

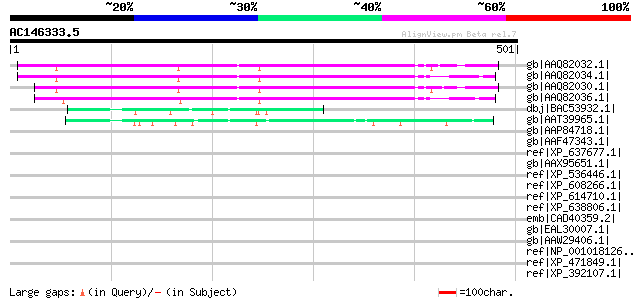
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146333.5 + phase: 0
(501 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAQ82032.1| unknown [Pisum sativum] 277 6e-73
gb|AAQ82034.1| unknown [Pisum sativum] 276 1e-72
gb|AAQ82030.1| unknown [Pisum sativum] 276 1e-72
gb|AAQ82036.1| unknown [Pisum sativum] 269 2e-70
dbj|BAC53932.1| hypothetical protein [Nicotiana tabacum] 59 3e-07
gb|AAT39965.1| hypothetical protein [Solanum demissum] 50 2e-04
gb|AAP84718.1| PHD zinc finger protein rhinoceros [Drosophila me... 40 0.22
gb|AAF47343.1| CG7036-PA, isoform A [Drosophila melanogaster] gi... 40 0.22
ref|XP_637677.1| hypothetical protein DDB0187005 [Dictyostelium ... 40 0.22
gb|AAX95651.1| hypothetical protein [Oryza sativa (japonica cult... 38 0.85
ref|XP_536446.1| PREDICTED: similar to step II splicing factor S... 38 0.85
ref|XP_608266.1| PREDICTED: similar to kinesin-like 7, partial [... 37 1.1
ref|XP_614710.1| PREDICTED: similar to kinesin-like 7, partial [... 37 1.1
ref|XP_638806.1| hypothetical protein DDB0218555 [Dictyostelium ... 37 1.1
emb|CAD40359.2| OSJNBa0093P23.5 [Oryza sativa (japonica cultivar... 37 1.5
gb|EAL30007.1| GA20049-PA [Drosophila pseudoobscura] 37 1.5
gb|AAW29406.1| P-209 [Borrelia turicatae] 37 1.5
ref|NP_001018126.1| hypothetical protein LOC347273 [Homo sapiens] 37 1.9
ref|XP_471849.1| OSJNBa0054D14.3 [Oryza sativa (japonica cultiva... 37 1.9
ref|XP_392107.1| PREDICTED: similar to CG4832-PA, isoform A [Api... 37 1.9
>gb|AAQ82032.1| unknown [Pisum sativum]
Length = 562
Score = 277 bits (708), Expect = 6e-73
Identities = 178/492 (36%), Positives = 256/492 (51%), Gaps = 28/492 (5%)
Query: 8 IHRRIGQIKNFVNITGMK-KTKSYKFKEVDLVSLRDLA--LKVKNQTGFRLRYGGLLTIL 64
+H R + + +K KT SY F L SL +L+ + NQ GF +YG +LT+L
Sbjct: 7 VHSRACSLIEMATVPELKRKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLL 66
Query: 65 RTNVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTGPGASLTP 124
+ V+ + TL+QFYDP CFTF D+QL PTLE YS LL P+ + PF +
Sbjct: 67 KMVVDPVPLQTLLQFYDPELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDF 126
Query: 125 LVIAKDLYLKTSDVSKHLTTKSHIRGFTSKYLLEQANLETT--CQDALEAILALLVYGLI 182
V+A+ L+L +VS + + G K+LL A E +A A LA+++YG++
Sbjct: 127 RVVARALHLGIKEVSDSWKSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIV 186
Query: 183 LFPNLDNFVDMNAIEIFHSRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHL 242
LFP++ NFVD A+ IF NPVPTLLAD Y+AIH R KG I CC+PLL RWF+S L
Sbjct: 187 LFPSMPNFVDYAAVSIFIGGNPVPTLLADTYYAIHSRHGKGGA-IRCCLPLLLRWFLSLL 245
Query: 243 PSS--FHDNSEDWSYSQRMMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINY 300
P S F D ++QR+M+L+ ++ W + V+ +I CG F NVPL+GTRG INY
Sbjct: 246 PVSGPFVDAQSTHKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINY 305
Query: 301 NPELAMRQFGFPMKTKPINLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVRSG 360
NP L++RQ GF MK +P+ + + + + E RAW + K LG +
Sbjct: 306 NPVLSLRQLGFVMKGRPLEAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFA 365
Query: 361 IAHEAYTQWVINRAEEIGLPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESR----- 415
+A YT WV R E + LPY M + P I P E Y++ L ME+R
Sbjct: 366 VAMPDYTDWVKERVETLLLPYDRMEPLQEQPPLILAESVPA--EHYKQAL-MENRRLREK 422
Query: 416 ----EKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMQDSK 471
+ +++KA+ ++ NL L G E+ L +R EE + +L + +
Sbjct: 423 EQDTQMELYKAK-ADSLNLAHQLRGVREEDA-------SRLRSKKRSYEEVESMLDAEHR 474
Query: 472 RARGRRNFYARY 483
+ A Y
Sbjct: 475 ECLRLQRVEASY 486
>gb|AAQ82034.1| unknown [Pisum sativum]
Length = 562
Score = 276 bits (705), Expect = 1e-72
Identities = 175/480 (36%), Positives = 250/480 (51%), Gaps = 31/480 (6%)
Query: 8 IHRRIGQIKNFVNITGMK-KTKSYKFKEVDLVSLRDLA--LKVKNQTGFRLRYGGLLTIL 64
+H R + + +K KT SY F L SL +L+ + NQ GF ++G +LT+L
Sbjct: 7 VHSRACSLIEMATVPELKRKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLL 66
Query: 65 RTNVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTGPGASLTP 124
+ V+ + TL+QFYDP CFTF D+QL PTLE YS LL P+ + PF +
Sbjct: 67 KMVVDPVPLQTLLQFYDPELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDF 126
Query: 125 LVIAKDLYLKTSDVSKHLTTKSHIRGFTSKYLLEQANLETT--CQDALEAILALLVYGLI 182
V+A+ L+L +VS + G K+LL A E +A A LA+++YG++
Sbjct: 127 GVVARALHLGIKEVSDSWKYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIV 186
Query: 183 LFPNLDNFVDMNAIEIFHSRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHL 242
LFP++ NFVD A+ IF NPVPTLLAD Y+AIH R KG I CC+PLL RWF+S L
Sbjct: 187 LFPSMPNFVDYAAVSIFIGGNPVPTLLADTYYAIHSRHGKGGA-IRCCLPLLLRWFLSLL 245
Query: 243 PSS--FHDNSEDWSYSQRMMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINY 300
P S F D ++QR+M+L+ ++ W + V+ +I CG F NVPL+GTRG INY
Sbjct: 246 PVSGPFVDAQSTHKWTQRVMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINY 305
Query: 301 NPELAMRQFGFPMKTKPINLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVRSG 360
NP L++RQ GF MK +P+ + ++ + + E RAW + K LG +
Sbjct: 306 NPVLSLRQLGFVMKGRPLEAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFV 365
Query: 361 IAHEAYTQWVINRAEEIGLPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMW 420
+A YT WV R E + LPY M + PSI P E Y++ L ME+R
Sbjct: 366 VAMPDYTDWVKERVETLLLPYDRMEPLQEQPPSILAESVPA--EHYKQAL-MENRR---- 418
Query: 421 KARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMQDSKRARGRRNFY 480
L GK++ E K + + + +L E +D+ R R ++ Y
Sbjct: 419 -------------LRGKEQDTQLELYKAKADKLNLAHQLRGVRE---EDASRLRSKKRSY 462
>gb|AAQ82030.1| unknown [Pisum sativum]
Length = 562
Score = 276 bits (705), Expect = 1e-72
Identities = 175/474 (36%), Positives = 250/474 (51%), Gaps = 27/474 (5%)
Query: 25 KKTKSYKFKEVDLVSLRDLA--LKVKNQTGFRLRYGGLLTILRTNVEEKLVHTLVQFYDP 82
+KT SY F L SL +L+ + NQ GF +YG +LT+L+ V+ + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 83 SFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTGPGASLTPLVIAKDLYLKTSDVSKHL 142
CFTF D+QL PTLE YS LL P+ + PF + V+A+ L+L +VS +
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDNW 144
Query: 143 TTKSHIRGFTSKYLLEQANLETT--CQDALEAILALLVYGLILFPNLDNFVDMNAIEIFH 200
+ + G K+LL A E +A A LA+++YG++LFP++ NFVD A+ IF
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 201 SRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHLPSS--FHDNSEDWSYSQR 258
NPVPTLLAD Y+AIH R KG I CC+PLL RWF+S LP S F D ++QR
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKGGA-IRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 259 MMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFPMKTKPI 318
+M+L+ ++ W + V+ +I CG F NVPL+GTRG INYNP L++RQ GF MK +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 319 NLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVRSGIAHEAYTQWVINRAEEIG 378
+ ++ + + E RAW + K LG + +A YT WV R E +
Sbjct: 324 EAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 379 LPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESR---------EKQMWKARYNEAEN 429
LPY M + P I P E Y++ L ME+R + +++KA+ N
Sbjct: 384 LPYDRMEPLQEQPPLILAESVPA--EHYKQAL-MENRRLREKEQDTQMELYKAKAGRL-N 439
Query: 430 LIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMQDSKRARGRRNFYARY 483
L L G E+ L +R EE + +L + + + A Y
Sbjct: 440 LAHQLRGVREEDA-------SRLRSKKRSYEEMESMLDAEHRECLRLQRVEASY 486
>gb|AAQ82036.1| unknown [Pisum sativum]
Length = 546
Score = 269 bits (687), Expect = 2e-70
Identities = 168/462 (36%), Positives = 241/462 (51%), Gaps = 30/462 (6%)
Query: 25 KKTKSYKFKEVDLVSLRDLALKVKNQT--GFRLRYGGLLTILRTNVEEKLVHTLVQFYDP 82
+KT SY F L SL +L + + GF ++G +LT+L+T V+ + TL+QFYDP
Sbjct: 9 RKTCSYSFYREPLTSLIELGSLMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYDP 68
Query: 83 SFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTGPGASLTPLVIAKDLYLKTSDVSKHL 142
RCFTF D+QL PTLE YS L+ PI + PF + VIA+ L + +V +
Sbjct: 69 ELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKEVCDNW 128
Query: 143 TTKSHIRGFTSKYLLEQANLETTCQ--DALEAILALLVYGLILFPNLDNFVDMNAIEIFH 200
+ G K+LL A E + A LA ++YG++LFP++ NF+D AI IF
Sbjct: 129 KPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIFI 188
Query: 201 SRNPVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHLPSS--FHDNSEDWSYSQR 258
NPVPTLLAD Y+AIH R KG I CC+PLL RWF+S LP+S F D ++QR
Sbjct: 189 GGNPVPTLLADTYYAIHSRHGKGGA-IRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQR 247
Query: 259 MMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINYNPELAMRQFGFPMKTKPI 318
+M+L+ ++ W + VK+++ CG+F NVPL+GT+G INYNP L++RQ GF M +P+
Sbjct: 248 VMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRPL 307
Query: 319 NLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVRSGIAHEAYTQWVINRAEEIG 378
++ + E RAW + K LG + IA YT WV R E +
Sbjct: 308 EAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETLL 367
Query: 379 LPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKD 438
LPY M + P I P E Y++ L ME+R L K+
Sbjct: 368 LPYDRMEPLQEQPPLILADSVPA--EHYKQAL-MENRR-----------------LREKE 407
Query: 439 EQKTHENLMLKKELVKVRRELEEKDELLMQDSKRARGRRNFY 480
+ E K + + + +L E+ +D+ RAR ++ Y
Sbjct: 408 QDTRMELYKAKADKLNLAHQLR---EMQGEDASRARSKKRSY 446
>dbj|BAC53932.1| hypothetical protein [Nicotiana tabacum]
Length = 464
Score = 59.3 bits (142), Expect = 3e-07
Identities = 70/307 (22%), Positives = 117/307 (37%), Gaps = 68/307 (22%)
Query: 58 GGLLTILRTNVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFTG 117
G L I++ + L+ LV F+DP F F DF+L PTLE E ++G
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLE-----------EIDGYSG 87
Query: 118 PGASLT------PLVIAKDLYLKTSDVSKHLTTKSHIRGFTSKYLL-------------- 157
G L P ++ + ++SK + + ++G S Y L
Sbjct: 88 FGRDLRNQELIFPRALSVHRFFDLLNISKQIRKTNVVKGCCSFYFLYSRFGQPNGFEMHE 147
Query: 158 ----EQANLET-TCQDALEAILALLVYGLILFPNLDNFVDMNAIEIF------HSRNPVP 206
+ N +T I+A L G+++FPN + +D + P
Sbjct: 148 KGLNNKQNKDTWHIHRCFAFIMAFL--GIMVFPNRERTIDTRIARVVQVLTTKEHHTLAP 205
Query: 207 TLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHL----------PS--SFHDNSED-- 252
+L+D+Y A+ G + C LL W I HL PS +F D+ E+
Sbjct: 206 IILSDIYRAL-TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYEERV 264
Query: 253 ---------WSYSQRMMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGINYNPE 303
++ + +L+ ++ W ++E+I ++ LLG R Y P
Sbjct: 265 KEYNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPYMPH 324
Query: 304 LAMRQFG 310
+RQ G
Sbjct: 325 RVLRQLG 331
>gb|AAT39965.1| hypothetical protein [Solanum demissum]
Length = 607
Score = 49.7 bits (117), Expect = 2e-04
Identities = 111/508 (21%), Positives = 180/508 (34%), Gaps = 105/508 (20%)
Query: 56 RYGGLLTILRTNVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPF 115
R G L ++L L+ LV F+DP F F DF+L PTLE E F
Sbjct: 47 RLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLE-----------EIGAF 95
Query: 116 TGPGASL---TPLV--------IAKDLYLKTSDVS--------------KHLTTKSHIRG 150
G G +L P++ + L++ +++ K K
Sbjct: 96 IGRGKNLHEGEPMIPKHINGRRFLELLHINENEIGGCLDNGWVPLEFLYKRYGRKDGFEL 155
Query: 151 FTSKYLLEQANL--ETTCQDALE-AILALLVY----GLILFPNLDNFVDMNAIEIFHSRN 203
+ K L ET DA+ A L ++V+ G I N++ + A E +
Sbjct: 156 YGKKLHNNGCRLTWETHSYDAITVAFLGIMVFLKRGGKI---NINLAAVITAFEKKPNIT 212
Query: 204 PVPTLLADMYHAIHDRTLKGRVYILCCVPLLYRWFISHL--------------------- 242
VP +LAD+Y A+ G Y C LL W I H+
Sbjct: 213 LVPMILADIYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHE 271
Query: 243 ----PSSFHDNSEDWSYSQRMMALSPNEVVWITPAAQVKEIITGCGDFLNVPLLGTRGGI 298
SF E W + + L+ +++VW E+I + L+G RG
Sbjct: 272 ERIKDHSFPKGIEAW--KKYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQ 329
Query: 299 NYNPELAMRQFGFPMKTKPINLATSPEFFYYSNAPTGQREAFTRAWSKVRWKSVKHLGVR 358
Y P MRQ G P + ++ P + E F + W S H V
Sbjct: 330 PYMPLRVMRQLGRRQFLPPNEDIREFMYEFHPEIPLRKSEIF-KIWGGCMLSS-PHDRVE 387
Query: 359 ---SGIAHEAYTQWVINRAEEIGLPYPAM----------------------RHVAASAPS 393
G +AY +WV ++ LP ++ R ++
Sbjct: 388 DRTKGEVDQAYLEWVHDQPSPKVLPEGSVKGPVDREAEIEVRIKQARLEVERSYRSTLDC 447
Query: 394 IPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENL---IMTLDGKDEQKTHENLMLKK 450
+ L +E+ Q E R ++ E+L I ++ ++E+ T E + +
Sbjct: 448 LSNDLKNAKEELAQRDAIFEVRVRKHRSTILTLQEDLGIVISAMEQQEEEYTKEKVQAEC 507
Query: 451 ELVKVRRELEEKDELLMQDSKRARGRRN 478
E+ + +R + E+D L +Q + R N
Sbjct: 508 EVERGQR-IAERDALHLQIEELQEQREN 534
>gb|AAP84718.1| PHD zinc finger protein rhinoceros [Drosophila melanogaster]
gi|45445719|gb|AAS64921.1| CG7036-PB, isoform B
[Drosophila melanogaster] gi|45552837|ref|NP_995944.1|
CG7036-PB, isoform B [Drosophila melanogaster]
Length = 3241
Score = 39.7 bits (91), Expect = 0.22
Identities = 29/89 (32%), Positives = 40/89 (44%), Gaps = 7/89 (7%)
Query: 385 RHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHE 444
R AA P+ P P P + EM + R + W++R DE+ TH
Sbjct: 1686 RPPAAPLPASPTPTPTSNDEMSDAGSDLSERRRMRWRSRRRRRRR----SHEPDEEHTHH 1741
Query: 445 NLMLKKELVKVRRELEE--KDELLMQDSK 471
L E+ ++ RELEE K+ELL SK
Sbjct: 1742 TQHLLNEM-EMARELEEERKNELLANASK 1769
>gb|AAF47343.1| CG7036-PA, isoform A [Drosophila melanogaster]
gi|24654556|ref|NP_612007.1| CG7036-PA, isoform A
[Drosophila melanogaster]
Length = 3201
Score = 39.7 bits (91), Expect = 0.22
Identities = 29/89 (32%), Positives = 40/89 (44%), Gaps = 7/89 (7%)
Query: 385 RHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHE 444
R AA P+ P P P + EM + R + W++R DE+ TH
Sbjct: 1646 RPPAAPLPASPTPTPTSNDEMSDAGSDLSERRRMRWRSRRRRRRR----SHEPDEEHTHH 1701
Query: 445 NLMLKKELVKVRRELEE--KDELLMQDSK 471
L E+ ++ RELEE K+ELL SK
Sbjct: 1702 TQHLLNEM-EMARELEEERKNELLANASK 1729
>ref|XP_637677.1| hypothetical protein DDB0187005 [Dictyostelium discoideum]
gi|60466105|gb|EAL64171.1| hypothetical protein
DDB0187005 [Dictyostelium discoideum]
Length = 907
Score = 39.7 bits (91), Expect = 0.22
Identities = 21/80 (26%), Positives = 41/80 (51%)
Query: 416 EKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELLMQDSKRARG 475
+K + + E E+L+ + +Q H+ +KK+ +E EEK+E + K A G
Sbjct: 266 DKTLDDIKVEETEDLLKEKQQQQQQHAHQKETIKKDQKDNNKEEEEKEEEFIIQKKSASG 325
Query: 476 RRNFYARYCGSDSESESEDH 495
NFY+ + ++S+ + D+
Sbjct: 326 NSNFYSYHELNNSKRKQPDN 345
>gb|AAX95651.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50918055|ref|XP_469424.1| putative growth factor
[Oryza sativa (japonica cultivar-group)]
Length = 479
Score = 37.7 bits (86), Expect = 0.85
Identities = 24/114 (21%), Positives = 53/114 (46%), Gaps = 8/114 (7%)
Query: 380 PYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDE 439
P P+++ A+S+ P P PT +E ++ A + EK K NE + + +
Sbjct: 331 PNPSIKSKASSSDQKPKPSKPTEEEREEKAAAAVAAEKPKKKKATNEIDKIFQATKSSGK 390
Query: 440 QKTHENLMLKKELVKVRRELEEKDELLMQDSKRARGRRNFYARYCGSDSESESE 493
++ + ++E V+ ++ +E+ E + +K +G + G D++ + E
Sbjct: 391 KRKQQQQQGEEESVRAKKP-KERSEGAKKSNKAKKGSK-------GRDTDDDDE 436
>ref|XP_536446.1| PREDICTED: similar to step II splicing factor SLU7 [Canis
familiaris]
Length = 617
Score = 37.7 bits (86), Expect = 0.85
Identities = 30/95 (31%), Positives = 48/95 (49%), Gaps = 10/95 (10%)
Query: 399 PPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKEL-VKVRR 457
P T EM+QE L E ++K+ K ++ ++ + D DE+K HE LKK L + R
Sbjct: 514 PQTLMEMHQEKLKEEKKKKKKKKKKHRKSSS-----DSDDEEKKHEK--LKKALNAEEAR 566
Query: 458 ELEEKDELLMQDSKRARGRRNFYARYCGSDSESES 492
L K+ +MQ +R R + Y ++ E E+
Sbjct: 567 LLHVKE--IMQIDERKRPYNSIYETREPTEEEMEA 599
>ref|XP_608266.1| PREDICTED: similar to kinesin-like 7, partial [Bos taurus]
Length = 353
Score = 37.4 bits (85), Expect = 1.1
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 15/118 (12%)
Query: 370 VINRAEEIGLPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAM----ESREKQMWKARYN 425
V + E + HV SA + P P T +Q HLA + +E + WKA
Sbjct: 75 VEQKKNEYNFKMRQLGHVMDSA--VNYPQSPKTPPHFQTHLAKLLETQEQEIEDWKASKI 132
Query: 426 EAENLIMTLDGKDEQKTHENLMLKKELVKVR------RELEEKDELL---MQDSKRAR 474
++LI L+ + E K E L +K++L ++ +L EK+ LL + D KR +
Sbjct: 133 SLQHLITKLNEEREVKNAEILRIKEQLCEMENLRLEAEQLREKNWLLQGQLDDIKRQK 190
>ref|XP_614710.1| PREDICTED: similar to kinesin-like 7, partial [Bos taurus]
Length = 1241
Score = 37.4 bits (85), Expect = 1.1
Identities = 34/118 (28%), Positives = 54/118 (44%), Gaps = 15/118 (12%)
Query: 370 VINRAEEIGLPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAM----ESREKQMWKARYN 425
V + E + HV SA + P P T +Q HLA + +E + WKA
Sbjct: 961 VEQKKNEYNFKMRQLGHVMDSA--VNYPQSPKTPPHFQTHLAKLLETQEQEIEDWKASKI 1018
Query: 426 EAENLIMTLDGKDEQKTHENLMLKKELVKVR------RELEEKDELL---MQDSKRAR 474
++LI L+ + E K E L +K++L ++ +L EK+ LL + D KR +
Sbjct: 1019 SLQHLITKLNEEREVKNAEILRIKEQLCEMENLRLEAEQLREKNWLLQGQLDDIKRQK 1076
>ref|XP_638806.1| hypothetical protein DDB0218555 [Dictyostelium discoideum]
gi|60467463|gb|EAL65486.1| hypothetical protein
DDB0218555 [Dictyostelium discoideum]
Length = 803
Score = 37.4 bits (85), Expect = 1.1
Identities = 23/94 (24%), Positives = 43/94 (45%), Gaps = 5/94 (5%)
Query: 2 HKFFA--CIHRRIGQIKNFVNITGMKKTKSYKFKEV--DLVSLRDLALKVKNQTGFRL-R 56
HKF + C ++ + ++N+TG F ++ + + + L K N GF +
Sbjct: 250 HKFLSTNCSFQQHHDLLVYLNLTGFHSFSKLNFTKICKEFLEINQLEFKKVNLEGFSIFE 309
Query: 57 YGGLLTILRTNVEEKLVHTLVQFYDPSFRCFTFP 90
+ TI N++ K +H +V DP++ FP
Sbjct: 310 INEIFTIALKNLDFKFLHLMVNCMDPNYFSVDFP 343
>emb|CAD40359.2| OSJNBa0093P23.5 [Oryza sativa (japonica cultivar-group)]
gi|50922621|ref|XP_471671.1| OSJNBa0041M21.10 [Oryza
sativa (japonica cultivar-group)]
gi|38343986|emb|CAD40452.2| OSJNBa0041M21.10 [Oryza
sativa (japonica cultivar-group)]
Length = 2074
Score = 37.0 bits (84), Expect = 1.5
Identities = 48/205 (23%), Positives = 78/205 (37%), Gaps = 20/205 (9%)
Query: 57 YGGLLTILRTNVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSHLLDSPIAEKTPFT 116
+GGLL I R NV L + +D F L T HLLD P + F
Sbjct: 493 FGGLLKIHRINVHRILCMWIANQFDTKAEAFKIQGSYLSLTNRDAEHLLDLPSQGEEIFE 552
Query: 117 GPGASLTPLVIAKDLYLKTSDVSKHLTTKSHIRGFTSKYLLEQANLETTCQDALEAILAL 176
P +DL+ + SK +HI+ +Y + D L
Sbjct: 553 PPQTK------NRDLFDEFKTTSKQ---GAHIKLSLQEYFQTNKGIH---DDNFIHRFVL 600
Query: 177 LVYGLILFPNLDNFVD---MNAIEIFHSRNPVPTLLADMYHAIHD----RTLKGRVYILC 229
V G+ L P +V +N +E ++ + + ++ H + T+KG V +
Sbjct: 601 FVIGVFLCPTTQRYVSSAYLNLVEDVNAISAINWTSLNLNHLMKSIKNISTIKG-VNLEG 659
Query: 230 CVPLLYRWFISHLPSSFHDNSEDWS 254
+PLL W+ L + D + D++
Sbjct: 660 NLPLLRLWYWEKLRADNLDPTIDYT 684
>gb|EAL30007.1| GA20049-PA [Drosophila pseudoobscura]
Length = 3238
Score = 37.0 bits (84), Expect = 1.5
Identities = 28/89 (31%), Positives = 40/89 (44%), Gaps = 7/89 (7%)
Query: 385 RHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHE 444
R AA P+ P P P + E+ + R + W++R DE+ TH
Sbjct: 1633 RPPAAPLPASPTPTPTSNDELSDAGSDLSERCRSRWRSRRRRRRR----SHEPDEEHTHH 1688
Query: 445 NLMLKKELVKVRRELEE--KDELLMQDSK 471
L E+ ++ RELEE K+ELL SK
Sbjct: 1689 TQHLLNEM-EMARELEEERKNELLANASK 1716
>gb|AAW29406.1| P-209 [Borrelia turicatae]
Length = 562
Score = 37.0 bits (84), Expect = 1.5
Identities = 21/92 (22%), Positives = 43/92 (45%), Gaps = 1/92 (1%)
Query: 403 QEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEK 462
+E QEH++ + +E + +N ++ KD++ T EN M KK+ K + + +K
Sbjct: 340 KENTQEHMSKKDKENTQEHMSKKDNKNTQENMNKKDKENTQEN-MSKKDNTKTQEHMNKK 398
Query: 463 DELLMQDSKRARGRRNFYARYCGSDSESESED 494
D+ Q+ + + N D+ + E+
Sbjct: 399 DKENTQEHMNKKDKENTQEHMSKKDNTNTQEN 430
Score = 35.0 bits (79), Expect = 5.5
Identities = 21/87 (24%), Positives = 37/87 (42%), Gaps = 1/87 (1%)
Query: 407 QEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEKDELL 466
QEH++ + +E + EN + KD KT E+ M KK+ + + +KD+
Sbjct: 248 QEHMSKKDKENTQEHMNKKDKENTQENMSKKDNTKTQEH-MNKKDKENTQEHMSKKDKEN 306
Query: 467 MQDSKRARGRRNFYARYCGSDSESESE 493
Q+ + +N D E+ E
Sbjct: 307 TQEHMSKKDNKNTQEHMSKKDKENTQE 333
Score = 34.7 bits (78), Expect = 7.2
Identities = 22/91 (24%), Positives = 37/91 (40%), Gaps = 1/91 (1%)
Query: 403 QEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEEK 462
+E QEH+ + +E + EN + KD + T EN M KK+ + + +K
Sbjct: 328 KENTQEHMNKKDKENTQEHMSKKDKENTQEHMSKKDNKNTQEN-MNKKDKENTQENMSKK 386
Query: 463 DELLMQDSKRARGRRNFYARYCGSDSESESE 493
D Q+ + + N D E+ E
Sbjct: 387 DNTKTQEHMNKKDKENTQEHMNKKDKENTQE 417
>ref|NP_001018126.1| hypothetical protein LOC347273 [Homo sapiens]
Length = 364
Score = 36.6 bits (83), Expect = 1.9
Identities = 21/76 (27%), Positives = 35/76 (45%)
Query: 399 PPTTQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRE 458
PP +E+ ESR ++ K+ +N+ ++ QKT +NL K ++ R
Sbjct: 167 PPVDLSSDEEYYVEESRSARLRKSGKEHIDNIKKAFSKENMQKTRQNLDKKVNRIRTRIV 226
Query: 459 LEEKDELLMQDSKRAR 474
E+ E L Q +R R
Sbjct: 227 TPERRERLRQSGERLR 242
>ref|XP_471849.1| OSJNBa0054D14.3 [Oryza sativa (japonica cultivar-group)]
gi|38347668|emb|CAE05602.2| OSJNBa0054D14.3 [Oryza
sativa (japonica cultivar-group)]
Length = 328
Score = 36.6 bits (83), Expect = 1.9
Identities = 34/125 (27%), Positives = 52/125 (41%), Gaps = 17/125 (13%)
Query: 379 LPYPAMRHVAASAPSIPLPLPPTTQEMYQEHLAMESREKQMWKA-RYNEAENLIMTLDGK 437
+P R + A P P+P P + MYQ+ + WK R +A +LI
Sbjct: 106 IPRHPKRKMRAEIPPKPMPHSPYPKPMYQQ-FRRQGTTPHWWKGKRKRKASSLI------ 158
Query: 438 DEQKTHENLMLKKELVKVRRELEEKDELLMQDSKR-----ARGRRNFYARYCGSDSESES 492
T+ L L K +++ EE +EL + KR + G+ YA C D+ E
Sbjct: 159 ----TYAELDLSKMGIEIGDLPEEVEELQLWQRKRDGCCFSCGQFGHYAIGCTQDTNEEQ 214
Query: 493 EDHPT 497
E P+
Sbjct: 215 ETLPS 219
>ref|XP_392107.1| PREDICTED: similar to CG4832-PA, isoform A [Apis mellifera]
Length = 1399
Score = 36.6 bits (83), Expect = 1.9
Identities = 24/70 (34%), Positives = 35/70 (49%), Gaps = 1/70 (1%)
Query: 402 TQEMYQEHLAMESREKQMWKARYNEAENLIMTLDGKDEQKTHENLMLKKELVKVRRELEE 461
T + Y++ L +E K R E M + DE +N+ LK E+ +R+EL E
Sbjct: 79 TMKEYEDQLEALKKENFNLKLRIYFLEER-MGITSADENAIKKNIELKVEIESLRKELVE 137
Query: 462 KDELLMQDSK 471
K ELL Q +K
Sbjct: 138 KQELLSQAAK 147
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.136 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 893,010,367
Number of Sequences: 2540612
Number of extensions: 39979938
Number of successful extensions: 107211
Number of sequences better than 10.0: 59
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 107097
Number of HSP's gapped (non-prelim): 119
length of query: 501
length of database: 863,360,394
effective HSP length: 132
effective length of query: 369
effective length of database: 527,999,610
effective search space: 194831856090
effective search space used: 194831856090
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146333.5


