
Sbjct: 296 KDKAYAQIHLDDMTVHQALQLGQDASPPYGIFNGQRCHMCLRSDSLVKVMERLANP 351
>pir||B86222 hypothetical protein [imported] - Arabidopsis thaliana
gi|2342682|gb|AAB70406.1| Contains similarity to Rattus
AMP-activated protein kinase (gb|X95577). [Arabidopsis
thaliana]
Length = 391
Score = 275 bits (703), Expect = 2e-72
Identities = 159/272 (58%), Positives = 187/272 (68%), Gaps = 22/272 (8%)
Query: 198 GIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEGKWTL-- 255
GIP+APLWDF KGQFVGVL LDFILILRELG HGSNLTEEELETHTI+AWKEGK +
Sbjct: 121 GIPLAPLWDFGKGQFVGVLGPLDFILILRELGTHGSNLTEEELETHTIAAWKEGKAHISR 180
Query: 256 --------FSRRFIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASLSG 307
+ R + GP DNLKDVALKILQN ++ VP+I+SS DGS+PQLLHLASLSG
Sbjct: 181 QYDGSGRPYPRPLVQVGPYDNLKDVALKILQNKVAAVPVIYSSLQDGSYPQLLHLASLSG 240
Query: 308 ILRCILGVVLVHCPYF--NFQFVQFLWAHGCP-KLGRQIAGL*QH*DQMLLLLQP*I-Y* 363
IL+CI C YF + + L C LG + + + + L L+P
Sbjct: 241 ILKCI-------CRYFRHSSSSLPILQQPICSIPLGTWVPRIGESSSKPLATLRPHASLG 293
Query: 364 FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQD 423
+ A+VSSIP+VD++DSL+DIY RSDITALAKD+AY I+LD+MTVHQALQL QD
Sbjct: 294 SALALLVQAEVSSIPVVDDNDSLIDIYSRSDITALAKDKAYAQIHLDDMTVHQALQLGQD 353
Query: 424 AFNP-NESRSQRCQMCLRTDSLHKVMERLANP 454
A P QRC MCLR+DSL KVMERLANP
Sbjct: 354 ASPPYGIFNGQRCHMCLRSDSLVKVMERLANP 385
>ref|XP_472364.1| OSJNBb0012E08.3 [Oryza sativa (japonica cultivar-group)]
gi|21740621|emb|CAD40779.1| OSJNBb0012E08.3 [Oryza
sativa (japonica cultivar-group)]
Length = 451
Score = 253 bits (646), Expect = 9e-66
Identities = 165/447 (36%), Positives = 236/447 (51%), Gaps = 48/447 (10%)
Query: 26 RFVWPYGGRTVYLSGSFTRWSELLQMSPVEGCPTVFQVIHNLAPGYHQYKFFVDGEWRHD 85
RF WPYGG+ GSFT W E P+ FQV+ +L PG +QY+F VDG WR D
Sbjct: 5 RFAWPYGGQRASFCGSFTGWREC----PMGLVGAEFQVVFDLPPGVYQYRFLVDGVWRCD 60
Query: 86 EHTPHITGDYGIVNTVLLATDPFVPVLPPDIVSGSNMDVDNETFQRVVRLTDGTLSEVMP 145
E P + +YG+++ +L D PV+ P ET RVV + +GT+ MP
Sbjct: 61 ETKPCVRDEYGLISNEVLV-DNTHPVVQP------------ETSIRVVSMDEGTILTTMP 107
Query: 146 -----RISDVDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIP 200
+ S V + R R+S L T Y+++P S K+ LD LPVKQAF I+H++G+
Sbjct: 108 PDQLSQNSGVQIAIFRHRVSEILLHNTIYDVVPVSSKIAVLDARLPVKQAFKIMHDEGLS 167
Query: 201 MAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEGKWTLFS--- 257
+ PLWD + G+L+ DF+LILR+L + L EELE H++SAWKE K +
Sbjct: 168 LVPLWDDQQQTVTGMLTASDFVLILRKLQRNIRTLGHEELEMHSVSAWKEAKLQFYGGPD 227
Query: 258 ------RRFIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHLASLSGILRC 311
R IH SDNL+DVAL I++N IS+VPI S+ P LL LA+L GI++
Sbjct: 228 VAAIQRRPLIHVKDSDNLRDVALAIIRNEISSVPIFKPSTDSSGMP-LLGLATLPGIVKF 286
Query: 312 ILGVVLVHCPYFNF---QFVQFLWAHGCPKLGRQIAGL*QH*DQMLLLLQP*I-Y*FKFN 367
I + ++F Q V P G+ ++ L +P +
Sbjct: 287 ICSKLQEQPEGYSFLQNQIVSMPIGTWSPHTGKAS-------NRQLRTSRPSTPLNSCLD 339
Query: 368 IFFAAQVSSIPIVDESDSLLDIYCRSDITALAKDRAYTHINLDEMTVHQALQLSQDAFNP 427
+ +VSSIPIVD++ +LLD+Y SDI AL K+ YT I L+++TV AL+L
Sbjct: 340 LLLEDRVSSIPIVDDNGALLDVYSLSDIMALGKNDVYTRIELEQVTVEHALELQYQV--- 396
Query: 428 NESRSQRCQMCLRTDSLHKVMERLANP 454
+ + C CL T + +V+E+L+ P
Sbjct: 397 --NGRRHCHTCLSTSTFLEVLEQLSAP 421
>emb|CAE00872.1| NF protein [Oryza sativa (japonica cultivar-group)]
Length = 245
Score = 139 bits (349), Expect = 2e-31
Identities = 80/190 (42%), Positives = 111/190 (58%), Gaps = 15/190 (7%)
Query: 137 DGTLSEVMP-----RISDVDVQTSRQRISTYLSMRTAYELLPESGKVVTLDVDLPVKQAF 191
+GT+ MP + S V + R R+S L T Y+++P S K+ LD LPVKQAF
Sbjct: 2 EGTILTTMPPDQLSQNSGVQIAIFRHRVSEILLHNTIYDVVPVSSKIAVLDARLPVKQAF 61
Query: 192 HILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHGSNLTEEELETHTISAWKEG 251
I+H++G+ + PLWD + G+L+ DF+LILR+L + L EELE H++SAWKE
Sbjct: 62 KIMHDEGLSLVPLWDDQQQTVTGMLTASDFVLILRKLQRNIRTLGHEELEMHSVSAWKEA 121
Query: 252 KWTLFS---------RRFIHAGPSDNLKDVALKILQNGISTVPIIHSSSADGSFPQLLHL 302
K + R IH SDNL+DVAL I++N IS+VPI S+ P LL L
Sbjct: 122 KLQFYGGPDVAAIQRRPLIHVKDSDNLRDVALAIIRNEISSVPIFKPSTDSSGMP-LLGL 180
Query: 303 ASLSGILRCI 312
A+L GI++ I
Sbjct: 181 ATLPGIVKFI 190
>gb|AAH97940.1| Unknown (protein for MGC:116060) [Rattus norvegicus]
gi|6981392|ref|NP_037142.1| AMP-activated protein
kinase, noncatalytic gamma-1 subunit [Rattus norvegicus]
gi|1185271|emb|CAA64831.1| AMP-activated protein kinase
gamma [Rattus norvegicus] gi|2507205|sp|P80385|AAKG1_RAT
5'-AMP-activated protein kinase, gamma-1 subunit (AMPK
gamma-1 chain) (AMPKg)
Length = 330
Score = 97.1 bits (240), Expect = 1e-18
Identities = 76/262 (29%), Positives = 128/262 (48%), Gaps = 26/262 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
+T++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 29 TTFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQSFVGMLTIT 88
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ +L D +++
Sbjct: 89 DFINILHRY--YKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIR 146
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L + P F + ++ L
Sbjct: 147 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA 201
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 202 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 247
Query: 398 LAKDRAYTHINLDEMTVHQALQ 419
LA ++ Y ++ +++V +ALQ
Sbjct: 248 LAAEKTYNNL---DVSVTKALQ 266
>gb|AAC52580.1| 5'-AMP-activated protein kinase, gamma-1 subunit
Length = 323
Score = 97.1 bits (240), Expect = 1e-18
Identities = 76/262 (29%), Positives = 128/262 (48%), Gaps = 26/262 (9%)
Query: 160 STYLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVL 219
+T++ Y+L+P S K+V D L VK+AF L G+ APLWD K FVG+L++
Sbjct: 22 TTFMKSHRCYDLIPTSSKLVVFDTSLQVKKAFFALVTNGVRAAPLWDSKKQSFVGMLTIT 81
Query: 220 DFILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQ 278
DFI IL + S L + ELE H I W+E + + P+ +L D +++
Sbjct: 82 DFINILHRY--YKSALVQIYELEEHKIETWREVYLQDSFKPLVCISPNASLFDAVSSLIR 139
Query: 279 NGISTVPIIHSSSADGSFPQLLHLASLSGILRCI-LGVVLVHCPYFNFQFVQFLWAHGCP 337
N I +P+I S + L++ + IL+ + L + P F + ++ L
Sbjct: 140 NKIHRLPVIDPESGN-----TLYILTHKRILKFLKLFITEFPKPEFMSKSLEELQIGTYA 194
Query: 338 KLGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITA 397
+ M+ P +Y IF +VS++P+VDE ++DIY + D+
Sbjct: 195 NIA------------MVRTTTP-VY-VALGIFVQHRVSALPVVDEKGRVVDIYSKFDVIN 240
Query: 398 LAKDRAYTHINLDEMTVHQALQ 419
LA ++ Y ++ +++V +ALQ
Sbjct: 241 LAAEKTYNNL---DVSVTKALQ 259
>ref|NP_999242.1| AMP-activated protein kinase gamma subunit [Sus scrofa]
gi|29812510|gb|AAF73988.2| AMP-activated protein kinase
gamma subunit [Sus scrofa]
gi|34223710|sp|Q9MYP4|AAKG3_PIG 5'-AMP-activated protein
kinase, gamma-3 subunit (AMPK gamma-3 chain) (AMPK
gamma3)
Length = 514
Score = 96.7 bits (239), Expect = 1e-18
Identities = 74/261 (28%), Positives = 129/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 212 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 271
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 272 ILVLHRY--YRSPLVQIYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNR 329
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 330 IHRLPVLDPVSG-----AVLHILTHKRLLKFLHIFGTLLPR-PSFLYRTIQDL------- 376
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E+ ++ +Y R D+ L
Sbjct: 377 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNETGQVVGLYSRFDVIHL 429
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M V +AL+
Sbjct: 430 AAQQTYNHL---DMNVGEALR 447
>gb|AAF73989.1| AMPK gamma subunit [Sus scrofa] gi|37956600|gb|AAP12533.1|
AMP-activated protein kinase gamma subunit [Sus scrofa]
Length = 464
Score = 96.7 bits (239), Expect = 1e-18
Identities = 74/261 (28%), Positives = 129/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 162 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 221
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 222 ILVLHRY--YRSPLVQIYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNR 279
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 280 IHRLPVLDPVSG-----AVLHILTHKRLLKFLHIFGTLLPR-PSFLYRTIQDL------- 326
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E+ ++ +Y R D+ L
Sbjct: 327 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNETGQVVGLYSRFDVIHL 379
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M V +AL+
Sbjct: 380 AAQQTYNHL---DMNVGEALR 397
>gb|AAH98255.1| Unknown (protein for MGC:119343) [Homo sapiens]
gi|67514236|gb|AAH98306.1| Unknown (protein for
MGC:119345) [Homo sapiens]
Length = 489
Score = 96.3 bits (238), Expect = 2e-18
Identities = 74/261 (28%), Positives = 130/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 187 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 246
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 247 ILVLHRY--YRSPLVQIYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNR 304
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S + +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 305 IHRLPVLDPVSGN-----VLHILTHKRLLKFLHIFGSLLPR-PSFLYRTIQDL------- 351
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E ++ +Y R D+ L
Sbjct: 352 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNECGQVVGLYSRFDVIHL 404
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 405 AAQQTYNHL---DMSVGEALR 422
>gb|AAH98277.1| Unknown (protein for MGC:119344) [Homo sapiens]
gi|66990060|gb|AAH98102.1| AMP-activated protein kinase,
non-catalytic gamma-3 subunit [Homo sapiens]
gi|47132577|ref|NP_059127.2| AMP-activated protein
kinase, non-catalytic gamma-3 subunit [Homo sapiens]
Length = 489
Score = 96.3 bits (238), Expect = 2e-18
Identities = 74/261 (28%), Positives = 130/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 187 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 246
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 247 ILVLHRY--YRSPLVQIYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNR 304
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S + +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 305 IHRLPVLDPVSGN-----VLHILTHKRLLKFLHIFGSLLPR-PSFLYRTIQDL------- 351
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E ++ +Y R D+ L
Sbjct: 352 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNECGQVVGLYSRFDVIHL 404
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 405 AAQQTYNHL---DMSVGEALR 422
>emb|CAB65117.1| AMP-activated protein kinase gamma 3 subunit [Homo sapiens]
Length = 492
Score = 96.3 bits (238), Expect = 2e-18
Identities = 74/261 (28%), Positives = 130/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 187 FIEEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 246
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 247 ILVLHRY--YRSPLVQIYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNR 304
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S + +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 305 IHRLPVLDPVSGN-----VLHILTHKRLLKFLHIFGSLLPR-PSFLYRTIQDL------- 351
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E ++ +Y R D+ L
Sbjct: 352 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNECGQVVGLYSRFDVIHL 404
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 405 AAQQTYNHL---DMSVGEALR 422
>gb|AAF73987.1| AMP-activated protein kinase gamma subunit [Homo sapiens]
gi|14194424|sp|Q9UGI9|AAKG3_HUMAN 5'-AMP-activated
protein kinase, gamma-3 subunit (AMPK gamma-3 chain)
(AMPK gamma3)
Length = 464
Score = 96.3 bits (238), Expect = 2e-18
Identities = 74/261 (28%), Positives = 130/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 162 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 221
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 222 ILVLHRY--YRSPLVQIYEIEQHKIETWREIYLQGCFKPLVSISPNDSLFEAVYTLIKNR 279
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S + +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 280 IHRLPVLDPVSGN-----VLHILTHKRLLKFLHIFGSLLPR-PSFLYRTIQDL------- 326
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E ++ +Y R D+ L
Sbjct: 327 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNECGQVVGLYSRFDVIHL 379
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 380 AAQQTYNHL---DMSVGEALR 397
>gb|AAP14907.1| AMP-activated protein kinase gamma subunit [Sus scrofa]
Length = 514
Score = 95.5 bits (236), Expect = 3e-18
Identities = 74/261 (28%), Positives = 129/261 (49%), Gaps = 28/261 (10%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ + S K+V D L +K+AF L G+ APLWD K FVG+L++ DF
Sbjct: 212 FMQEHTCYDAMATSSKLVIFDTMLEIKKAFFALVANGVRAAPLWDSKKQSFVGMLTITDF 271
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
IL+L + S L + E+E H I W+E + + P+D+L + +++N
Sbjct: 272 ILVLHRY--YRSPLVQIYEIEEHKIETWREIYLQGCFKPLVSISPNDSLFEAVYALIKNR 329
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +P++ S +LH+ + +L+ I G +L P F ++ +Q L
Sbjct: 330 IHRLPVLDPVSG-----AVLHILTHKRLLKFLHIFGTLLPR-PSFLYRTIQDL------- 376
Query: 339 LGRQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITAL 398
G+ D ++L I +IF +VS++P+V+E+ + +Y R D+ L
Sbjct: 377 ------GIGTFRDLAVVLETAPIL-TALDIFVDRRVSALPVVNETGQVEGLYSRFDVIHL 429
Query: 399 AKDRAYTHINLDEMTVHQALQ 419
A + Y H+ +M+V +AL+
Sbjct: 430 AAQQTYNHL---DMSVGEALR 447
>gb|AAN13855.1| CG17299-PH, isoform H [Drosophila melanogaster]
gi|24648663|ref|NP_732602.1| CG17299-PH, isoform H
[Drosophila melanogaster]
Length = 585
Score = 95.1 bits (235), Expect = 4e-18
Identities = 88/351 (25%), Positives = 156/351 (44%), Gaps = 57/351 (16%)
Query: 114 PDIVSGSNMDVDNETFQRVVRLTDGTLSEVMPRISDVDVQTSRQRIST-YLSMRTAYELL 172
P + SG N D + + LT S ++ R + + + +I + Y+L+
Sbjct: 65 PSLPSGKNKTKDGKKLKAT--LTAAMSSSLLRRKAHSNHKEDDSQIFVKFFRFHKCYDLI 122
Query: 173 PESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDFILILRELGNHG 232
P S K+V D L VK+AF+ L G+ APLWD K QFVG+L++ DFI IL ++
Sbjct: 123 PTSAKLVVFDTQLLVKKAFYALVYNGVRAAPLWDSEKQQFVGMLTITDFIKIL-QMYYKS 181
Query: 233 SNLTEEELETHTISAWKEGKWTLFSR--RFIHAGPSDNLKDVALKILQNGISTVPIIHSS 290
N + E+LE H + W+ L ++ + GP +L D ++ + I +P+I
Sbjct: 182 PNASMEQLEEHKLDTWRS---VLHNQVMPLVSIGPDASLYDAIKILIHSRIHRLPVI--- 235
Query: 291 SADGSFPQLLHLASLSGILRCILGVVLVHCPYFNFQFVQFLWAHGCPKLGRQIAGL---- 346
D + +L++ + ILR + FL+ + PK L
Sbjct: 236 --DPATGNVLYILTHKRILRFL-----------------FLYINELPKPAYMQKSLRELK 276
Query: 347 ------*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDITALAK 400
+ D+ ++ F +VS++P+VD L+DIY + D+ LA
Sbjct: 277 IGTYNNIETADETTSIIT------ALKKFVERRVSALPLVDSDGRLVDIYAKFDVINLAA 330
Query: 401 DRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQMCLRTDSLHKVMERL 451
++ Y + ++++ +A + + F + Q C +SL+ +MER+
Sbjct: 331 EKTYNDL---DVSLRKANEHRNEWF-------EGVQKCNLDESLYTIMERI 371
>emb|CAG08201.1| unnamed protein product [Tetraodon nigroviridis]
Length = 297
Score = 82.4 bits (202), Expect = 3e-14
Identities = 83/301 (27%), Positives = 135/301 (44%), Gaps = 59/301 (19%)
Query: 162 YLSMRTAYELLPESGKVVTLDVDLPVKQAFHILHEQGIPMAPLWDFCKGQFVGVLSVLDF 221
++ T Y+ +P S K+V D L VK+AF L G+ APLWD FVG+L++ DF
Sbjct: 9 FMKSHTCYDAIPTSSKLVIFDTTLQVKKAFFALVANGLRAAPLWDNKLKCFVGMLTITDF 68
Query: 222 ILILRELGNHGSNLTE-EELETHTISAWKEGKWTLFSRRFIHAGPSDNLKDVALKILQNG 280
I IL + S L + ELE H I W+E + + I P +L D +L+N
Sbjct: 69 INILHRY--YKSPLVQIYELEEHKIETWREIYLEYSTNKLISITPECSLFDAIYSLLKNK 126
Query: 281 ISTVPIIHSSSADGSFPQLLHLASLSGILRC--ILGVVLVHCPYFNFQFVQFLWAHGCPK 338
I +PII S D +LH+ + IL+ I G ++ + Q G
Sbjct: 127 IHRLPIIDPVSGD-----VLHILTHKRILKFLHIFGSMIPKPRFLQRQI-------GDVA 174
Query: 339 LG--RQIAGL*QH*DQMLLLLQP*IY*FKFNIFFAAQVSSIPIVDESDSLLDIYCRSDIT 396
+G RQ+A + + L+ IF +VS++P+V++ +L
Sbjct: 175 IGTFRQVATVQESASVYDALM----------IFVERRVSALPVVNKEGTL---------- 214
Query: 397 ALAKDRAYTHINLDEMTVHQALQLSQDAFNPNESRSQRCQM-----CLRTDSLHKVMERL 451
LA + Y ++N MT+ +A+ S+ C + C R ++L +++R+
Sbjct: 215 NLAAQKTYNNLN---MTMREAI------------ASRACCVEGVLKCYRHETLETIIDRI 259
Query: 452 A 452
A
Sbjct: 260 A 260
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.328 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 770,782,226
Number of Sequences: 2540612
Number of extensions: 32539919
Number of successful extensions: 87099
Number of sequences better than 10.0: 269
Number of HSP's better than 10.0 without gapping: 205
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 86481
Number of HSP's gapped (non-prelim): 448
length of query: 454
length of database: 863,360,394
effective HSP length: 131
effective length of query: 323
effective length of database: 530,540,222
effective search space: 171364491706
effective search space used: 171364491706
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146552.9


