

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146551.12 + phase: 0 /pseudo
(447 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
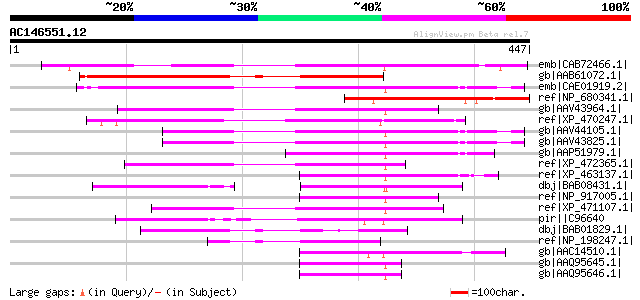
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB72466.1| putative protein [Arabidopsis thaliana] gi|11358... 288 3e-76
gb|AAB61072.1| contains similarity to a DNAJ-like domain [Arabid... 227 5e-58
emb|CAE01919.2| OSJNBb0078D11.1 [Oryza sativa (japonica cultivar... 189 1e-46
ref|NP_680341.1| hypothetical protein [Arabidopsis thaliana] 176 1e-42
gb|AAV43964.1| putative polyprotein [Oryza sativa (japonica cult... 171 6e-41
ref|XP_470247.1| Unknown protein [Oryza sativa (japonica cultiva... 163 9e-39
gb|AAV44105.1| unknown protein [Oryza sativa (japonica cultivar-... 160 7e-38
gb|AAV43825.1| putative polyprotein [Oryza sativa (japonica cult... 160 7e-38
gb|AAP51979.1| putative transposase [Oryza sativa (japonica cult... 147 7e-34
ref|XP_472365.1| OSJNBb0012E08.4 [Oryza sativa (japonica cultiva... 144 7e-33
ref|XP_463137.1| expressed protein [Oryza sativa (japonica culti... 139 2e-31
dbj|BAB08431.1| unnamed protein product [Arabidopsis thaliana] g... 129 2e-28
ref|NP_917005.1| P0519D04.27 [Oryza sativa (japonica cultivar-gr... 124 6e-27
ref|XP_471107.1| OSJNBa0042N22.14 [Oryza sativa (japonica cultiv... 121 4e-26
pir||C96640 hypothetical protein T25B24.14 [imported] - Arabidop... 117 1e-24
dbj|BAB01829.1| unnamed protein product [Arabidopsis thaliana] 107 1e-21
ref|NP_198247.1| hypothetical protein [Arabidopsis thaliana] 103 1e-20
gb|AAC14510.1| En/Spm-like transposon protein [Arabidopsis thali... 100 9e-20
gb|AAQ95645.1| transposase [Pharus latifolius] 91 1e-16
gb|AAQ95646.1| transposase [Coix aquatica] 90 2e-16
>emb|CAB72466.1| putative protein [Arabidopsis thaliana] gi|11358076|pir||T47439
hypothetical protein T18B22.40 - Arabidopsis thaliana
Length = 377
Score = 288 bits (736), Expect = 3e-76
Identities = 187/461 (40%), Positives = 229/461 (49%), Gaps = 130/461 (28%)
Query: 28 DEELYEAIYMYMMAIHALMHVVN------QFLNMMRGEHVERPLTRRRITSKGYDYIHKS 81
D Y+ + M+ + M V N + N+ E +E RR ++ GY+YI +
Sbjct: 4 DLSFYDVFFNIMLCATSNMMVFNMENIGDEEENLNDREKIEAEDVRRSVSRVGYNYIQTA 63
Query: 82 LNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNTRYCII 141
L + FRQ+YRM P+VF KLC +I +
Sbjct: 64 LTVNPAHFRQLYRMEPEVFHKLCDVI-------------------------------LLY 92
Query: 142 RNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPYFKILYN*KYI 201
+ FHK LKAL +AP+ MAKP VPAKIREST FYPYFK
Sbjct: 93 NGHISEIKLFYKHKFHKALKALAFVAPNWMAKPEVRVPAKIRESTSFYPYFK-------- 144
Query: 202 WPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAIDGTHIPALVR 261
+ AIDGTHI A+V
Sbjct: 145 --------------------------------------------DCIGAIDGTHIFAMVP 160
Query: 262 GRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDALKRK-NGLKVPH 320
D +S+R+R G ISQNVLAACNFDL+F+Y+LSGW+GSAHDSK+LNDAL R N L+VP
Sbjct: 161 TCDAASFRNRKGFISQNVLAACNFDLQFIYILSGWKGSAHDSKVLNDALTRNTNRLQVPE 220
Query: 321 G-------------------------------HGNDPENEKELFNLRHASLRNVVERIFG 349
G G DP+ + ELFNLRHASLRNV+ERIFG
Sbjct: 221 GKFYLVDCGYANRRKFLAPFRRTRYHLQDFRGQGRDPKTQNELFNLRHASLRNVIERIFG 280
Query: 350 IFKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRKECRSDEFPIEPS-NESSSSSSMLP 408
IFKSRF IFKSAPP+ FKTQ ELVLACA LHNFL +ECRSDEFP E S NE +S
Sbjct: 281 IFKSRFLIFKSAPPYPFKTQTELVLACAGLHNFLHQECRSDEFPPETSINEENSDD---- 336
Query: 409 IHEDNNDELNVQT---QEQEREDANIWRTNLGLDMWRDVQG 446
+E+NN E N + Q+RE AN WR + +MW D G
Sbjct: 337 -NEENNYEENGDVGILESQQREYANNWRDTISHNMWMDATG 376
>gb|AAB61072.1| contains similarity to a DNAJ-like domain [Arabidopsis thaliana]
gi|7485307|pir||T01797 hypothetical protein A_TM021B04.9
- Arabidopsis thaliana
Length = 1609
Score = 227 bits (579), Expect = 5e-58
Identities = 128/263 (48%), Positives = 167/263 (62%), Gaps = 54/263 (20%)
Query: 61 VERPLTRRRITSKGYDYIHKSLNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTRFIC 120
+ RP+ RR+IT KG+ YI K+L D FRQ+YRM P+VF +LC +++ KT L T +C
Sbjct: 1088 IPRPM-RRQITKKGHAYIQKALKDDPIHFRQLYRMNPEVFAELCHLLQMKTGLKGTPHVC 1146
Query: 121 IEEMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPA 180
+EEM+A+FL VGQN+RYC +TF RS+F+ S NFHK+L+ALN +AP LMAK T++VP+
Sbjct: 1147 VEEMVATFLITVGQNSRYCHTMDTFKRSKFSTSINFHKVLRALNMLAPTLMAKVTNTVPS 1206
Query: 181 KIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKG 240
KI ++TRFYP + +D G
Sbjct: 1207 KISKTTRFYP---------------------YFKDCVG---------------------- 1223
Query: 241 KHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSA 300
AIDGTHI A+V+G + +SYR+R G ISQNVLAACNFDLEF+YVLSGWEGSA
Sbjct: 1224 ---------AIDGTHINAMVQGPEKASYRNRKGVISQNVLAACNFDLEFIYVLSGWEGSA 1274
Query: 301 HDSKLLNDAL-KRKNGLKVPHGH 322
HDSK+L DAL +R N L+VP G+
Sbjct: 1275 HDSKVLQDALTRRTNRLQVPEGN 1297
>emb|CAE01919.2| OSJNBb0078D11.1 [Oryza sativa (japonica cultivar-group)]
gi|38345857|emb|CAD41055.2| OSJNBa0084K11.21 [Oryza
sativa (japonica cultivar-group)]
gi|50927943|ref|XP_473499.1| OSJNBa0084K11.21 [Oryza
sativa (japonica cultivar-group)]
Length = 377
Score = 189 bits (481), Expect = 1e-46
Identities = 139/421 (33%), Positives = 193/421 (45%), Gaps = 107/421 (25%)
Query: 58 GEHVERPLTRRRITSKGYDYIHKSLNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTR 117
G VER R+RI +Y+ + + + R+ F + C + R++ L DT
Sbjct: 22 GPMVERD--RKRI-----EYLETKIWWNDTTCINMLRLRRASFFRFCKLFRDRGLLEDTI 74
Query: 118 FICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSS 177
+CIEE +A FL VG N R ++R F RS+ S F+K+L A+ + +L+ P+
Sbjct: 75 HMCIEEQVAMFLHTVGHNLRNRLVRTNFDRSRETVSRYFNKVLHAIGELRDELIRPPSLD 134
Query: 178 VPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACK 237
P KI + R+ PYFK
Sbjct: 135 TPTKIAGNPRWDPYFK-------------------------------------------- 150
Query: 238 NKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWE 297
+ AIDGTHI A VR SS+R R +QNV+AA +FDL F YVL+GWE
Sbjct: 151 --------DCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWE 202
Query: 298 GSAHDSKLLNDALKRKNGLKVPHGH---------------------------------GN 324
G+AHD+ +L DAL+R+NGL+VP G+ GN
Sbjct: 203 GTAHDAVVLRDALERENGLRVPQGNRLQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEWGN 262
Query: 325 DP-ENEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPFL-FKTQAELVLACAALHNF 382
+P +NEKELFNLRH+SLR VER FG K RF + A PF F+TQ ++V+AC +HN+
Sbjct: 263 NPVQNEKELFNLRHSSLRVTVERAFGSLKRRFKVLDDATPFFPFQTQVDIVVACCIIHNW 322
Query: 383 LRKECRSDEFPIEPSNESSSSSSMLPIHEDNNDELNVQTQEQEREDANIWRTNLGLDMWR 442
+ + DE I P + SS + N+ L VQ +R L MW
Sbjct: 323 VVND-GIDEL-IAPPDWSSEDIDESLTGQANDHALMVQ-----------FRQGLADQMWA 369
Query: 443 D 443
D
Sbjct: 370 D 370
>ref|NP_680341.1| hypothetical protein [Arabidopsis thaliana]
Length = 211
Score = 176 bits (446), Expect = 1e-42
Identities = 95/185 (51%), Positives = 122/185 (65%), Gaps = 27/185 (14%)
Query: 289 FMYVLSGWEGSAHDSKLLNDALKR---------------------KNGLKVPHGHGNDPE 327
F+YVLSGWEGSAHDS++L+DAL++ + L+ G DPE
Sbjct: 25 FIYVLSGWEGSAHDSRVLSDALRKFYLVDCGFANRLNFLAPFRGVRYHLQEFAGQRRDPE 84
Query: 328 NEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRKEC 387
ELFNLRH SLRNV+ERIFGIFKSRF IFKSAPPF +K QA LVL CAALHNFLRKEC
Sbjct: 85 TPHELFNLRHVSLRNVIERIFGIFKSRFAIFKSAPPFSYKKQAGLVLTCAALHNFLRKEC 144
Query: 388 RSDE--FPIEPSNES---SSSSSMLPIHEDNNDELNVQTQEQEREDANIWRTNLGLDMWR 442
RSDE FP E NE ++ + + +E +N+E ++ Q+Q+RE+ N+WR ++ DMW+
Sbjct: 145 RSDEADFPDEVGNEGDVVNNEGNAMNTNEIDNEE-PLEAQKQDRENTNMWRKSMAEDMWK 203
Query: 443 DVQGI 447
D +
Sbjct: 204 DATNL 208
>gb|AAV43964.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 561
Score = 171 bits (432), Expect = 6e-41
Identities = 110/306 (35%), Positives = 145/306 (46%), Gaps = 82/306 (26%)
Query: 94 RMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFAAS 153
R+ F + C + R++ L DT +CIEE +A FL VG N R ++R + RS S
Sbjct: 28 RLRKGSFFRFCKLFRDRGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTNYSRSGETVS 87
Query: 154 ENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFS 213
F+K+L A+ + +L+ P+ P KI + R+ PYFK
Sbjct: 88 RYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPYFK-------------------- 127
Query: 214 QDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHG 273
+ AIDGTHI A VR SS+R R
Sbjct: 128 --------------------------------DCIGAIDGTHIRASVRKNVESSFRGRKS 155
Query: 274 RISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDALKRKNGLKVPHGH----------- 322
+QNV+AA +FDL F YVL+GWEG+AHD+ +L DAL+R+NGL VP G
Sbjct: 156 HATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDALERENGLHVPQGKFYLVDAGYGAK 215
Query: 323 -----------------GNDP-ENEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPF 364
GN+P +NEKELFNLRH+SLR VER FG K RF + A PF
Sbjct: 216 QGFLPPFRAVRYHLKEWGNNPVQNEKELFNLRHSSLRITVERAFGSLKRRFKVLDDATPF 275
Query: 365 L-FKTQ 369
F+TQ
Sbjct: 276 FPFRTQ 281
>ref|XP_470247.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
gi|21397269|gb|AAM51833.1| Unknown protein [Oryza sativa
(japonica cultivar-group)]
Length = 1202
Score = 163 bits (413), Expect = 9e-39
Identities = 125/376 (33%), Positives = 168/376 (44%), Gaps = 103/376 (27%)
Query: 67 RRRITSKGYDY---IHKSLNGDSEIFR----------QVYRMYPDVFRKLCMIIREKTPL 113
RRR + KG Y +H+ + SE+ R Q RM VF KLC +R K L
Sbjct: 557 RRRSSRKGVKYGPLVHRDVWRTSELIRLINISDRTCTQQLRMSRAVFYKLCARLRNKGLL 616
Query: 114 VDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAK 173
VDT + +EE +A FL+ VGQ+ + +F RS S F +L+A+ IA +L+
Sbjct: 617 VDTFHVSVEEQVAMFLKKVGQHHSVSCVGFSFWRSGETVSRYFRIVLRAMCEIARELIYI 676
Query: 174 PTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLK 233
+++ +KI
Sbjct: 677 RSTNTHSKITSK------------------------------------------------ 688
Query: 234 RACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVL 293
KNK + A+DGTHI A V + V +R R +QNVLAA +FDL F+Y+L
Sbjct: 689 ---KNKFYPYFKDCIGALDGTHIRASVPAKKVDRFRGRKPYPTQNVLAAVDFDLRFIYIL 745
Query: 294 SGWEGSAHDSKLLNDALKRKNGLKV----------------------------------- 318
+GWEGSAHDS +L DAL R NGLK+
Sbjct: 746 AGWEGSAHDSLVLQDALSRPNGLKILEGKFFLADAGYAARPGILPSYRRVRYHLKEFRGP 805
Query: 319 --PHGHGNDPENEKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPFLFKTQAELVLAC 376
PHG P+ KELFN RH+SLR +ER FG K+RF I + P K Q+ +V+AC
Sbjct: 806 QGPHG-PQGPKCPKELFNYRHSSLRTTIERAFGALKNRFKILMNKPFIPLKAQSMVVIAC 864
Query: 377 AALHNFLRKECRSDEF 392
ALHN++ E SDEF
Sbjct: 865 CALHNWI-LENGSDEF 879
>gb|AAV44105.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 1220
Score = 160 bits (405), Expect = 7e-38
Identities = 115/342 (33%), Positives = 154/342 (44%), Gaps = 95/342 (27%)
Query: 132 VGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPY 191
VG N R ++R F RS S F+K+L A+ + +L+ P+ P KI + R+ PY
Sbjct: 171 VGHNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPY 230
Query: 192 FKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAI 251
FK + AI
Sbjct: 231 FK----------------------------------------------------DCIGAI 238
Query: 252 DGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDALK 311
DGTHI A VR SS+R R +QNV+AA +FDL F YVL+GWEG+ HD+ +L DAL+
Sbjct: 239 DGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDAVVLRDALE 298
Query: 312 RKNGLKVPHGH----------------------------GNDP-ENEKELFNLRHASLRN 342
R+N L+VP G GN+P +NEKELFNLRH+SLR
Sbjct: 299 RENSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEWGNNPVQNEKELFNLRHSSLRV 358
Query: 343 VVERIFGIFKSRFTIFKSAPPFL-FKTQAELVLACAALHNFLRKECRSDEFPIEPSNESS 401
VER FG K RF + A PF F+TQ ++V+AC +HN++ + DE I P + SS
Sbjct: 359 TVERAFGSLKRRFKVLDDATPFFPFRTQVDIVVACCIIHNWVVND-GIDEL-IAPPDWSS 416
Query: 402 SSSSMLPIHEDNNDELNVQTQEQEREDANIWRTNLGLDMWRD 443
+ N+ L VQ +R L MW D
Sbjct: 417 EDIDESLTGQANDHALMVQ-----------FRQGLADQMWAD 447
>gb|AAV43825.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1067
Score = 160 bits (405), Expect = 7e-38
Identities = 115/342 (33%), Positives = 154/342 (44%), Gaps = 95/342 (27%)
Query: 132 VGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPY 191
VG N R ++R F RS S F+K+L A+ + +L+ P+ P KI + R+ PY
Sbjct: 18 VGHNLRNRLVRTNFDRSGETVSRYFNKVLHAIGELRDELIRPPSLDTPTKIAGNPRWDPY 77
Query: 192 FKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAI 251
FK + AI
Sbjct: 78 FK----------------------------------------------------DCIGAI 85
Query: 252 DGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDALK 311
DGTHI A VR SS+R R +QNV+AA +FDL F YVL+GWEG+ HD+ +L DAL+
Sbjct: 86 DGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTTHDAVVLRDALE 145
Query: 312 RKNGLKVPHGH----------------------------GNDP-ENEKELFNLRHASLRN 342
R+N L+VP G GN+P +NEKELFNLRH+SLR
Sbjct: 146 RENSLRVPQGKYYLVDAGYGAKQGFLPPFRAVRYHLNEWGNNPVQNEKELFNLRHSSLRV 205
Query: 343 VVERIFGIFKSRFTIFKSAPPFL-FKTQAELVLACAALHNFLRKECRSDEFPIEPSNESS 401
VER FG K RF + A PF F+TQ ++V+AC +HN++ + DE I P + SS
Sbjct: 206 TVERAFGSLKRRFKVLDDATPFFPFRTQVDIVVACCIIHNWVVND-GIDEL-IAPPDWSS 263
Query: 402 SSSSMLPIHEDNNDELNVQTQEQEREDANIWRTNLGLDMWRD 443
+ N+ L VQ +R L MW D
Sbjct: 264 EDIDESLTGQANDHALMVQ-----------FRQGLADQMWAD 294
>gb|AAP51979.1| putative transposase [Oryza sativa (japonica cultivar-group)]
gi|37530780|ref|NP_919692.1| putative transposase [Oryza
sativa (japonica cultivar-group)]
gi|21326484|gb|AAM47612.1| Putative transposase [Oryza
sativa (japonica cultivar-group)]
gi|20514796|gb|AAM23241.1| Putative transposase [Oryza
sativa (japonica cultivar-group)]
Length = 535
Score = 147 bits (371), Expect = 7e-34
Identities = 90/210 (42%), Positives = 118/210 (55%), Gaps = 32/210 (15%)
Query: 238 NKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWE 297
N G+ + AIDGTHI A VR SS+R R +QNV+AA +FDL F YVL+GWE
Sbjct: 47 NYGRSGETDCIGAIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWE 106
Query: 298 GSAHDSKLLNDALKRKNGLKVPHGH----------------------------GNDP-EN 328
G+AHD+ +L DAL+R+NGL VP G GN+P +N
Sbjct: 107 GTAHDAVVLRDALERENGLHVPQGKFYLVDGGYGAKQGFLPPFRAVRYHLKEWGNNPVQN 166
Query: 329 EKELFNLRHASLRNVVERIFGIFKSRFTIFKSAPPFL-FKTQAELVLACAALHNFLRKEC 387
EKELFNLRH+SLR VER FG K RF + A PF F+TQ ++V+AC +HN++ +
Sbjct: 167 EKELFNLRHSSLRITVERAFGSLKRRFKVLDDATPFFPFRTQVDIVVACCIIHNWVIND- 225
Query: 388 RSDEFPIEPSNESSSSSSMLPIHEDNNDEL 417
DE + PS+ SS P + N+ L
Sbjct: 226 GIDEL-VAPSDWSSEDIDESPTWQANDHAL 254
Score = 38.5 bits (88), Expect = 0.43
Identities = 21/51 (41%), Positives = 29/51 (56%), Gaps = 5/51 (9%)
Query: 103 LCMIIREK-----TPLVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRS 148
+ MIIR + + DT +CIEE +A FL VG N R ++R +GRS
Sbjct: 1 MAMIIRSRKRTRGSRREDTIHMCIEEQVAMFLHTVGHNLRNRLVRTNYGRS 51
>ref|XP_472365.1| OSJNBb0012E08.4 [Oryza sativa (japonica cultivar-group)]
gi|38345208|emb|CAD40780.2| OSJNBb0012E08.4 [Oryza
sativa (japonica cultivar-group)]
Length = 261
Score = 144 bits (362), Expect = 7e-33
Identities = 93/271 (34%), Positives = 125/271 (45%), Gaps = 81/271 (29%)
Query: 100 FRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKI 159
F + C + R+ L DT +CIEE +A FL VG N R ++R + RS S F+K+
Sbjct: 34 FFRFCKLFRDCGLLEDTIHMCIEEQVAMFLHTVGHNLRNRLVRTNYDRSGETVSRYFNKV 93
Query: 160 LKALNSIAPDLMAKPTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGF 219
L A+ + +L+ P+ P KI + R+ PYFK
Sbjct: 94 LHAIGELRDELIRPPSLDTPTKIAGNPRWDPYFK-------------------------- 127
Query: 220 ELNST*FDG*TNLKRACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNV 279
+ AIDGTHI A +R SS+R R +QNV
Sbjct: 128 --------------------------DCIGAIDGTHIRASIRKNVESSFRGRKSHATQNV 161
Query: 280 LAACNFDLEFMYVLSGWEGSAHDSKLLNDALKRKNGLKVPHGH----------------- 322
+AA +FDL F YVL+GWEG+ HD+ +L DAL+R+NGL VP G
Sbjct: 162 MAAVDFDLRFTYVLAGWEGTTHDAVVLRDALERENGLHVPQGKFYLVDAGYGAKQGFLPP 221
Query: 323 -----------GNDP-ENEKELFNLRHASLR 341
GN+P +NEKELFNLRH+SLR
Sbjct: 222 FRAVRYHLKEWGNNPVQNEKELFNLRHSSLR 252
>ref|XP_463137.1| expressed protein [Oryza sativa (japonica cultivar-group)]
gi|40786599|gb|AAR89874.1| expressed protein [Oryza
sativa (japonica cultivar-group)]
Length = 608
Score = 139 bits (349), Expect = 2e-31
Identities = 88/202 (43%), Positives = 113/202 (55%), Gaps = 40/202 (19%)
Query: 250 AIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDA 309
AIDGTH+ A V SS+R R +QNV+AA +FDL F YVL+GWEG+AHD +L DA
Sbjct: 143 AIDGTHVRASVPKNMESSFRGRKNHATQNVMAAVDFDLRFTYVLAGWEGAAHDVVVLRDA 202
Query: 310 LKRKNGLKVPHGH----------------------------GNDP-ENEKELFNLRHASL 340
L+R+NGL VP G GN+P +NEKELFNLRH+SL
Sbjct: 203 LERENGLHVPQGKFYLVDVGYGAKPGFLPPFRSTRYHLNEWGNNPVQNEKELFNLRHSSL 262
Query: 341 RNVVERIFGIFKSRFTIFKSAPPFL-FKTQAELVLACAALHNFLRKECRSDEFPIEPSNE 399
R VER FG K RF I A PF F+TQ +V+AC +HN++ + DE + P NE
Sbjct: 263 RITVERAFGSLKRRFKILDDATPFFPFQTQVNIVVACCIIHNWVIND-DIDELVMGP-NE 320
Query: 400 SSSSSSMLPIHEDNNDELNVQT 421
S I +DN+ ++ T
Sbjct: 321 S--------IIQDNDTSTDMDT 334
>dbj|BAB08431.1| unnamed protein product [Arabidopsis thaliana]
gi|28416671|gb|AAO42866.1| At5g41980 [Arabidopsis
thaliana] gi|15238244|ref|NP_199013.1| expressed protein
[Arabidopsis thaliana]
Length = 374
Score = 129 bits (324), Expect = 2e-28
Identities = 71/162 (43%), Positives = 93/162 (56%), Gaps = 22/162 (13%)
Query: 251 IDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDAL 310
+D HIP +V + +R+ +G ++QNVLAA +FDL F YVL+GWEGSA D ++LN AL
Sbjct: 147 VDSFHIPVMVGVDEQGPFRNGNGLLTQNVLAASSFDLRFNYVLAGWEGSASDQQVLNAAL 206
Query: 311 KRKNGLKVPHG-------------------HG---NDPENEKELFNLRHASLRNVVERIF 348
R+N L+VP G HG N E KE+FN RH L + R F
Sbjct: 207 TRRNKLQVPQGKYYIVDNKYPNLPGFIAPYHGVSTNSREEAKEMFNERHKLLHRAIHRTF 266
Query: 349 GIFKSRFTIFKSAPPFLFKTQAELVLACAALHNFLRKECRSD 390
G K RF I SAPP+ +TQ +LV+A ALHN++R E D
Sbjct: 267 GALKERFPILLSAPPYPLQTQVKLVIAACALHNYVRLEKPDD 308
Score = 57.4 bits (137), Expect = 9e-07
Identities = 40/122 (32%), Positives = 63/122 (50%), Gaps = 5/122 (4%)
Query: 72 SKGYDYIHKSLNGDSEIFRQVYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQI 131
S G ++++ LNG +E + +RM VF KLC +++ + L T I IE LA FL I
Sbjct: 25 SDGNKFVYQILNGPNEQCFENFRMDKPVFYKLCDLLQTRGLLRHTNRIKIEAQLAIFLFI 84
Query: 132 VGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPY 191
+G N R ++ F S S +F+ +L A+ +I+ D +P S+ + PY
Sbjct: 85 IGHNLRTRAVQELFCYSGETISRHFNNVLNAVIAISKDFF-QPNSNSDTLENDD----PY 139
Query: 192 FK 193
FK
Sbjct: 140 FK 141
>ref|NP_917005.1| P0519D04.27 [Oryza sativa (japonica cultivar-group)]
Length = 538
Score = 124 bits (311), Expect = 6e-27
Identities = 70/149 (46%), Positives = 86/149 (56%), Gaps = 29/149 (19%)
Query: 250 AIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDA 309
AIDGTHI A VR SS+R R +QNV+AA +FDL F YVL+GWEG+AHD+ +L DA
Sbjct: 113 AIDGTHIRASVRKNVESSFRGRKSHATQNVMAAVDFDLRFTYVLAGWEGTAHDAVVLRDA 172
Query: 310 LKRKNGLKVPHGH----------------------------GNDPENEKELFNLRHASLR 341
L+R+NG+ VP G GN+P +ELFNLRH+SLR
Sbjct: 173 LERENGIHVPQGKFYLVDAGYGAKQGFLPPFRAVRYHLKEWGNNPVQNEELFNLRHSSLR 232
Query: 342 NVVERIFGIFKSRFTIFKSAPPFL-FKTQ 369
VER FG K RF + A PF F+TQ
Sbjct: 233 ITVERAFGSLKRRFKVLDDATPFFPFRTQ 261
>ref|XP_471107.1| OSJNBa0042N22.14 [Oryza sativa (japonica cultivar-group)]
gi|38569160|emb|CAE03671.3| OSJNBa0042N22.14 [Oryza
sativa (japonica cultivar-group)]
Length = 401
Score = 121 bits (304), Expect = 4e-26
Identities = 86/281 (30%), Positives = 124/281 (43%), Gaps = 82/281 (29%)
Query: 123 EMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMAKPTSSVPAKI 182
E +A FL G N R ++ F RS S F+ +L A+ + +L+ P+ + P+KI
Sbjct: 45 EQVAMFLHTFGHNVRNRVVATNFYRSGETISRYFNLVLHAVGELRKELIRPPSITTPSKI 104
Query: 183 RESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNLKRACKNKGKH 242
+ + PYFK
Sbjct: 105 LGNPMWDPYFK------------------------------------------------- 115
Query: 243 KILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHD 302
+ AIDGTH+ V S+R R +QNV+AA +FDL+F YVL+GWE +AHD
Sbjct: 116 ---DCIGAIDGTHVRVSVTKDMELSFRGRKDHATQNVMAAIDFDLKFTYVLAGWETTAHD 172
Query: 303 SKLLNDALKRKNGLKVPHGH----------------------------GNDP-ENEKELF 333
+ +L D+++R +GL+VP G G++P +NEKELF
Sbjct: 173 AVVLRDSIERTDGLRVPQGKFYLVDVGYGAKPGFLPPFRGVRYHLNEWGSNPVQNEKELF 232
Query: 334 NLRHASLRNVVERIFGIFKSRFTIFKSAPPFL-FKTQAELV 373
NLRH+SLR V R F K R+ I A PF + TQ ++V
Sbjct: 233 NLRHSSLRVTVARAFRSLKRRWKILDDASPFFPYPTQVDIV 273
>pir||C96640 hypothetical protein T25B24.14 [imported] - Arabidopsis thaliana
gi|4585884|gb|AAD25557.1| Hypothetical protein
[Arabidopsis thaliana]
Length = 404
Score = 117 bits (292), Expect = 1e-24
Identities = 89/331 (26%), Positives = 139/331 (41%), Gaps = 81/331 (24%)
Query: 92 VYRMYPDVFRKLCMIIREKTPLVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFA 151
+ RM+ FR LC I+ E+ L ++ I +EE +A F+++V Q+ +I + S
Sbjct: 71 ILRMHQSTFRTLCKILSEQYKLEESCNIYLEESVAMFIEMVAQDLTVRVIAERYQHSLET 130
Query: 152 ASENFHKILKALNSIAPDLMAKPTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*K 211
++L AL +A D++ KPT + T P+ L N K P I C
Sbjct: 131 VKRKLDEVLSALLKLAADIV-KPTRD------DVTGISPF---LVNDKRYMPYFIDC--- 177
Query: 212 FSQDFEGFELNST*FDG*TNLKRACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDR 271
+ A+DGTH+ DV YR R
Sbjct: 178 ------------------------------------IGALDGTHVSVRPPSGDVERYRGR 201
Query: 272 HGRISQNVLAACNFDLEFMYVLSGWEGSAHDSK---------------------LLNDAL 310
+ N+LA CNF ++F G G AHD+K L++
Sbjct: 202 KSEATMNILAVCNFSMKFNIAYVGVPGRAHDTKVLTYCATHEASFPHPPAGKYYLVDSGY 261
Query: 311 KRKNGLKVPH----------GHGNDPENEKELFNLRHASLRNVVERIFGIFKSRFTIF-K 359
++G PH G P N +ELFN RH+SLR+V+ER FG++K+++ I +
Sbjct: 262 PTRSGYLGPHRRTRYHLELFNRGGPPTNSRELFNRRHSSLRSVIERTFGVWKAKWRILDR 321
Query: 360 SAPPFLFKTQAELVLACAALHNFLRKECRSD 390
P + K ++V + ALHN++R + D
Sbjct: 322 KHPKYEVKKWIKIVTSTMALHNYIRDSQQED 352
>dbj|BAB01829.1| unnamed protein product [Arabidopsis thaliana]
Length = 194
Score = 107 bits (266), Expect = 1e-21
Identities = 78/230 (33%), Positives = 102/230 (43%), Gaps = 78/230 (33%)
Query: 113 LVDTRFICIEEMLASFLQIVGQNTRYCIIRNTFGRSQFAASENFHKILKALNSIAPDLMA 172
L DT + +EEMLASFL I+GQN RY ++ F RS+F+ S +F+ ILK LN AP MA
Sbjct: 3 LRDTMNVSVEEMLASFLFIIGQNLRYIQAQDRFKRSRFSISTSFNTILKVLNVFAPSYMA 62
Query: 173 KPTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*TNL 232
KPT VP KI+++TRFYP + +D G
Sbjct: 63 KPTLVVPTKIKDNTRFYP---------------------YFKDCVG-------------- 87
Query: 233 KRACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYV 292
AIDGTHIPA++ G++ +SY D C F
Sbjct: 88 -----------------AIDGTHIPAMITGKETASYLD------------CGF------- 111
Query: 293 LSGWEGSAHDSKLLNDALKRKNGLKVPHGHGNDPENEKELFNLRHASLRN 342
A+ + L+ G G DP N+ ELFN RH+SLRN
Sbjct: 112 -------ANCRNCFALFRSTRYHLQDFRGQGKDPNNQNELFNHRHSSLRN 154
>ref|NP_198247.1| hypothetical protein [Arabidopsis thaliana]
Length = 148
Score = 103 bits (256), Expect = 1e-20
Identities = 67/150 (44%), Positives = 78/150 (51%), Gaps = 53/150 (35%)
Query: 171 MAKPTSSVPAKIRESTRFYPYFKILYN*KYIWPITICCK*KFSQDFEGFELNST*FDG*T 230
MAKP +VP KIRESTR YP + +D G
Sbjct: 1 MAKPEIAVPRKIRESTRLYP---------------------YFKDCVG------------ 27
Query: 231 NLKRACKNKGKHKILSLLQAIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFM 290
AID THI A+V + + S+R+R G ISQN+LAACNFD+EFM
Sbjct: 28 -------------------AIDDTHIFAMVSQKKMPSFRNRKGDISQNMLAACNFDVEFM 68
Query: 291 YVLSGWEGSAHDSKLLNDALKR-KNGLKVP 319
YVLSGWEGSAHDSK+LNDAL R N L VP
Sbjct: 69 YVLSGWEGSAHDSKVLNDALTRNSNRLPVP 98
Score = 39.3 bits (90), Expect = 0.25
Identities = 20/55 (36%), Positives = 33/55 (59%), Gaps = 3/55 (5%)
Query: 389 SDEFPIEPSNESSSSSSMLPIHEDNNDELNVQTQEQEREDANIWRTNLGLDMWRD 443
S+ P+ +ES+ ++ DNNDE+ + TQ+Q+RE AN WR + +MW +
Sbjct: 92 SNRLPVPEEDESAEE--VVEEVNDNNDEV-LTTQDQQREYANQWRETIASNMWNN 143
>gb|AAC14510.1| En/Spm-like transposon protein [Arabidopsis thaliana]
gi|7487888|pir||T00996 En/Spm-like transposon protein
[imported] - Arabidopsis thaliana
Length = 292
Score = 100 bits (249), Expect = 9e-20
Identities = 64/210 (30%), Positives = 97/210 (45%), Gaps = 39/210 (18%)
Query: 250 AIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLN-- 307
A+DGTH+P + Y+ R + NVLA CNFD++F+Y G G AHD+K+LN
Sbjct: 47 ALDGTHVPVRPPSQTAKKYKGRKLEPTMNVLAICNFDMKFIYAYVGVPGRAHDTKVLNYC 106
Query: 308 -------------------DALKRKNGLKVPH----------GHGNDPENEKELFNLRHA 338
+ G PH G G P +ELFN +H+
Sbjct: 107 ATNEPYFSHPPNGKYYLVDSGYPTRTGYLGPHRRMRYHLGQFGRGGPPVTARELFNRKHS 166
Query: 339 SLRNVVERIFGIFKSRFTIF-KSAPPFLFKTQAELVLACAALHNFLRKECRSDEFPIEPS 397
LR+V+ER FG++K+++ I + P + ++V A ALHNF+R R D
Sbjct: 167 GLRSVIERTFGVWKAKWRIVDRKHPKYGLAKWIKIVTATMALHNFIRDSHRED------- 219
Query: 398 NESSSSSSMLPIHEDNNDELNVQTQEQERE 427
++ S+ H D+ +E + E+E E
Sbjct: 220 HDFLQWQSIEEYHVDDEEEEEEEEGEEEEE 249
>gb|AAQ95645.1| transposase [Pharus latifolius]
Length = 119
Score = 90.5 bits (223), Expect = 1e-16
Identities = 53/118 (44%), Positives = 63/118 (52%), Gaps = 30/118 (25%)
Query: 250 AIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDA 309
A+DGTHIPA V + R +QNVLAA +FDL F+YVL+GWEGSAHDS +L+DA
Sbjct: 2 ALDGTHIPASVPTDMQDRFIGRKLYPTQNVLAAVDFDLRFVYVLAGWEGSAHDSYVLDDA 61
Query: 310 LKRKNGLKVPHGH------------------------------GNDPENEKELFNLRH 337
L R GLK+P GH PE+EKELFN RH
Sbjct: 62 LSRPTGLKIPEGHYFLADAGYATRKGILPPYRGVRYHLNEYRGTRQPEDEKELFNPRH 119
>gb|AAQ95646.1| transposase [Coix aquatica]
Length = 119
Score = 89.7 bits (221), Expect = 2e-16
Identities = 51/118 (43%), Positives = 61/118 (51%), Gaps = 30/118 (25%)
Query: 250 AIDGTHIPALVRGRDVSSYRDRHGRISQNVLAACNFDLEFMYVLSGWEGSAHDSKLLNDA 309
A+DGTHIPA V +R R +QNVLA +FDL F+YVL+GWEGSAHDS +L DA
Sbjct: 2 ALDGTHIPACVPFHMQDRFRGRKSITTQNVLAVVDFDLRFIYVLAGWEGSAHDSYVLQDA 61
Query: 310 LKRKNGLKVPHGH------------------------------GNDPENEKELFNLRH 337
L R NGL +P G +PE+ KELFN RH
Sbjct: 62 LSRPNGLNIPEGKYFLADAGYAARPGVLPPYRGTRYHLQEYRGTREPESPKELFNPRH 119
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.326 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 753,917,753
Number of Sequences: 2540612
Number of extensions: 31808603
Number of successful extensions: 424169
Number of sequences better than 10.0: 1756
Number of HSP's better than 10.0 without gapping: 1550
Number of HSP's successfully gapped in prelim test: 224
Number of HSP's that attempted gapping in prelim test: 366084
Number of HSP's gapped (non-prelim): 25131
length of query: 447
length of database: 863,360,394
effective HSP length: 131
effective length of query: 316
effective length of database: 530,540,222
effective search space: 167650710152
effective search space used: 167650710152
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146551.12


