

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.11 - phase: 0
(366 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
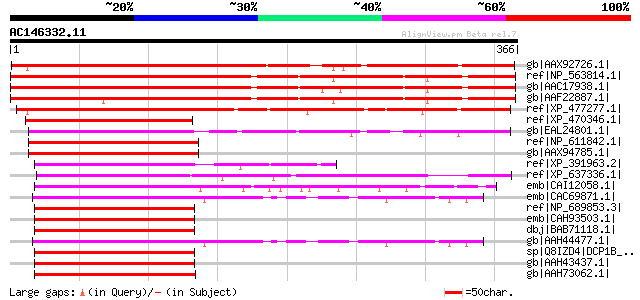
Score E
Sequences producing significant alignments: (bits) Value
gb|AAX92726.1| Dcp1-like decapping family, putative [Oryza sativ... 452 e-126
ref|NP_563814.1| hydroxyproline-rich glycoprotein family protein... 451 e-125
gb|AAC17938.1| similar to yeast dcp1 [Arabidopsis thaliana] 447 e-124
gb|AAF22887.1| T27G7.7 [Arabidopsis thaliana] 435 e-121
ref|XP_477277.1| putative transcription factor [Oryza sativa (ja... 365 1e-99
ref|XP_470346.1| hypothetical protein [Oryza sativa (japonica cu... 181 3e-44
gb|EAL24801.1| GA10823-PA [Drosophila pseudoobscura] 122 2e-26
ref|NP_611842.1| CG11183-PA [Drosophila melanogaster] gi|7291667... 115 3e-24
gb|AAX94785.1| GH04763p [Drosophila melanogaster] 115 3e-24
ref|XP_391963.2| PREDICTED: similar to GA10823-PA [Apis mellifera] 115 3e-24
ref|XP_637336.1| hypothetical protein DDB0187387 [Dictyostelium ... 114 5e-24
emb|CAI12058.1| MAD homolog 4 interacting transcription coactiva... 111 4e-23
emb|CAC69871.1| transcription factor [Danio rerio] 109 2e-22
ref|NP_689853.3| decapping enzyme Dcp1b [Homo sapiens] 107 7e-22
emb|CAH93503.1| hypothetical protein [Pongo pygmaeus] gi|6038982... 107 7e-22
dbj|BAB71118.1| unnamed protein product [Homo sapiens] 107 7e-22
gb|AAH44477.1| Decapping enzyme [Danio rerio] gi|47271431|ref|NP... 107 7e-22
sp|Q8IZD4|DCP1B_HUMAN mRNA decapping enzyme 1B gi|24756831|gb|AA... 107 7e-22
gb|AAH43437.1| Decapping enzyme Dcp1b [Homo sapiens] 107 7e-22
gb|AAH73062.1| LOC443617 protein [Xenopus laevis] 105 2e-21
>gb|AAX92726.1| Dcp1-like decapping family, putative [Oryza sativa (japonica
cultivar-group)]
Length = 380
Score = 452 bits (1163), Expect = e-126
Identities = 242/375 (64%), Positives = 288/375 (76%), Gaps = 26/375 (6%)
Query: 2 SQNGKLMPNL--DQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVE 59
+ GK+ PNL D + T+LLNLTVLQR+DP VE+ILITAAHVT Y+FNIDL+QWSRKDVE
Sbjct: 19 ANGGKVTPNLTMDAEGTRLLNLTVLQRLDPAVEDILITAAHVTLYDFNIDLNQWSRKDVE 78
Query: 60 GSLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYN 119
GSLFVVKRNTQPRFQFIVMNRRNT+NLVE+LL DFEYE+Q PYLLYRNAAQEVNGIWFYN
Sbjct: 79 GSLFVVKRNTQPRFQFIVMNRRNTDNLVEDLLSDFEYELQPPYLLYRNAAQEVNGIWFYN 138
Query: 120 ARECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNV 179
+CE VA+LF RILNAYAKVPPK KV STKSEFEELEAVPT A +DGPLEPS + T+ V
Sbjct: 139 QHDCEAVASLFGRILNAYAKVPPKPKVPSTKSEFEELEAVPTSAAIDGPLEPSPATTTIV 198
Query: 180 ADVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPL------P 233
+D PD+ S +N+FS+A IG+ SNAP+ G+ + SS ++ ATP VPL P
Sbjct: 199 SDAPDE-SLVNYFSSAASIGSVSNAPMAGRAHPSSESV--------ATPHVPLIIPSATP 249
Query: 234 TLQIPS--LPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPT 291
T QIP + +S P P HD ++++ NLV P+FF PP SS+ M+PP SS +PT
Sbjct: 250 THQIPPPLVGSSAPPLPLHDTNV---HTARSANLVTPAFFTPPSSSSTSMVPPASSMMPT 306
Query: 292 APPLHPT-GTVQR-PYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQD 349
APPLHPT + QR YGTP+LQPFPPPTPPPSL P S P ISR+KV++ALL LVQ+
Sbjct: 307 APPLHPTSASAQRATYGTPLLQPFPPPTPPPSLTP--SYNEGPIISRDKVKEALLRLVQN 364
Query: 350 NQFIDMVYKALLNAH 364
+QFID+VY+ L NAH
Sbjct: 365 DQFIDLVYRELQNAH 379
>ref|NP_563814.1| hydroxyproline-rich glycoprotein family protein [Arabidopsis
thaliana] gi|67460429|sp|Q9SJF3|DCP1_ARATH mRNA
decapping enzyme-like protein (DCP1 homolog)
Length = 367
Score = 451 bits (1159), Expect = e-125
Identities = 244/375 (65%), Positives = 287/375 (76%), Gaps = 19/375 (5%)
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQNGK++PNLDQ ST+LLNLTVLQRIDP++EEILITAAHVTFYEFNI+LSQWSRKDVEG
Sbjct: 1 MSQNGKIIPNLDQNSTRLLNLTVLQRIDPYIEEILITAAHVTFYEFNIELSQWSRKDVEG 60
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 120
SLFVVKR+TQPRFQFIVMNRRNT+NLVENLLGDFEYE+Q PYLLYRNA+QEVNGIWFYN
Sbjct: 61 SLFVVKRSTQPRFQFIVMNRRNTDNLVENLLGDFEYEVQGPYLLYRNASQEVNGIWFYNK 120
Query: 121 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 180
RECEEVA LF+RIL+AY+KV K K SS+KSEFEELEA PTMAVMDGPLEPSS+
Sbjct: 121 RECEEVATLFNRILSAYSKVNQKPKASSSKSEFEELEAKPTMAVMDGPLEPSST----AR 176
Query: 181 DVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPL-----PTL 235
D PDDP+F+NFFS+ M +GNT++ +G PYQSSA H T P+ P +
Sbjct: 177 DAPDDPAFVNFFSSTMNLGNTASGSASG-PYQSSAIPHQPHQPHQPTIAPPVAAAAPPQI 235
Query: 236 QI-PSLPTSTPFTPQHD-APESINSSSQA-TNLVKPSFFVPPLSSAAMMMPPVSSSIPTA 292
Q P L +S+P D PE I+S+S T+LV PSFF PP A + P S+PTA
Sbjct: 236 QSPPPLQSSSPLMTLFDNNPEVISSNSNIHTDLVTPSFFGPPRMMAQPHLIP-GVSMPTA 294
Query: 293 PPLHPTGT--VQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLVQDN 350
PPL+P QR YGTP+LQPFPPPTPPPSLAP + P ISR+KV++ALL L+Q++
Sbjct: 295 PPLNPNNASHQQRSYGTPVLQPFPPPTPPPSLAPAPT---GPVISRDKVKEALLSLLQED 351
Query: 351 QFIDMVYKALLNAHQ 365
+FID + + L NA Q
Sbjct: 352 EFIDKITRTLQNALQ 366
>gb|AAC17938.1| similar to yeast dcp1 [Arabidopsis thaliana]
Length = 370
Score = 447 bits (1150), Expect = e-124
Identities = 244/378 (64%), Positives = 286/378 (75%), Gaps = 22/378 (5%)
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQNGK++PNLDQ ST+LLNLTVLQRIDP++EEILITAAHVTFYEFNI+LSQWSRKDVEG
Sbjct: 1 MSQNGKIIPNLDQNSTRLLNLTVLQRIDPYIEEILITAAHVTFYEFNIELSQWSRKDVEG 60
Query: 61 SLFVVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNA 120
SLFVVKR+TQPRFQFIVMNRRNT+NLVENLLGDFEYE+Q PYLLYRNA+QEVNGIWFYN
Sbjct: 61 SLFVVKRSTQPRFQFIVMNRRNTDNLVENLLGDFEYEVQGPYLLYRNASQEVNGIWFYNK 120
Query: 121 RECEEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVA 180
RECEEVA LF+RIL+AY+KV K K SS+KSEFEELEA PTMAVMDGPLEPSS+
Sbjct: 121 RECEEVATLFNRILSAYSKVNQKPKASSSKSEFEELEAKPTMAVMDGPLEPSST----AR 176
Query: 181 DVPDDPSFINFFSAAMGIGNTSNAPITGQPYQSSATISSSGPTH------AATPVVPLPT 234
D PDDP+F+NFFS+ M +GNT++ +G PYQSSA H A PV
Sbjct: 177 DAPDDPAFVNFFSSTMNLGNTASGSASG-PYQSSAIPHQPHQPHQPHQPTIAPPVAAAAP 235
Query: 235 LQI---PSLPTSTPFTPQHD-APESINSSSQA-TNLVKPSFFVPPLSSAAMMMPPVSSSI 289
QI P L +S+P D PE I+S+S T+LV PSFF PP A + P S+
Sbjct: 236 PQIQSPPPLQSSSPLMTLFDNNPEVISSNSNIHTDLVTPSFFGPPRMMAQPHLIP-GVSM 294
Query: 290 PTAPPLHPTGT--VQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDALLVLV 347
P+APPL+P QR YGTP+LQPFPPPTPPPSLAP + P ISR+KV++ALL L+
Sbjct: 295 PSAPPLNPNNASHQQRSYGTPVLQPFPPPTPPPSLAPAPT---GPVISRDKVKEALLSLL 351
Query: 348 QDNQFIDMVYKALLNAHQ 365
Q+++FID + + L NA Q
Sbjct: 352 QEDEFIDKITRTLQNALQ 369
>gb|AAF22887.1| T27G7.7 [Arabidopsis thaliana]
Length = 396
Score = 435 bits (1119), Expect = e-121
Identities = 244/404 (60%), Positives = 287/404 (70%), Gaps = 48/404 (11%)
Query: 1 MSQNGKLMPNLDQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEG 60
MSQNGK++PNLDQ ST+LLNLTVLQRIDP++EEILITAAHVTFYEFNI+LSQWSRKDVEG
Sbjct: 1 MSQNGKIIPNLDQNSTRLLNLTVLQRIDPYIEEILITAAHVTFYEFNIELSQWSRKDVEG 60
Query: 61 SLFVVK-----------------------------RNTQPRFQFIVMNRRNTENLVENLL 91
SLFVVK R+TQPRFQFIVMNRRNT+NLVENLL
Sbjct: 61 SLFVVKRLLLSIFNYVYLIFNRLLKSDWILFFVSFRSTQPRFQFIVMNRRNTDNLVENLL 120
Query: 92 GDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNAYAKVPPKLKVSSTKS 151
GDFEYE+Q PYLLYRNA+QEVNGIWFYN RECEEVA LF+RIL+AY+KV K K SS+KS
Sbjct: 121 GDFEYEVQGPYLLYRNASQEVNGIWFYNKRECEEVATLFNRILSAYSKVNQKPKASSSKS 180
Query: 152 EFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPITGQPY 211
EFEELEA PTMAVMDGPLEPSS+ D PDDP+F+NFFS+ M +GNT++ +G PY
Sbjct: 181 EFEELEAKPTMAVMDGPLEPSST----ARDAPDDPAFVNFFSSTMNLGNTASGSASG-PY 235
Query: 212 QSSATISSSGPTHAATPVVPL-----PTLQI-PSLPTSTPFTPQHD-APESINSSSQA-T 263
QSSA H T P+ P +Q P L +S+P D PE I+S+S T
Sbjct: 236 QSSAIPHQPHQPHQPTIAPPVAAAAPPQIQSPPPLQSSSPLMTLFDNNPEVISSNSNIHT 295
Query: 264 NLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLHPTGT--VQRPYGTPMLQPFPPPTPPPS 321
+LV PSFF PP A + P S+PTAPPL+P QR YGTP+LQPFPPPTPPPS
Sbjct: 296 DLVTPSFFGPPRMMAQPHLIP-GVSMPTAPPLNPNNASHQQRSYGTPVLQPFPPPTPPPS 354
Query: 322 LAPVSSPLPNPAISREKVRDALLVLVQDNQFIDMVYKALLNAHQ 365
LAP + P ISR+KV++ALL L+Q+++FID + + L NA Q
Sbjct: 355 LAPAPT---GPVISRDKVKEALLSLLQEDEFIDKITRTLQNALQ 395
>ref|XP_477277.1| putative transcription factor [Oryza sativa (japonica
cultivar-group)] gi|51963346|ref|XP_506243.1| PREDICTED
P0021G06.113 gene product [Oryza sativa (japonica
cultivar-group)] gi|33146979|dbj|BAC80052.1| putative
transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 382
Score = 365 bits (937), Expect = 1e-99
Identities = 201/379 (53%), Positives = 261/379 (68%), Gaps = 31/379 (8%)
Query: 6 KLMPNL--DQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLF 63
K+ PNL D++ T++LN+TVLQR+DP VE+ILITA HVT Y+F+ +L+QWSRKDVEGSLF
Sbjct: 10 KVTPNLAMDEEGTRVLNITVLQRLDPAVEDILITAGHVTLYDFDTNLNQWSRKDVEGSLF 69
Query: 64 VVKRNTQPRFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNAREC 123
VVKRN QPRFQF+VMNRRNT+NLVE+LLGDFEY++QVPY++YRNAAQEV GIWFYN++EC
Sbjct: 70 VVKRNAQPRFQFVVMNRRNTDNLVEDLLGDFEYQLQVPYIMYRNAAQEVIGIWFYNSQEC 129
Query: 124 EEVANLFSRILNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEP-SSSITSNVADV 182
EEVANLFSRILNA++K PK K S KSEFEELEA PT+ ++GPLEP +S+I V
Sbjct: 130 EEVANLFSRILNAFSKATPKPKAPSIKSEFEELEAAPTL--VEGPLEPQTSNIIPATTHV 187
Query: 183 PDDPSFINFFSAAMGIGNTSNAPITGQPYQS--SATISSSGPTHAATPVVPLPTLQIPSL 240
+DP FFS A+ +G+ S + GQ QS S +SS PT + P +PS
Sbjct: 188 QEDP-LSAFFSGAINVGSASGLSVAGQLNQSFGSTPLSSHAPTSISISQPPAVHHLLPSQ 246
Query: 241 PTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPLH---- 296
+S H ++ +++ +L+ PS F PL+S+ M + PTAPP H
Sbjct: 247 TSSVISPDVHGGTGAV--VNRSASLLNPSLF-SPLTSSQTTMARTNPVAPTAPPQHPRIT 303
Query: 297 --------------PTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDA 342
PT + PYGTP+LQPFPPP P PSLA S+P+ +P +SREKVRDA
Sbjct: 304 QQPHSAPLLQPFPLPTASPSPPYGTPLLQPFPPPNPSPSLA--SAPVYSPVLSREKVRDA 361
Query: 343 LLVLVQDNQFIDMVYKALL 361
LL LV+++ FID+VY+ ++
Sbjct: 362 LLRLVENDDFIDLVYREIV 380
>ref|XP_470346.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|28376714|gb|AAO41144.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 143
Score = 181 bits (459), Expect = 3e-44
Identities = 83/121 (68%), Positives = 102/121 (83%)
Query: 12 DQQSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQP 71
DQ+ T+ LNLTVL+R+DP V +ILI AAHV Y F+ ++ QWSR+ VEGSLFVVKRNTQP
Sbjct: 23 DQEGTRTLNLTVLRRLDPAVADILIIAAHVVLYSFDDNIHQWSRRPVEGSLFVVKRNTQP 82
Query: 72 RFQFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFS 131
RFQFIVMNR+NTENL E+LLG FEY++QVPY++Y NAA E+ GIWFY+ +ECE+V LFS
Sbjct: 83 RFQFIVMNRKNTENLTEDLLGGFEYQVQVPYIMYHNAADEITGIWFYDPQECEQVGYLFS 142
Query: 132 R 132
R
Sbjct: 143 R 143
>gb|EAL24801.1| GA10823-PA [Drosophila pseudoobscura]
Length = 336
Score = 122 bits (306), Expect = 2e-26
Identities = 95/359 (26%), Positives = 163/359 (44%), Gaps = 37/359 (10%)
Query: 14 QSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRF 73
+S +NL+ +++IDP+ +EI+ +++HV FY FN + ++W + DVEG+ F+ RN +P
Sbjct: 4 ESITRMNLSAIKKIDPYAKEIVDSSSHVAFYTFNSEQNEWEKTDVEGAFFIYHRNAEPFH 63
Query: 74 QFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
+ NR NT + VE + G E + Q P+LLYRN + G WFYN+ EC+ + + + +
Sbjct: 64 SIFINNRLNTTSFVEPITGSLELQSQPPFLLYRNERSRIRGFWFYNSEECDRMNGVVNSL 123
Query: 134 LNAYAKVPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDD-PSFINFF 192
L+ + A PT + P ++SI + ++ + + N
Sbjct: 124 ----------LQNKESNGLVANPNAAPTFVI---PKHDNASIFNMLSKAQKEYNAQCNNQ 170
Query: 193 SAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLP-TSTP---FTP 248
S +M N + + + ++S+ ++ T V PL Q+ +TP P
Sbjct: 171 SKSMQPENVTPGNVL-KFFESAKQATAESGTVQYNRVQPLSVDQLEKQQRAATPNEGLIP 229
Query: 249 QHDAPESINSSSQATNLVKPSFFVPPLSSAAMMMPPVSSSIPTAPPL---HPTGTVQRPY 305
+AP S + + KPS PL P+ S + PL G R
Sbjct: 230 TREAP----SECRLSPFQKPSLNAQPL--------PLLSELKIGSPLMANEDAGNCLRRL 277
Query: 306 GTPMLQPFPPPTPPPSL--APVSSPLPNPAISREKVRDALLVLVQ-DNQFIDMVYKALL 361
P PPS+ AP +P P +S + A L+Q D +F++ +++A L
Sbjct: 278 LVGDQPEAKPALMPPSMFDAPNGNPEPVQPLSSSRFVQAFTYLIQNDKEFVNKLHRAYL 336
>ref|NP_611842.1| CG11183-PA [Drosophila melanogaster] gi|7291667|gb|AAF47089.1|
CG11183-PA [Drosophila melanogaster]
Length = 372
Score = 115 bits (287), Expect = 3e-24
Identities = 47/123 (38%), Positives = 77/123 (62%)
Query: 14 QSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRF 73
+S +NL +++IDP+ +EI+ +++HV FY FN ++W + DVEG+ F+ RN +P
Sbjct: 4 ESITRMNLAAIKKIDPYAKEIVDSSSHVAFYTFNSSQNEWEKTDVEGAFFIYHRNAEPFH 63
Query: 74 QFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
+ NR NT + VE + G E + Q P+LLYRN + G WFYN+ EC+ ++ L + +
Sbjct: 64 SIFINNRLNTTSFVEPITGSLELQSQPPFLLYRNERSRIRGFWFYNSEECDRISGLVNGL 123
Query: 134 LNA 136
L +
Sbjct: 124 LKS 126
>gb|AAX94785.1| GH04763p [Drosophila melanogaster]
Length = 387
Score = 115 bits (287), Expect = 3e-24
Identities = 47/123 (38%), Positives = 77/123 (62%)
Query: 14 QSTKLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRF 73
+S +NL +++IDP+ +EI+ +++HV FY FN ++W + DVEG+ F+ RN +P
Sbjct: 19 ESITRMNLAAIKKIDPYAKEIVDSSSHVAFYTFNSSQNEWEKTDVEGAFFIYHRNAEPFH 78
Query: 74 QFIVMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
+ NR NT + VE + G E + Q P+LLYRN + G WFYN+ EC+ ++ L + +
Sbjct: 79 SIFINNRLNTTSFVEPITGSLELQSQPPFLLYRNERSRIRGFWFYNSEECDRISGLVNGL 138
Query: 134 LNA 136
L +
Sbjct: 139 LKS 141
>ref|XP_391963.2| PREDICTED: similar to GA10823-PA [Apis mellifera]
Length = 496
Score = 115 bits (287), Expect = 3e-24
Identities = 70/220 (31%), Positives = 112/220 (50%), Gaps = 18/220 (8%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
+N+ L+R+DP+V++IL TA HV Y FN ++W + ++EG+LFV RN +P ++M
Sbjct: 42 MNVAALKRVDPYVKDILETATHVALYTFNAINNEWEKTNIEGALFVYSRNGEPYNSVLIM 101
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNAYA 138
NR NT NLVE + + ++Q P+LLYRN+ + GIWFY+ EC + + ++++
Sbjct: 102 NRLNTNNLVEPVTQGLDLQLQEPFLLYRNSRCNIYGIWFYDKEECVRIGAMLNKLI---- 157
Query: 139 KVPPKLKVSSTKSEFEELEAVPTMAVM--DGPLEPSSSITSNVADVPDDPSFINFFSAAM 196
E + PT+ V GP + I S ++ +D + N +++
Sbjct: 158 ---------KESEENRKTYNKPTVNVKKDSGPNVNNVDIFSMLSKAQEDFN-TNRNNSSS 207
Query: 197 GIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQ 236
G G T P + A SGP A P+ P T Q
Sbjct: 208 GSGGIKEVLNTKSPLSTPAADDVSGP--LAAPLGPDVTSQ 245
>ref|XP_637336.1| hypothetical protein DDB0187387 [Dictyostelium discoideum]
gi|60465746|gb|EAL63823.1| hypothetical protein
DDB0187387 [Dictyostelium discoideum]
Length = 358
Score = 114 bits (285), Expect = 5e-24
Identities = 90/380 (23%), Positives = 156/380 (40%), Gaps = 69/380 (18%)
Query: 20 NLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVMN 79
NL+ LQR+D + +L T+ HVT Y+F+ + QW RKD+EGSLF+V P + IVMN
Sbjct: 10 NLSALQRLDSKICGVLGTSTHVTAYKFDESVKQWFRKDIEGSLFIVNTTEYPYCKLIVMN 69
Query: 80 RRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNAYAK 139
R +T+NLVE + IQ YL+Y+N + E+NGIWFY + E++ NL + L
Sbjct: 70 RLSTKNLVEEINEKIVIRIQGQYLIYKNKSGEINGIWFYEVTDQEKIYNLL-KDLQRNPP 128
Query: 140 VPPKLKVSSTKSE--------------------FEELEAVPTMAVMDGPLEPSSSITSNV 179
PP+++ + ++++ P + + P
Sbjct: 129 QPPQIQQQPQPQQPQQQQQQQQQQQPPQPQQQPPQQMQQQPPGTMSPQGMIPPPGFIIPQ 188
Query: 180 ADVPDDPSFI-----------------NFFSAAMGIGNTSNAPITGQPYQSSATISSSGP 222
+P P F+ N+ + + N +N SS +S+ P
Sbjct: 189 GGIPPHPQFMPQSHVYPYMMQPQQIQPNYNNNSSNNNNNNN---NSSNNNSSNNNNSNPP 245
Query: 223 THAATPVVPLPTLQIPSLPTSTPFTPQHDAPESINSSSQATNLVKPSFFVPPLSSAAMMM 282
T + P + + Q + +IN+++ + N+ S P ++ +
Sbjct: 246 TPNKQNINDTPITPNTIINQIQQNSTQSNNSNNINNNNNSNNINNNSKTTPYSTTPQQLS 305
Query: 283 PPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPNPAISREKVRDA 342
P + +P L + V+ I+++++R+
Sbjct: 306 PTIKKEVPLLSVLMNSVVVK----------------------------ENTITKDQIRET 337
Query: 343 LLVLVQDNQFIDMVYKALLN 362
L LV D FID+VY + +N
Sbjct: 338 LKRLVNDENFIDLVYNSYIN 357
>emb|CAI12058.1| MAD homolog 4 interacting transcription coactivator 1 [Danio rerio]
Length = 496
Score = 111 bits (277), Expect = 4e-23
Identities = 98/387 (25%), Positives = 173/387 (44%), Gaps = 65/387 (16%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L+ L+R+DP++ I A+ V Y FN + ++W + +VEG+LFV R PR F ++
Sbjct: 14 MSLSALKRLDPYISSITDLASQVALYTFNNNTNEWEKTNVEGTLFVYSRLASPRHGFTIL 73
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNA-- 136
NR + +NL E + D ++++Q P+LLYRNA + GIWFY+ +C+ +A L + + A
Sbjct: 74 NRLSMDNLTEPITKDLDFQLQHPFLLYRNARLSIYGIWFYDKEDCQRIAELMKKYVLAGQ 133
Query: 137 -----------------------YAKVPPKLKVSSTKSEFEELEAVPTMAVMDG------ 167
++ K + K + E + + + ++ G
Sbjct: 134 EQQLQAQIATGCLSPRSAEQGVDIMQMLTKARNDYDKGKLSEPKEIGSGGIIYGQTPHLI 193
Query: 168 ---PLEPSSSITSNVADVPDDP---SFINFFSA--AMGIGNTSNAPITGQ--PYQSSA-- 215
PL+P+ +++ D D S + F A +G+T + G+ P +SS
Sbjct: 194 KPIPLKPAQD--THIQDAEMDSKHLSVVTLFGAQPKPDLGSTQPSLSVGRMGPARSSVAR 251
Query: 216 TISSSGPTHAATPVVPLPTLQ----IPSLPTSTPFTPQHDAPESINSSSQATNL----VK 267
++S P+ A VV +Q I L + + P ++PES + A + ++
Sbjct: 252 SLSYDDPSPAGGAVVSSTPMQHCPAIHKLMSGSLLQPLSESPESRLCENGAVQIQPDPIQ 311
Query: 268 PSFFVPPLSSAAMMMPPV---SSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAP 324
PP + +MP + + P +H ++ +P G Q P PS A
Sbjct: 312 KLLMNPPPVAGTTVMPSIQLKQGEVGMGPQIH---SMAKPSGPFPAQHLFVPNKVPS-AG 367
Query: 325 VSSPLPNPAISREKVRDALLVLVQDNQ 351
V+S + +P +K L LVQ Q
Sbjct: 368 VTSNVVSPHELLQK-----LTLVQQEQ 389
>emb|CAC69871.1| transcription factor [Danio rerio]
Length = 439
Score = 109 bits (272), Expect = 2e-22
Identities = 89/336 (26%), Positives = 146/336 (42%), Gaps = 37/336 (11%)
Query: 17 KLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFI 76
+L++L LQ+ DP++ ++L V Y FN ++W + ++EG+LFV R+ P F
Sbjct: 9 QLMSLAALQQHDPYIVKLLDVTGQVALYTFNPKANEWEKNEIEGTLFVYARSASPHHGFT 68
Query: 77 VMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNA 136
+MNR +TENLVE + D E+++Q P+LLYRN + IWFY+ +C+ +A L +I+
Sbjct: 69 IMNRLSTENLVEPINKDLEFQLQDPFLLYRNGNLGIYSIWFYDKADCQRIAQLMLQIVKQ 128
Query: 137 YAK----VPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFF 192
+ V P VS + + + ++ E S DVP +N
Sbjct: 129 ESLRAQCVSPDRSVSIWTNGCIQNRPAGILELLSKAKEEYQRSRSTERDVP-----VNCE 183
Query: 193 SAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDA 252
S +T ++ S +H+ + + L SLP P
Sbjct: 184 S------QEKREDVTAAQHEKS--------SHSVVKQITVEELFGSSLPKDPP------P 223
Query: 253 PESINSSSQATNLVKPSF--FVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPML 310
SI+ + L F + P S+ PP+ S + P P G+
Sbjct: 224 LSSISVVGEGLGLRGIPFNPALAPRLSSDPGGPPLLSLLQPRDPRTSAEETSAPRGS--T 281
Query: 311 QPFPPP--TPPPSLAPVSSP--LPNPAISREKVRDA 342
PFPP + PVS P +P+P ++ + RD+
Sbjct: 282 SPFPPAFISADAHGMPVSLPGFMPSPLVTPQSFRDS 317
>ref|NP_689853.3| decapping enzyme Dcp1b [Homo sapiens]
Length = 617
Score = 107 bits (266), Expect = 7e-22
Identities = 46/115 (40%), Positives = 71/115 (61%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L LQR DP++ I+ A+ V Y F ++W + DVEG+LFV R+ P+ F +M
Sbjct: 16 ISLAALQRHDPYINRIVDVASQVALYTFGHRANEWEKTDVEGTLFVYTRSASPKHGFTIM 75
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
NR + EN E + D ++++Q P+LLYRNA + GIWFY+ EC+ +A L +
Sbjct: 76 NRLSMENRTEPITKDLDFQLQDPFLLYRNARLSIYGIWFYDKEECQRIAELMKNL 130
Score = 38.9 bits (89), Expect = 0.25
Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 9/95 (9%)
Query: 272 VPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN 331
+P ++ +++M P+ + PT+ P + P G PP L P+ SP P+
Sbjct: 515 IPATAAPSLLMSPMVFAQPTSVPPKERESGLLPVGGQE----PPAAATSLLLPIQSPEPS 570
Query: 332 ----PAISREKVRDALLVLVQ-DNQFIDMVYKALL 361
+++ ++++ALL L+Q D+ F++++Y+A L
Sbjct: 571 VITSSPLTKLQLQEALLYLIQNDDNFLNIIYEAYL 605
>emb|CAH93503.1| hypothetical protein [Pongo pygmaeus]
gi|60389822|sp|Q5R413|DCP1B_PONPY mRNA decapping enzyme
1B
Length = 609
Score = 107 bits (266), Expect = 7e-22
Identities = 46/115 (40%), Positives = 71/115 (61%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L LQR DP++ I+ A+ V Y F ++W + DVEG+LFV R+ P+ F +M
Sbjct: 16 ISLAALQRHDPYINRIVDVASQVALYTFGHRANEWEKTDVEGTLFVYTRSASPKHGFTIM 75
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
NR + EN E + D ++++Q P+LLYRNA + GIWFY+ EC+ +A L +
Sbjct: 76 NRLSMENRTEPITKDLDFQLQDPFLLYRNARLSIYGIWFYDKEECQRIAELMKNL 130
Score = 38.1 bits (87), Expect = 0.42
Identities = 57/256 (22%), Positives = 103/256 (39%), Gaps = 46/256 (17%)
Query: 147 SSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPI 206
S T++ FE+L++ P A P P+ + +S + P+ + + G+ A
Sbjct: 347 SRTQNLFEKLQSTPGAANKCDPSTPAPA-SSAALNRSRAPTSVTPQAPGKGLAQPPQAYF 405
Query: 207 TGQ-PYQSSATISSS-------------GPTHAATPVVPLPTLQI------------PSL 240
G P Q+ S+ G + +P L LQI P+L
Sbjct: 406 NGSLPPQAHGREQSTLPRQTLPISGNQTGSSGVISPQELLKKLQIVQQEQQLHASNRPAL 465
Query: 241 PTSTPFTPQHDAP----ESINSSSQATNLVKPSFFV------PPLSSAAMMMPPVSSSIP 290
P Q ES + + +T P F V P ++ +++ P+ + P
Sbjct: 466 AAKFPVLSQSSGTGKPLESWINKTSSTEQQTPLFQVISPQRIPATAAPSLLTSPMVFAQP 525
Query: 291 TAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN----PAISREKVRDALLVL 346
T+ P + P G PP L P+ SP P+ +++ ++++ALL L
Sbjct: 526 TSVPPKERESGLLPVGGQE----PPAAATSLLLPIQSPEPSMITSSPLTKLQLQEALLYL 581
Query: 347 VQ-DNQFIDMVYKALL 361
+Q D+ F++++Y+A L
Sbjct: 582 IQNDDNFLNIIYEAYL 597
>dbj|BAB71118.1| unnamed protein product [Homo sapiens]
Length = 618
Score = 107 bits (266), Expect = 7e-22
Identities = 46/115 (40%), Positives = 71/115 (61%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L LQR DP++ I+ A+ V Y F ++W + DVEG+LFV R+ P+ F +M
Sbjct: 16 ISLAALQRHDPYINRIVDVASQVALYTFGHRANEWEKTDVEGTLFVYTRSASPKHGFTIM 75
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
NR + EN E + D ++++Q P+LLYRNA + GIWFY+ EC+ +A L +
Sbjct: 76 NRLSMENRTEPITKDLDFQLQDPFLLYRNARLSIYGIWFYDKEECQRIAELMKNL 130
Score = 38.9 bits (89), Expect = 0.25
Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 9/95 (9%)
Query: 272 VPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN 331
+P ++ +++M P+ + PT+ P + P G PP L P+ SP P+
Sbjct: 516 IPATAAPSLLMSPMVFAQPTSVPPKERESGLLPVGGQE----PPAAATSLLLPIQSPEPS 571
Query: 332 ----PAISREKVRDALLVLVQ-DNQFIDMVYKALL 361
+++ ++++ALL L+Q D+ F++++Y+A L
Sbjct: 572 VITSSPLTKLQLQEALLYLIQNDDNFLNIIYEAYL 606
>gb|AAH44477.1| Decapping enzyme [Danio rerio] gi|47271431|ref|NP_878313.2|
decapping enzyme [Danio rerio]
Length = 439
Score = 107 bits (266), Expect = 7e-22
Identities = 88/336 (26%), Positives = 145/336 (42%), Gaps = 37/336 (11%)
Query: 17 KLLNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFI 76
+L++L LQ+ DP++ ++L V Y FN ++W + ++EG+LFV R+ P F
Sbjct: 9 QLMSLAALQQHDPYIVKLLDVTGQVALYTFNPKANEWEKNEIEGTLFVYARSASPHHGFT 68
Query: 77 VMNRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRILNA 136
+MNR +TENLVE + D E+++Q P+LLYRN + IWFY+ +C+ +A L +I+
Sbjct: 69 IMNRLSTENLVEPINKDLEFQLQDPFLLYRNGNLGIYSIWFYDKADCQRIAQLMLQIVKQ 128
Query: 137 YAK----VPPKLKVSSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFF 192
+ V P VS + + + ++ E S DV +N
Sbjct: 129 ESLRVQCVSPDRSVSIRTNGCVQNRPAGILELLSKAKEEYQRSRSTERDVS-----VNCE 183
Query: 193 SAAMGIGNTSNAPITGQPYQSSATISSSGPTHAATPVVPLPTLQIPSLPTSTPFTPQHDA 252
S +T ++ S +H+ + + L SLP P
Sbjct: 184 S------QEKREDVTAAQHEKS--------SHSVVKQITVEELFGSSLPKDPP------P 223
Query: 253 PESINSSSQATNLVKPSF--FVPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPML 310
SI+ + L F + P S+ PP+ S + P P G+
Sbjct: 224 LSSISVVGEGLGLRGIPFNPALAPRLSSDPGGPPLLSLLQPRDPRTAAEETSAPRGS--T 281
Query: 311 QPFPPP--TPPPSLAPVSSP--LPNPAISREKVRDA 342
PFPP + PVS P +P+P ++ + RD+
Sbjct: 282 SPFPPAFISADAHGMPVSLPGFMPSPLVTPQSFRDS 317
>sp|Q8IZD4|DCP1B_HUMAN mRNA decapping enzyme 1B gi|24756831|gb|AAN62764.1| decapping
enzyme hDcp1b [Homo sapiens]
Length = 618
Score = 107 bits (266), Expect = 7e-22
Identities = 46/115 (40%), Positives = 71/115 (61%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L LQR DP++ I+ A+ V Y F ++W + DVEG+LFV R+ P+ F +M
Sbjct: 16 ISLAALQRHDPYINRIVDVASQVALYTFGHRANEWEKTDVEGTLFVYTRSASPKHGFTIM 75
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
NR + EN E + D ++++Q P+LLYRNA + GIWFY+ EC+ +A L +
Sbjct: 76 NRLSMENRTEPITKDLDFQLQDPFLLYRNARLSIYGIWFYDKEECQRIAELMKNL 130
Score = 38.9 bits (89), Expect = 0.25
Identities = 25/95 (26%), Positives = 51/95 (53%), Gaps = 9/95 (9%)
Query: 272 VPPLSSAAMMMPPVSSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN 331
+P ++ +++M P+ + PT+ P + P G PP L P+ SP P+
Sbjct: 516 IPATAAPSLLMSPMVFAQPTSVPPKERESGLLPVGGQE----PPAAATSLLLPIQSPEPS 571
Query: 332 ----PAISREKVRDALLVLVQ-DNQFIDMVYKALL 361
+++ ++++ALL L+Q D+ F++++Y+A L
Sbjct: 572 VITSSPLTKLQLQEALLYLIQNDDNFLNIIYEAYL 606
>gb|AAH43437.1| Decapping enzyme Dcp1b [Homo sapiens]
Length = 618
Score = 107 bits (266), Expect = 7e-22
Identities = 46/115 (40%), Positives = 71/115 (61%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L LQR DP++ I+ A+ V Y F ++W + DVEG+LFV R+ P+ F +M
Sbjct: 16 ISLAALQRHDPYINRIVDVASQVALYTFGHRANEWEKTDVEGTLFVYTRSASPKHGFTIM 75
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRI 133
NR + EN E + D ++++Q P+LLYRNA + GIWFY+ EC+ +A L +
Sbjct: 76 NRLSMENRTEPITKDLDFQLQDPFLLYRNARLSIYGIWFYDKEECQRIAELMKNL 130
Score = 39.7 bits (91), Expect = 0.15
Identities = 59/261 (22%), Positives = 104/261 (39%), Gaps = 51/261 (19%)
Query: 147 SSTKSEFEELEAVPTMAVMDGPLEPSSSITSNVADVPDDPSFINFFSAAMGIGNTSNAPI 206
S T++ FE+L++ P A P P+ + +S + P+ + + G+ A
Sbjct: 351 SRTQNLFEKLQSTPGAANKCDPSTPAPA-SSAALNRSRAPTSVTPVAPGKGLAQPPQAYF 409
Query: 207 TGQ------------------PYQSSATISS-SGPTHAATPVVPLPTLQI---------- 237
G P Q+ A S +G + +P L LQI
Sbjct: 410 NGSLPPQTVGHQAHGREQSTLPRQTLAISGSQTGSSGVISPQELLKKLQIVQQEQQLHAS 469
Query: 238 --PSLPTSTPFTPQHDAP----ESINSSSQATNLVKPSFFV------PPLSSAAMMMPPV 285
P+L P Q ES + + T P F V P ++ +++M P+
Sbjct: 470 NRPALAAKFPVLAQSSGTGKPLESWINKTPNTEQQTPLFQVISPQRIPATAAPSLLMSPM 529
Query: 286 SSSIPTAPPLHPTGTVQRPYGTPMLQPFPPPTPPPSLAPVSSPLPN----PAISREKVRD 341
+ PT+ P + P G PP L P+ SP P+ +++ ++++
Sbjct: 530 VFAQPTSVPPKERESGLLPVGGQE----PPAAATSLLLPIQSPEPSVITSSPLTKLQLQE 585
Query: 342 ALLVLVQ-DNQFIDMVYKALL 361
ALL L+Q D+ F++++Y+A L
Sbjct: 586 ALLYLIQNDDNFLNIIYEAYL 606
>gb|AAH73062.1| LOC443617 protein [Xenopus laevis]
Length = 535
Score = 105 bits (263), Expect = 2e-21
Identities = 44/116 (37%), Positives = 71/116 (60%)
Query: 19 LNLTVLQRIDPFVEEILITAAHVTFYEFNIDLSQWSRKDVEGSLFVVKRNTQPRFQFIVM 78
++L L + DP++ I+ HV Y F+ ++W + DVEG+LFV R+ P F +M
Sbjct: 14 MSLAALHQSDPYISSIVDVTGHVALYRFSPQANEWEKTDVEGTLFVYTRSASPHHGFTIM 73
Query: 79 NRRNTENLVENLLGDFEYEIQVPYLLYRNAAQEVNGIWFYNARECEEVANLFSRIL 134
NR N NLVE + D E ++ P+LLYRN++ + IWFYN +C+ +A L ++++
Sbjct: 74 NRLNMHNLVEPMNKDLELQLHEPFLLYRNSSLAIYSIWFYNKSDCQRIAKLMTQVV 129
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.315 0.131 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 662,765,993
Number of Sequences: 2540612
Number of extensions: 31716883
Number of successful extensions: 284741
Number of sequences better than 10.0: 7520
Number of HSP's better than 10.0 without gapping: 981
Number of HSP's successfully gapped in prelim test: 6954
Number of HSP's that attempted gapping in prelim test: 203688
Number of HSP's gapped (non-prelim): 35569
length of query: 366
length of database: 863,360,394
effective HSP length: 129
effective length of query: 237
effective length of database: 535,621,446
effective search space: 126942282702
effective search space used: 126942282702
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146332.11


