

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.5 + phase: 0
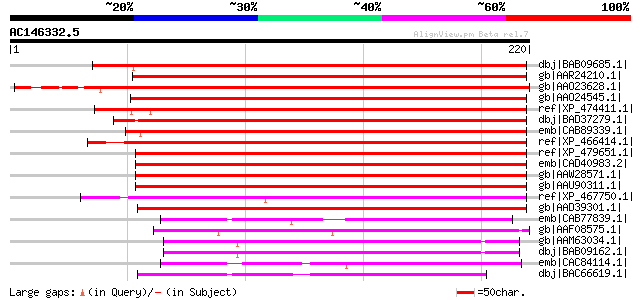
(220 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB09685.1| unnamed protein product [Arabidopsis thaliana] g... 210 2e-53
gb|AAR24210.1| At3g11550 [Arabidopsis thaliana] gi|46931254|gb|A... 205 8e-52
gb|AAO23628.1| At2g27370 [Arabidopsis thaliana] gi|5306261|gb|AA... 202 5e-51
gb|AAO24545.1| At2g36100 [Arabidopsis thaliana] gi|4678222|gb|AA... 195 9e-49
ref|XP_474411.1| OSJNBa0088H09.7 [Oryza sativa (japonica cultiva... 174 2e-42
dbj|BAD37279.1| integral membrane family protein-like [Oryza sat... 172 5e-42
emb|CAB89339.1| putative protein [Arabidopsis thaliana] gi|26449... 171 1e-41
ref|XP_466414.1| integral membrane-like protein [Oryza sativa (j... 164 2e-39
ref|XP_479651.1| hypothetical protein [Oryza sativa (japonica cu... 164 2e-39
emb|CAD40983.2| OSJNBa0072F16.8 [Oryza sativa (japonica cultivar... 163 4e-39
gb|AAW28571.1| hypothetical protein PGEC132D05.14 [Solanum demis... 162 8e-39
gb|AAU90311.1| hypothetical protein [Solanum demissum] 160 2e-38
ref|XP_467750.1| integral membrane family protein-like [Oryza sa... 151 1e-35
gb|AAD39301.1| Hypothetical protein [Arabidopsis thaliana] gi|30... 140 3e-32
emb|CAB77839.1| hypothetical protein [Arabidopsis thaliana] gi|4... 80 5e-14
gb|AAF08575.1| unknown protein [Arabidopsis thaliana] gi|2897363... 79 9e-14
gb|AAM63034.1| unknown [Arabidopsis thaliana] 77 3e-13
dbj|BAB09162.1| unnamed protein product [Arabidopsis thaliana] g... 77 3e-13
emb|CAC84114.1| hypothetical protein [Gossypium hirsutum] 77 3e-13
dbj|BAC66619.1| hypothetical protein [Oryza sativa (japonica cul... 77 3e-13
>dbj|BAB09685.1| unnamed protein product [Arabidopsis thaliana]
gi|15239955|ref|NP_196238.1| integral membrane family
protein [Arabidopsis thaliana]
Length = 202
Score = 210 bits (535), Expect = 2e-53
Identities = 109/191 (57%), Positives = 139/191 (72%), Gaps = 7/191 (3%)
Query: 36 PPHAATVVTTKATPLQ-------KGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQIL 88
P +++V+ KA L GG K+G++I DF+LRL AI AAL AA MGT+++ L
Sbjct: 11 PAESSSVIKGKAPLLGLARDHTGSGGYKRGLSIFDFLLRLAAIVAALAAAATMGTSDETL 70
Query: 89 PFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVD 148
PFFTQFLQF A +DD P F+FFVVA AG+L+LSLPFS+V IVRPLA PR LL+++D
Sbjct: 71 PFFTQFLQFEASYDDLPTFQFFVVAIAIVAGYLVLSLPFSVVTIVRPLAVAPRLLLLVLD 130
Query: 149 LVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLAC 208
+AL AAAS+AAA+VYLAHNG+ + NW ICQQF DFCQ +S AVV++F + FLA
Sbjct: 131 TAALALDTAAASAAAAIVYLAHNGNTNTNWLPICQQFGDFCQKTSGAVVSAFASVTFLAI 190
Query: 209 LVVVSSVALKR 219
LVV+S V+LKR
Sbjct: 191 LVVISGVSLKR 201
>gb|AAR24210.1| At3g11550 [Arabidopsis thaliana] gi|46931254|gb|AAT06431.1|
At3g11550 [Arabidopsis thaliana]
gi|12322903|gb|AAG51441.1| unknown protein; 80797-81587
[Arabidopsis thaliana] gi|15229791|ref|NP_187762.1|
integral membrane family protein [Arabidopsis thaliana]
Length = 204
Score = 205 bits (521), Expect = 8e-52
Identities = 102/167 (61%), Positives = 128/167 (76%)
Query: 53 GGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVV 112
GG +G+AI DF+LRL AI AAL AA MGT+++ LPFFTQFLQF A +DD P F+FFV+
Sbjct: 37 GGYNRGLAIFDFLLRLAAIVAALAAAATMGTSDETLPFFTQFLQFEASYDDLPTFQFFVI 96
Query: 113 ANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNG 172
A G+L+LSLP S+V I+RPLA PR LL+++D ++AL AAASSAAA+ YLAH+G
Sbjct: 97 AMALVGGYLVLSLPISVVTILRPLATAPRLLLLVLDTGVLALNTAAASSAAAISYLAHSG 156
Query: 173 SQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
+Q+ NW ICQQF DFCQ SS AVV++FV+ VF LVV+S VALKR
Sbjct: 157 NQNTNWLPICQQFGDFCQKSSGAVVSAFVSVVFFTILVVISGVALKR 203
>gb|AAO23628.1| At2g27370 [Arabidopsis thaliana] gi|5306261|gb|AAD41993.1| unknown
protein [Arabidopsis thaliana]
gi|20197657|gb|AAM15182.1| unknown protein [Arabidopsis
thaliana] gi|25407887|pir||B84672 hypothetical protein
At2g27370 [imported] - Arabidopsis thaliana
gi|15225873|ref|NP_180305.1| integral membrane family
protein [Arabidopsis thaliana]
Length = 221
Score = 202 bits (514), Expect = 5e-51
Identities = 111/220 (50%), Positives = 144/220 (65%), Gaps = 9/220 (4%)
Query: 3 SRREVEESSTAPILESKRTRSNGKGKSIDGDHSPP--HAATVVTTKATPLQKGGMKKGIA 60
SRRE EE PI++ + GKGK+ +PP + + K + G K+G+A
Sbjct: 8 SRREEEE----PIVQRPKL-DKGKGKA--HVFAPPMNYNRIMDKHKQEKMSPAGWKRGVA 60
Query: 61 ILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGF 120
I DF+LRL A A+ AA M T E+ LPFFTQFLQF A + D P FV+ N G+
Sbjct: 61 IFDFVLRLIAAITAMAAAAKMATTEETLPFFTQFLQFQADYTDLPTMSSFVIVNSIVGGY 120
Query: 121 LILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNA 180
L LSLPFSIVCI+RPLA PR L++ D V+M L + AAS++AA+VYLAHNG+ +NW
Sbjct: 121 LTLSLPFSIVCILRPLAVPPRLFLILCDTVMMGLTLMAASASAAIVYLAHNGNSSSNWLP 180
Query: 181 ICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
+CQQF DFCQG+S AVVASF+A+ L LV++S+ ALKRT
Sbjct: 181 VCQQFGDFCQGTSGAVVASFIAATLLMFLVILSAFALKRT 220
>gb|AAO24545.1| At2g36100 [Arabidopsis thaliana] gi|4678222|gb|AAD26967.1| unknown
protein [Arabidopsis thaliana] gi|25408448|pir||H84776
hypothetical protein At2g36100 [imported] - Arabidopsis
thaliana gi|15227576|ref|NP_181154.1| integral membrane
family protein [Arabidopsis thaliana]
Length = 206
Score = 195 bits (495), Expect = 9e-49
Identities = 93/168 (55%), Positives = 124/168 (73%)
Query: 52 KGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFV 111
+GG K+G+AI DF+LRL AI +GAA +M T E+ LPFFTQFLQF A +DD P F++FV
Sbjct: 38 RGGAKRGLAIFDFLLRLAAIAVTIGAASVMYTAEETLPFFTQFLQFQAGYDDLPAFQYFV 97
Query: 112 VANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHN 171
+A A +L+LSLPFSIV IVRP A PR +L+I D +++ L +AA++AA++ YLAHN
Sbjct: 98 IAVAVVASYLVLSLPFSIVSIVRPHAVAPRLILLICDTLVVTLNTSAAAAAASITYLAHN 157
Query: 172 GSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
G+Q NW ICQQF DFCQ S AVVA +A +F L+++S++ALKR
Sbjct: 158 GNQSTNWLPICQQFGDFCQNVSTAVVADSIAILFFIVLIIISAIALKR 205
>ref|XP_474411.1| OSJNBa0088H09.7 [Oryza sativa (japonica cultivar-group)]
gi|32488706|emb|CAE03449.1| OSJNBa0088H09.7 [Oryza
sativa (japonica cultivar-group)]
Length = 224
Score = 174 bits (440), Expect = 2e-42
Identities = 90/189 (47%), Positives = 130/189 (68%), Gaps = 6/189 (3%)
Query: 37 PHAATVVTTKATPL---QKGGMKKG---IAILDFILRLGAIGAALGAAVIMGTNEQILPF 90
P AA V + + ++G +G +A LDFILR+ A G AL AA+ GT+++ L
Sbjct: 35 PAAAPAVAPRKVGIPFFRRGDHHRGSRCLAFLDFILRIAAFGPALAAAISTGTSDETLSV 94
Query: 91 FTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLV 150
FT+F QF A++DDFP F FF+VAN AG+L+LSLPFS V ++RP G R LL++ D++
Sbjct: 95 FTEFYQFRARFDDFPAFLFFLVANAIVAGYLVLSLPFSAVLVIRPQTIGLRLLLLVCDMI 154
Query: 151 LMALVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLV 210
+ A++ AAAS+AAA+V LAHNG+ ANW AIC QF FCQ +S +VVASF+ V L LV
Sbjct: 155 MAAMLTAAASAAAAIVDLAHNGNLRANWVAICMQFHGFCQRTSGSVVASFLTVVILMFLV 214
Query: 211 VVSSVALKR 219
++++ ++++
Sbjct: 215 ILAACSIRK 223
>dbj|BAD37279.1| integral membrane family protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 184
Score = 172 bits (437), Expect = 5e-42
Identities = 89/175 (50%), Positives = 117/175 (66%), Gaps = 1/175 (0%)
Query: 45 TKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDF 104
T PL +G + KG++ILD ILR AI L +A+ MGT Q LPFFTQF++F AQ+ D
Sbjct: 10 TSKAPLSRG-VSKGVSILDVILRFVAIIGTLASAIAMGTTNQTLPFFTQFIRFKAQYSDL 68
Query: 105 PMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAA 164
P FFVVAN + +LILSLP SIV ++R A R +L+ D ++ALV A AS+AAA
Sbjct: 69 PTLTFFVVANSIVSAYLILSLPLSIVHVIRSRAKYSRLILIFFDAAMLALVTAGASAAAA 128
Query: 165 VVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
+VYLAH G+ ANW AICQQF FC+ S +++ SF A V L L+ +S++AL R
Sbjct: 129 IVYLAHKGNARANWLAICQQFDSFCERISGSLIGSFAAMVVLVLLIFLSAIALAR 183
>emb|CAB89339.1| putative protein [Arabidopsis thaliana] gi|26449778|dbj|BAC42012.1|
unknown protein [Arabidopsis thaliana]
gi|28372942|gb|AAO39953.1| At5g15290 [Arabidopsis
thaliana] gi|15242268|ref|NP_197033.1| integral membrane
family protein [Arabidopsis thaliana]
gi|11357888|pir||T49964 hypothetical protein F8M21.180 -
Arabidopsis thaliana
Length = 187
Score = 171 bits (434), Expect = 1e-41
Identities = 84/171 (49%), Positives = 121/171 (70%), Gaps = 1/171 (0%)
Query: 50 LQKGG-MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFK 108
+QK G M + IAIL+FILR+ A +G+A++MGT + LPFFTQF++F A+++D P
Sbjct: 15 IQKSGLMSRRIAILEFILRIVAFFNTIGSAILMGTTHETLPFFTQFIRFQAEYNDLPALT 74
Query: 109 FFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYL 168
FFVVAN +G+LILSL + V IV+ R LL+I+D+ ++ L+ + ASSAAA+VYL
Sbjct: 75 FFVVANAVVSGYLILSLTLAFVHIVKRKTQNTRILLIILDVAMLGLLTSGASSAAAIVYL 134
Query: 169 AHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
AHNG+ NW AICQQF FC+ S +++ SF+A V L L+++S++AL R
Sbjct: 135 AHNGNNKTNWFAICQQFNSFCERISGSLIGSFIAIVLLILLILLSAIALSR 185
>ref|XP_466414.1| integral membrane-like protein [Oryza sativa (japonica
cultivar-group)] gi|50253293|dbj|BAD29562.1| integral
membrane-like protein [Oryza sativa (japonica
cultivar-group)] gi|50725265|dbj|BAD34267.1| integral
membrane-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 201
Score = 164 bits (415), Expect = 2e-39
Identities = 82/186 (44%), Positives = 117/186 (62%), Gaps = 7/186 (3%)
Query: 34 HSPPHAATVVTTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQ 93
H PPH P++ G+ +++ D +LR AIG G+A+ M T + LPF
Sbjct: 21 HPPPHLPP-------PMRSSGVSLVLSVADLVLRFVAIGGTAGSAIAMATTSETLPFAAP 73
Query: 94 FLQFHAQWDDFPMFKFFVVANGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMA 153
F++F A++ D P FFVVA+ +L+LSLP S+V +VRP A R +L +D V++A
Sbjct: 74 FVRFRAEYSDLPTLMFFVVASSVVCAYLVLSLPASVVHVVRPGARSSRAILAFLDTVMLA 133
Query: 154 LVVAAASSAAAVVYLAHNGSQDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
L+ A+AS+AAA+VYLAH GS ANW ICQQFT FCQ + ++V SF A+V L LV +S
Sbjct: 134 LLTASASAAAAIVYLAHRGSARANWLGICQQFTSFCQRITASLVGSFAAAVVLVALVFLS 193
Query: 214 SVALKR 219
+++L R
Sbjct: 194 ALSLAR 199
>ref|XP_479651.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|38637294|dbj|BAD03557.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|50725687|dbj|BAD33153.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 237
Score = 164 bits (415), Expect = 2e-39
Identities = 78/166 (46%), Positives = 118/166 (70%)
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVA 113
G+++ + ++DF+LR+ A G L AA+ +GT+++ L FT + QF A++DDFP F+FF+VA
Sbjct: 65 GLRRCLGLIDFVLRVAAFGPTLAAAISIGTSDERLSVFTNYFQFRARFDDFPAFEFFIVA 124
Query: 114 NGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGS 173
N AAG+++LSLPFS I+ A G + LL+I D +++ L+ AAAS+AAA+VY+AH G+
Sbjct: 125 NAIAAGYMVLSLPFSAATIMSSKATGVKLLLLICDTIMVGLLTAAASAAAAMVYVAHEGN 184
Query: 174 QDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
ANW IC QF FCQ +S AV+ASF+A L L+V+++ + R
Sbjct: 185 LRANWVPICLQFHGFCQRTSGAVIASFLAVFVLMVLIVMAAFTMPR 230
>emb|CAD40983.2| OSJNBa0072F16.8 [Oryza sativa (japonica cultivar-group)]
gi|50924804|ref|XP_472751.1| OSJNBa0072F16.8 [Oryza
sativa (japonica cultivar-group)]
Length = 186
Score = 163 bits (412), Expect = 4e-39
Identities = 80/166 (48%), Positives = 110/166 (66%)
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVA 113
G+ + +A++D LR AI +G+A+ MGT + LPFFTQF+QF A++ D P F FFV A
Sbjct: 19 GVNRAVAVVDTFLRFIAIIGTIGSAIAMGTTNETLPFFTQFIQFEAKYSDLPSFTFFVAA 78
Query: 114 NGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGS 173
N +L+LS+P SIV I+RP A R LV D ++AL+ A AS+AAA+VYLAH G+
Sbjct: 79 NAVVCTYLVLSIPLSIVHILRPRARYSRLFLVFFDTAMLALLTAGASAAAAIVYLAHKGN 138
Query: 174 QDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
ANW +ICQQF FC+ S +++ SF A V L L+ +S+ AL R
Sbjct: 139 VRANWFSICQQFDSFCERISGSLIGSFAAMVLLVVLITLSAFALAR 184
>gb|AAW28571.1| hypothetical protein PGEC132D05.14 [Solanum demissum]
Length = 185
Score = 162 bits (409), Expect = 8e-39
Identities = 76/166 (45%), Positives = 113/166 (67%)
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVA 113
G+ +GI++ D +LR+ A+ L +AV MGT +Q L F TQ + F AQ+DD FKFFVV+
Sbjct: 19 GVNRGISVFDLVLRIVALVGTLASAVAMGTADQALSFSTQIVNFEAQYDDIDAFKFFVVS 78
Query: 114 NGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGS 173
N +L LS+P SI I+R A R LL+++D +++ + + AS+AAA+VYLAHNG+
Sbjct: 79 NSITCVYLALSIPISIFHIIRSRAGKSRVLLIVLDAIMLVFLTSGASAAAAIVYLAHNGN 138
Query: 174 QDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
NW +ICQQ+TDFCQ S+ +++ SF A + L+++SS+AL R
Sbjct: 139 TSTNWFSICQQYTDFCQRSAGSLIGSFGAMALMVLLIILSSIALSR 184
>gb|AAU90311.1| hypothetical protein [Solanum demissum]
Length = 185
Score = 160 bits (405), Expect = 2e-38
Identities = 76/166 (45%), Positives = 112/166 (66%)
Query: 54 GMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVA 113
G+ +GI++ D +LR+ A+ L +AV MGT Q L F TQ + F AQ+DD FKFFVV+
Sbjct: 19 GVNRGISVFDLVLRIVALVGTLASAVAMGTAGQALSFSTQIVNFEAQYDDIDAFKFFVVS 78
Query: 114 NGAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGS 173
N +L LS+P SI I+R A R LL+++D +++ + + AS+AAA+VYLAHNG+
Sbjct: 79 NSITCVYLALSIPISIFHIIRSRAGKSRVLLIVLDAIMLVFLTSGASAAAAIVYLAHNGN 138
Query: 174 QDANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
NW +ICQQ+TDFCQ S+ +++ SF A + L+++SS+AL R
Sbjct: 139 TSTNWFSICQQYTDFCQRSAGSLIGSFGAMALMVLLIILSSIALSR 184
>ref|XP_467750.1| integral membrane family protein-like [Oryza sativa (japonica
cultivar-group)] gi|46390633|dbj|BAD16116.1| integral
membrane family protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 230
Score = 151 bits (381), Expect = 1e-35
Identities = 86/218 (39%), Positives = 124/218 (56%), Gaps = 32/218 (14%)
Query: 31 DGDHSPPHAATVVTTKATPLQKGGMKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPF 90
DG+ S A TVV+ + + G + +++ D ILR+ A+ A + +A+ MGT Q LPF
Sbjct: 15 DGEASRTAARTVVSGSS---RGGAASRALSVADLILRVVAVVAIVDSAIAMGTTNQTLPF 71
Query: 91 FTQFLQFHAQWDDFPMF-----------------------------KFFVVANGAAAGFL 121
FTQFL+F AQ+ D P + FVVAN A +L
Sbjct: 72 FTQFLRFKAQYSDLPTLTCDHSCLLATIVNLHYFRLQARLFLPVVTRLFVVANSAVTAYL 131
Query: 122 ILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAI 181
+LS+P S+V I+R A+ R +L+ +D V++ALV A AS++AA+VYLAH G+ ANW A+
Sbjct: 132 VLSIPLSVVHIIRSRASYSRLVLIFLDSVMLALVAAVASASAAIVYLAHKGNVRANWFAV 191
Query: 182 CQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
CQQF FC+ S ++ SF A L LV++S+ AL R
Sbjct: 192 CQQFDSFCERISGPLIGSFAAMAVLLLLVLLSAAALAR 229
>gb|AAD39301.1| Hypothetical protein [Arabidopsis thaliana]
gi|30017253|gb|AAP12860.1| At1g14160 [Arabidopsis
thaliana] gi|26449701|dbj|BAC41974.1| unknown protein
[Arabidopsis thaliana] gi|15223101|ref|NP_172868.1|
integral membrane family protein [Arabidopsis thaliana]
gi|25513451|pir||B86275 F7A19.24 protein - Arabidopsis
thaliana
Length = 209
Score = 140 bits (352), Expect = 3e-32
Identities = 70/165 (42%), Positives = 105/165 (63%)
Query: 55 MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVAN 114
+ KGI++L F+LRL A+ +G+A+ MGT + + +Q + ++ D P FFVVAN
Sbjct: 44 INKGISVLGFVLRLFAVFGTIGSALAMGTTHESVVSLSQLVLLKVKYSDLPTLMFFVVAN 103
Query: 115 GAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQ 174
+ G+L+LSLP SI I A R +L++VD V++ALV + AS+A A VYLAH G+
Sbjct: 104 AISGGYLVLSLPVSIFHIFSTQAKTSRIILLVVDTVMLALVSSGASAATATVYLAHEGNT 163
Query: 175 DANWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKR 219
ANW ICQQF FC+ S +++ SF A + L +V+ S+++L R
Sbjct: 164 TANWPPICQQFDGFCERISGSLIGSFCAVILLMLIVINSAISLSR 208
>emb|CAB77839.1| hypothetical protein [Arabidopsis thaliana]
gi|4206208|gb|AAD11596.1| hypothetical protein
[Arabidopsis thaliana] gi|4263041|gb|AAD15310.1|
hypothetical protein [Arabidopsis thaliana]
gi|25407207|pir||H85044 hypothetical protein AT4g03540
[imported] - Arabidopsis thaliana
gi|15236329|ref|NP_192263.1| integral membrane family
protein [Arabidopsis thaliana]
Length = 164
Score = 79.7 bits (195), Expect = 5e-14
Identities = 50/151 (33%), Positives = 79/151 (52%), Gaps = 13/151 (8%)
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAA--GFLI 122
+LRL A GAAL A ++M T+ + F + A++ D FK+FV+AN + FL+
Sbjct: 12 VLRLAAFGAALAALIVMITSRERASFLA--ISLEAKYTDMAAFKYFVIANAVVSVYSFLV 69
Query: 123 LSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAIC 182
L LP + +V++DLV+ L+ ++ S+A AV + G+ +A W IC
Sbjct: 70 LFLPKESLLWK---------FVVVLDLVMTMLLTSSLSAALAVAQVGKKGNANAGWLPIC 120
Query: 183 QQFTDFCQGSSLAVVASFVASVFLACLVVVS 213
Q FC + A++A FVA V L++ S
Sbjct: 121 GQVPKFCDQITGALIAGFVALVLYVLLLLYS 151
>gb|AAF08575.1| unknown protein [Arabidopsis thaliana] gi|28973636|gb|AAO64140.1|
unknown protein [Arabidopsis thaliana]
gi|28393130|gb|AAO41998.1| unknown protein [Arabidopsis
thaliana] gi|15230708|ref|NP_187290.1| integral membrane
family protein [Arabidopsis thaliana]
Length = 199
Score = 79.0 bits (193), Expect = 9e-14
Identities = 49/164 (29%), Positives = 89/164 (53%), Gaps = 6/164 (3%)
Query: 62 LDFILRLGAIGAALGAAVIMGTNEQI----LPFFTQFLQFHAQWDDFPMFKFFVVANGAA 117
+D I R+ A L A ++M T++Q LP + A+++D P F +FVVA A
Sbjct: 37 IDIITRVLLFSATLTALIVMVTSDQTEMTQLPGVSSPAPVSAEFNDSPAFIYFVVALVVA 96
Query: 118 AGFLILSLPFSIVCIVRP-LAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDA 176
+ + ++S SI +++P A L +D+V++ ++ +A +A V Y+A G+++
Sbjct: 97 SFYALISTLVSISLLLKPEFTAQFSIYLASLDMVMLGILASATGTAGGVAYIALKGNEEV 156
Query: 177 NWNAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVALKRT 220
WN IC + FC+ + ++ S AS+ L L + S+++ KRT
Sbjct: 157 GWNKICNVYDKFCRYIATSLALSLFASLLLLVLSIWSALS-KRT 199
>gb|AAM63034.1| unknown [Arabidopsis thaliana]
Length = 197
Score = 77.4 bits (189), Expect = 3e-13
Identities = 48/158 (30%), Positives = 83/158 (52%), Gaps = 8/158 (5%)
Query: 66 LRLGAIGAALGAAVIMGTNEQILPFFTQFL-------QFHAQWDDFPMFKFFVVANGAAA 118
LRL A+ A L AA++MG N++ F + F A++D P F FFVVAN +
Sbjct: 22 LRLLALSATLSAAIVMGLNKETKTFVVGKVGNTPIQATFTAKFDHTPAFVFFVVANAMVS 81
Query: 119 GFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANW 178
+L + I + I+D++ + L+ AAA++AA + + NG++ A W
Sbjct: 82 FHNLLMIALQIFGGKMEFTGFRLLSVAILDMLNVTLISAAANAAAFMAEVGKNGNKHARW 141
Query: 179 NAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVA 216
+ IC +F +C + A++A+F A V L ++ +S++
Sbjct: 142 DKICDRFATYCDHGAGALIAAF-AGVILMLIISAASIS 178
>dbj|BAB09162.1| unnamed protein product [Arabidopsis thaliana]
gi|21386917|gb|AAM47862.1| unknown protein [Arabidopsis
thaliana] gi|17473557|gb|AAL38255.1| unknown protein
[Arabidopsis thaliana] gi|15241524|ref|NP_199268.1|
integral membrane family protein [Arabidopsis thaliana]
Length = 197
Score = 77.0 bits (188), Expect = 3e-13
Identities = 48/158 (30%), Positives = 82/158 (51%), Gaps = 8/158 (5%)
Query: 66 LRLGAIGAALGAAVIMGTNEQILPFFTQFL-------QFHAQWDDFPMFKFFVVANGAAA 118
LRL A A L AA++MG N++ F + F A++D P F FFVVAN +
Sbjct: 22 LRLLAFSATLSAAIVMGLNKETKTFIVGKVGNTPIQATFTAKFDHTPAFVFFVVANAMVS 81
Query: 119 GFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANW 178
+L + I + I+D++ + L+ AAA++AA + + NG++ A W
Sbjct: 82 FHNLLMIALQIFGGKMEFTGFRLLSVAILDMLNVTLISAAANAAAFMAEVGKNGNKHARW 141
Query: 179 NAICQQFTDFCQGSSLAVVASFVASVFLACLVVVSSVA 216
+ IC +F +C + A++A+F A V L ++ +S++
Sbjct: 142 DKICDRFATYCDHGAGALIAAF-AGVILMLIISAASIS 178
>emb|CAC84114.1| hypothetical protein [Gossypium hirsutum]
Length = 207
Score = 77.0 bits (188), Expect = 3e-13
Identities = 50/154 (32%), Positives = 81/154 (52%), Gaps = 10/154 (6%)
Query: 65 ILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVANGAAAGFLILS 124
+LRL + + A V+M N Q F + + D F++ V ANG AG+ +LS
Sbjct: 41 MLRLVPMALGVAALVVMLKNSQSNDFGS------VSYSDLGAFRYLVHANGICAGYSLLS 94
Query: 125 LPFSIVCIVRPLAAGPR-FLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQDANWNAICQ 183
+I+ V + PR + ++D +L +++ AA+ + V+YLA+ G W+A C
Sbjct: 95 ---AIIAAVPSPSTMPRAWTFFLLDQILTYVILGAAAVSTEVLYLANKGDSAITWSAACG 151
Query: 184 QFTDFCQGSSLAVVASFVASVFLACLVVVSSVAL 217
F FC +++AVV +FVA + A L +VSS L
Sbjct: 152 TFAGFCHKATIAVVITFVAVICYAVLSLVSSYRL 185
>dbj|BAC66619.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 162
Score = 77.0 bits (188), Expect = 3e-13
Identities = 49/148 (33%), Positives = 76/148 (51%), Gaps = 9/148 (6%)
Query: 55 MKKGIAILDFILRLGAIGAALGAAVIMGTNEQILPFFTQFLQFHAQWDDFPMFKFFVVAN 114
M K +++ +LRL A AA AAV+M T+ + FF +Q A++ P F FFVVA
Sbjct: 1 MAKVHRLMNAVLRLAAAAAAATAAVVMVTSRETTSFFG--IQMEAKYSYTPSFIFFVVAY 58
Query: 115 GAAAGFLILSLPFSIVCIVRPLAAGPRFLLVIVDLVLMALVVAAASSAAAVVYLAHNGSQ 174
AA + S++ + P + L + D+VL ++ A +SA A+ +A NG+
Sbjct: 59 AVAAAY-------SLLVLAVPAGSALSRLALTTDVVLGMVLAGAVASAGAISDIAKNGNS 111
Query: 175 DANWNAICQQFTDFCQGSSLAVVASFVA 202
A W +C Q +C A++A FVA
Sbjct: 112 HAGWLPVCGQIHAYCNHVMAALIAGFVA 139
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.325 0.136 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 337,987,891
Number of Sequences: 2540612
Number of extensions: 12827869
Number of successful extensions: 46982
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 46885
Number of HSP's gapped (non-prelim): 71
length of query: 220
length of database: 863,360,394
effective HSP length: 123
effective length of query: 97
effective length of database: 550,865,118
effective search space: 53433916446
effective search space used: 53433916446
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146332.5


