

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146332.10 - phase: 0
(468 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
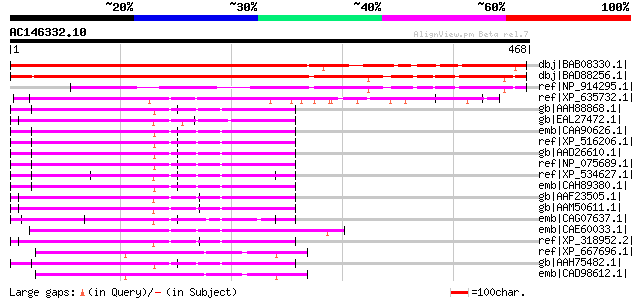
dbj|BAB08330.1| unnamed protein product [Arabidopsis thaliana] g... 381 e-104
dbj|BAD88256.1| leucine-rich repeat family protein-like [Oryza s... 336 1e-90
ref|NP_914295.1| P0458E05.24 [Oryza sativa (japonica cultivar-gr... 207 6e-52
ref|XP_635732.1| hypothetical protein DDB0188882 [Dictyostelium ... 180 7e-44
gb|AAH88868.1| Protein phosphatase 1, regulatory (inhibitor) sub... 125 3e-27
gb|EAL27472.1| GA19179-PA [Drosophila pseudoobscura] 124 5e-27
emb|CAA90626.1| yeast sds22 homolog [Homo sapiens] gi|54697128|g... 124 8e-27
ref|XP_516206.1| PREDICTED: similar to protein phosphatase 1, re... 124 8e-27
gb|AAD26610.1| protein phosphatase-1 regulatory subunit 7 alpha2... 124 8e-27
ref|NP_075689.1| protein phosphatase-1 regulatory subunit 7 [Mus... 122 2e-26
ref|XP_534627.1| PREDICTED: similar to protein phosphatase 1, re... 122 3e-26
emb|CAH89380.1| hypothetical protein [Pongo pygmaeus] 121 5e-26
gb|AAF23505.1| putative mitotic protein phosphatase 1 regulator ... 119 3e-25
gb|AAM50611.1| GH07711p [Drosophila melanogaster] gi|7300250|gb|... 118 5e-25
emb|CAG07637.1| unnamed protein product [Tetraodon nigroviridis] 117 8e-25
emb|CAE60033.1| Hypothetical protein CBG03541 [Caenorhabditis br... 117 1e-24
ref|XP_318952.2| ENSANGP00000015763 [Anopheles gambiae str. PEST... 116 2e-24
ref|XP_667696.1| leucine rich repeat protein [Cryptosporidium ho... 114 7e-24
gb|AAH75482.1| Protein phosphatase 1, regulatory subunit 7 [Xeno... 114 9e-24
emb|CAD98612.1| conserved hypothetical Leucine Rich Repeat prote... 112 2e-23
>dbj|BAB08330.1| unnamed protein product [Arabidopsis thaliana]
gi|19699236|gb|AAL90984.1| AT5g22320/MWD9_11
[Arabidopsis thaliana] gi|18420455|ref|NP_568416.1|
leucine-rich repeat family protein [Arabidopsis
thaliana] gi|15912299|gb|AAL08283.1| AT5g22320/MWD9_11
[Arabidopsis thaliana]
Length = 452
Score = 381 bits (979), Expect = e-104
Identities = 219/476 (46%), Positives = 310/476 (65%), Gaps = 35/476 (7%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L+ EQVLK+ +P S+ L+L HKAL+DVSCL+ F LEKLDL+FNNLT L+GL+
Sbjct: 1 MNSLTVEQVLKEKKTNDPDSVKELNLGHKALTDVSCLSKFKNLEKLDLRFNNLTDLQGLK 60
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
+CV LKWLSVVENKL+SL GI+ LTKLTVLNAGKNKLKSM+EI SL +RALILNDNEI
Sbjct: 61 SCVNLKWLSVVENKLQSLNGIEALTKLTVLNAGKNKLKSMNEISSLVNLRALILNDNEIS 120
Query: 121 SICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTEL 180
SIC LD +K+LN+LVLS+NPI +IG++L K+K+++K+SLS C+++ I +SLK C +L EL
Sbjct: 121 SICKLDLLKDLNSLVLSRNPISEIGDSLSKLKNLSKISLSDCRIKAIGSSLKSCSDLKEL 180
Query: 181 RLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEK 240
RLA+N+IK+LP EL N +L NLD+GNNVI + S ++VL +L+ LRNLN++GNP++ N+K
Sbjct: 181 RLANNEIKALPAELAVNKRLLNLDVGNNVITQLSGLEVLGTLSCLRNLNIRGNPISDNDK 240
Query: 241 VIRKIKN-ALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFD--HVDQNEDDHLEAADKR 297
+K++ LP + VFNA+P++K ++N K H+ D D +FD H E++ + KR
Sbjct: 241 SAKKVRTLLLPSVNVFNAQPLEKSSRNAK-HIRLDTDDETFDAYHNKSAEEEQSKEDRKR 299
Query: 298 KSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDD 357
K + KR ++ +E +N+D+ +KKK + T + K KK+ K+
Sbjct: 300 KKSSKRNKS------------EEEEVNNEDHKSKKKKSKSNTNVDQVETK--KKEEHKEK 345
Query: 358 NKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKDQENLNHGGEM 417
PS ++ + EKK+K KE E D IDDAE SFAEIF+ +EN+ G E
Sbjct: 346 TIPSN-----NDDDDDAEKKQKRATPKE--ELDAIDDAETSFAEIFS---RENVPKGSED 395
Query: 418 KLQDQVPKDLKLVSSIETLPVKHKSAKMHNVESLSSP-------GTEIGMGGPSTW 466
++ + ++ ++ + K K + + S EIG+GG S W
Sbjct: 396 GIEKKKKSSVQETGLVKVIDTKANKKKKKSEKKQSKSVVIDLPMEVEIGLGGESKW 451
>dbj|BAD88256.1| leucine-rich repeat family protein-like [Oryza sativa (japonica
cultivar-group)]
Length = 463
Score = 336 bits (861), Expect = 1e-90
Identities = 207/480 (43%), Positives = 292/480 (60%), Gaps = 32/480 (6%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M RL+ EQ ++ + + +SL+L+H+ALSDVSCL+SF LE+LDL +N L +LEGL
Sbjct: 1 MARLTVEQAKREAGSAGTLA-TSLNLSHRALSDVSCLSSFVNLERLDLGYNCLLTLEGLS 59
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
C LKWLSV+ENKL SL+G++GL+KL VLNAGKNKLK+MDE+ SL+++ ALILNDN I
Sbjct: 60 NCANLKWLSVIENKLVSLKGVEGLSKLQVLNAGKNKLKTMDEVKSLTSLGALILNDNNIS 119
Query: 121 SICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTEL 180
SIC LDQ+ +LNTLVLSKNPI IG+AL K K++ KLSLSHCQ+E I +SL CVEL EL
Sbjct: 120 SICKLDQLHQLNTLVLSKNPIFTIGDALMKAKAMKKLSLSHCQIEKIGSSLTACVELKEL 179
Query: 181 RLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEK 240
RLAHN I ++P +L N K+ NLDLGNN+I + SD++VL L LRNLNLQGNP+A +
Sbjct: 180 RLAHNKITTIPSDLAKNVKILNLDLGNNLIERRSDLEVLSELHYLRNLNLQGNPIAEKDG 239
Query: 241 VIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAADKRK-S 299
+ +K+K +P L++FN+KP++ +K++ + + D +D + K+K
Sbjct: 240 LAKKVKKLVPNLRIFNSKPMEASSKSKNSR----EENLPINDADTPDDGPTDIYTKKKGK 295
Query: 300 NKKRKETADASEKEAGVYDKENT-------GHNKDNGNKKKDKLTGTVDPDTKNKSTKKK 352
K K+ + E+ AG + + D KK+K ++ KK
Sbjct: 296 GKHSKQQIKSPEEPAGQSTRPDVTIAAPAKSELLDGKEMKKEKAA------VEHVKNKKS 349
Query: 353 LKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKDQENLN 412
+KDDN + ++ V++ K+ K+ K KE+ D IDD E FA++ + E N
Sbjct: 350 KRKDDNSSVDHT---DKKVSKGAKRTKSAK-KEEKNADGIDDTEMPFADL--VFSGEGNN 403
Query: 413 HGGEMKLQDQ-VPKDLKLVSSIETLPVKHKSAK-----MHNVESLSSPGTEIGMGGPSTW 466
E+K ++Q + +D K + K K AK +E LSS E+G G S W
Sbjct: 404 PELELKGKNQEIARDGKFGGLVIDHTKKKKKAKGTVFGSSALEQLSSV-PEVGSGALSGW 462
>ref|NP_914295.1| P0458E05.24 [Oryza sativa (japonica cultivar-group)]
Length = 362
Score = 207 bits (527), Expect = 6e-52
Identities = 153/425 (36%), Positives = 216/425 (50%), Gaps = 79/425 (18%)
Query: 56 LEGLRACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILN 115
L+GL C LKWLSV+ENKL SL+G++GL+KL VLNAGKNKLK+MDE+ SL+++ ALILN
Sbjct: 2 LKGLSNCANLKWLSVIENKLVSLKGVEGLSKLQVLNAGKNKLKTMDEVKSLTSLGALILN 61
Query: 116 DNEIVSICNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCV 175
VLSKNPI IG+AL K K++ KLSLSHCQ+E I +SL CV
Sbjct: 62 -------------------VLSKNPIFTIGDALMKAKAMKKLSLSHCQIEKIGSSLTACV 102
Query: 176 ELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPV 235
EL ELRLAHN I VL L LRNLNLQGNP+
Sbjct: 103 ELKELRLAHNKI-----------------------------TVLSELHYLRNLNLQGNPI 133
Query: 236 ATNEKVIRKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFDHVDQNEDDHLEAAD 295
A + + +K+K +P L++FN+KP++ +K++ + + D +D +
Sbjct: 134 AEKDGLAKKVKKLVPNLRIFNSKPMEASSKSKNSR----EENLPINDADTPDDGPTDIYT 189
Query: 296 KRK-SNKKRKETADASEKEAGVYDKENT-------GHNKDNGNKKKDKLTGTVDPDTKNK 347
K+K K K+ + E+ AG + + D KK+K ++
Sbjct: 190 KKKGKGKHSKQQIKSPEEPAGQSTRPDVTIAAPAKSELLDGKEMKKEKAA------VEHV 243
Query: 348 STKKKLKKDDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKD 407
KK +KDDN + ++ V++ K+ K+ K KE+ D IDD E FA++ +
Sbjct: 244 KNKKSKRKDDNSSVDHT---DKKVSKGAKRTKSAK-KEEKNADGIDDTEMPFADL--VFS 297
Query: 408 QENLNHGGEMKLQDQ-VPKDLKLVSSIETLPVKHKSAK-----MHNVESLSSPGTEIGMG 461
E N E+K ++Q + +D K + K K AK +E LSS E+G G
Sbjct: 298 GEGNNPELELKGKNQEIARDGKFGGLVIDHTKKKKKAKGTVFGSSALEQLSSV-PEVGSG 356
Query: 462 GPSTW 466
S W
Sbjct: 357 ALSGW 361
>ref|XP_635732.1| hypothetical protein DDB0188882 [Dictyostelium discoideum]
gi|60464064|gb|EAL62226.1| hypothetical protein
DDB0188882 [Dictyostelium discoideum]
Length = 693
Score = 180 bits (457), Expect = 7e-44
Identities = 135/448 (30%), Positives = 227/448 (50%), Gaps = 35/448 (7%)
Query: 19 SSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESL 78
++ ++ +L++K ++ L+S L+K+DL N LTS+ GL+ ++L+W+++ NKLE +
Sbjct: 18 TTATTFNLSNKGYENIEDLSSCIDLKKIDLSKNELTSIVGLKDNLSLEWINLSGNKLERM 77
Query: 79 EGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSK 138
++ L+KL VLN NKLK ++ + L ++AL+LN+NEI I N++ + LNTLVLSK
Sbjct: 78 NDLKLLSKLKVLNISHNKLKRIEGLMKLGDLKALVLNNNEITKIENMEYVPHLNTLVLSK 137
Query: 139 NPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNS 198
N + I L+ +K +TKLSL++ ++ I + V L E++L+HN I S+ + +
Sbjct: 138 NQLEDI-SGLRFLKELTKLSLTNNNIKHI-PDISQNVLLKEIKLSHNKIFSIDPKFSNLH 195
Query: 199 KLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNAK 258
L LDL NN++ +SDI+VL SL L+ L+L GNP+A E KI+ P L + K
Sbjct: 196 NLFILDLSNNLLKTYSDIQVLSSLKNLKTLSLIGNPIAALEDYKEKIREMFPNLDNLDGK 255
Query: 259 PID---------KDTKNEKGHMTDDAHDFSFDHVDQNE--------------DDHLEAAD 295
P K +EK M + + + E D++LE+++
Sbjct: 256 PFSEKSVKRLTKKKENSEKREMIEKDKERKLIRKENEEKGIPNKPRKTFNKIDNNLESSE 315
Query: 296 KRKSNKKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKK 355
+ KKR + D E+ KE T ++ + +++ + K +S K +
Sbjct: 316 R----KKRTDRPDRPERTDRPERKERT--DRPERTDRIERIERKQPFERKERSDKPERAD 369
Query: 356 DDNKPSEKALALEENVNRTEKKKKNRKNKEQSEFDIIDDAEASFAEIFNIKDQENL--NH 413
+ +E+ +E R E+ K K + DIID + E + +D EN N
Sbjct: 370 RPERSNERKQFNKEISERNERPKTFDKKNQNRNQDIIDKSNKRKYENYQDEDNENQADNE 429
Query: 414 GGEMKLQDQVPKDLKLVSSIETLPVKHK 441
++ PKD + SI+T K K
Sbjct: 430 DRPEEVSTSKPKDFR--DSIKTKNSKVK 455
Score = 63.2 bits (152), Expect = 2e-08
Identities = 91/388 (23%), Positives = 163/388 (41%), Gaps = 52/388 (13%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
L++ ++ K N +++L L+ L D+S L +L KL L NN+ + + V
Sbjct: 113 LNNNEITKIENMEYVPHLNTLVLSKNQLEDISGLRFLKELTKLSLTNNNIKHIPDISQNV 172
Query: 64 TLKWLSVVENKLESLE-GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSI 122
LK + + NK+ S++ L L +L+ N LK+ +I LS+
Sbjct: 173 LLKEIKLSHNKIFSIDPKFSNLHNLFILDLSNNLLKTYSDIQVLSS-------------- 218
Query: 123 CNLDQMKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRL 182
+K L TL L NPI + + +K++ + L+ +D K E + RL
Sbjct: 219 -----LKNLKTLSLIGNPIAALEDYKEKIREMFP------NLDNLDG--KPFSEKSVKRL 265
Query: 183 AHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVI 242
S E++ K R L N K K K+ K+ N NL+ +
Sbjct: 266 TKKKENSEKREMIEKDKERKLIRKENE-EKGIPNKPRKTFNKIDN-NLESSE-------- 315
Query: 243 RKIKNALPKLQVFNAKPIDKDTKNEKGHMTDDAHDFSFDHVDQNED--DHLEAADK-RKS 299
RK + P +P ++ + ++ TD + ++ D E AD+ +S
Sbjct: 316 RKKRTDRPDRPERTDRP-ERKERTDRPERTDRIERIERKQPFERKERSDKPERADRPERS 374
Query: 300 NKKRKETADASEK--EAGVYDKENTGHNKDNGNK-KKDKLTGTVDPDTKNKSTKKKLKKD 356
N++++ + SE+ +DK+N N+D +K K K D D +N++ +
Sbjct: 375 NERKQFNKEISERNERPKTFDKKNQNRNQDIIDKSNKRKYENYQDEDNENQA------DN 428
Query: 357 DNKPSEKALALEENVNRTEKKKKNRKNK 384
+++P E + + ++ R K KN K K
Sbjct: 429 EDRPEEVSTSKPKDF-RDSIKTKNSKVK 455
Score = 60.8 bits (146), Expect = 9e-08
Identities = 98/493 (19%), Positives = 197/493 (39%), Gaps = 85/493 (17%)
Query: 4 LSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACV 63
LS ++ + N+ S + L+++H L + L L+ L L N +T +E +
Sbjct: 69 LSGNKLERMNDLKLLSKLKVLNISHNKLKRIEGLMKLGDLKALVLNNNEITKIENMEYVP 128
Query: 64 TLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSIC 123
L L + +N+LE + G++ L +LT L+ N +K + +I ++ + L+ N+I SI
Sbjct: 129 HLNTLVLSKNQLEDISGLRFLKELTKLSLTNNNIKHIPDISQNVLLKEIKLSHNKIFSID 188
Query: 124 N--------------------------LDQMKELNTLVLSKNPIRKIGEALKKVKSITKL 157
L +K L TL L NPI + + +K++ +
Sbjct: 189 PKFSNLHNLFILDLSNNLLKTYSDIQVLSSLKNLKTLSLIGNPIAALEDYKEKIREMFP- 247
Query: 158 SLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSKLRNLDLGNNVIAKWSDIK 217
L+ +D K E + RL S E++ K R L N K K
Sbjct: 248 -----NLDNLDG--KPFSEKSVKRLTKKKENSEKREMIEKDKERKLIRKENE-EKGIPNK 299
Query: 218 VLKSLTKLRNLNLQGN-----------PVATNEKVIRKIKNALPKL-----QVFNAKPID 261
K+ K+ N NL+ + P T ++ RK + P+ ++ +P +
Sbjct: 300 PRKTFNKIDN-NLESSERKKRTDRPDRPERT-DRPERKERTDRPERTDRIERIERKQPFE 357
Query: 262 KDTKNEKGHMTD------DAHDFSFDHVDQNEDDHLEAADKRKSNKKRKETADASEKEAG 315
+ +++K D + F+ + ++NE + DK+ N+ + +++++
Sbjct: 358 RKERSDKPERADRPERSNERKQFNKEISERNERP--KTFDKKNQNRNQDIIDKSNKRKYE 415
Query: 316 VYDKENTGHNKDNGNKKKDKLTGTVDP-----DTKNKSTKKKLKK--------------- 355
Y E+ + DN ++ ++ T TKN K K+
Sbjct: 416 NYQDEDNENQADNEDRPEEVSTSKPKDFRDSIKTKNSKVKGGDKRQRTATTTSTIIPTST 475
Query: 356 -DDNKPSEKALALEENVNRTEKKKKNRKNK-EQSEFDIIDDAEASFAEIFNIKDQENLNH 413
+++ S++A ++ + + KK +N + I+ + S +E+ + +N ++
Sbjct: 476 FNNSTVSKQAETPKKGFSLMDHYKKEAENDFSDDDMKIVGENPISLSEL--LDSNKNKDN 533
Query: 414 GGEMKLQDQVPKD 426
E K ++ KD
Sbjct: 534 NNEKKTNNKDKKD 546
Score = 42.7 bits (99), Expect = 0.024
Identities = 39/152 (25%), Positives = 62/152 (40%), Gaps = 16/152 (10%)
Query: 255 FNAKPIDKDTKNEK------GHMTDDA-HDFSFDH---VDQNEDDHLEAADKRKS----N 300
FN + K + K H +A +DFS D V +N E D K+ N
Sbjct: 476 FNNSTVSKQAETPKKGFSLMDHYKKEAENDFSDDDMKIVGENPISLSELLDSNKNKDNNN 535
Query: 301 KKRKETADASEKEAGVYDKENTGHNKDNGNKKKDKLTGTVDPDTKNKSTKKKLKKDDNKP 360
+K+ D + N +NK K+ + + T P +K TK+++KK NK
Sbjct: 536 EKKTNNKDKKDNNNNKNKNNNENNNKSINKKENQEKSKTPTPKSKEPQTKEEVKKTPNKK 595
Query: 361 SEKALALEENVNRTEKKKKNRKNKEQSEFDII 392
S +E+ T +K + ++SE D I
Sbjct: 596 STP--TSKESTTTTTVPQKQQTKVQKSETDDI 625
>gb|AAH88868.1| Protein phosphatase 1, regulatory (inhibitor) subunit 7 (predicted)
[Rattus norvegicus] gi|57634526|ref|NP_001009825.1|
protein phosphatase 1, regulatory (inhibitor) subunit 7
(predicted) [Rattus norvegicus]
Length = 360
Score = 125 bits (314), Expect = 3e-27
Identities = 75/238 (31%), Positives = 133/238 (55%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 121 SLRELDLYDNQIKKIENLEALTELEVLDISFNLLRNIEGIDKLTQLKKLFLVNNKINKIE 180
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
I L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 181 NISTLQQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALSNLTVLSMQSN 240
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 241 RLTKI-EGLQNLVNLRELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 297
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 298 LQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 355
Score = 80.5 bits (197), Expect = 1e-13
Identities = 46/154 (29%), Positives = 86/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + + L L ++ N LT +EGL+
Sbjct: 190 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALSNLTVLSMQSNRLTKIEGLQ 249
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ ++ +NDN +
Sbjct: 250 NLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTELQEFWMNDNLLE 309
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 310 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 343
>gb|EAL27472.1| GA19179-PA [Drosophila pseudoobscura]
Length = 326
Score = 124 bits (312), Expect = 5e-27
Identities = 89/250 (35%), Positives = 135/250 (53%), Gaps = 5/250 (2%)
Query: 9 VLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWL 68
+ K N ++ L L ++ V L KLE LDL FN LT +E L A V L+ L
Sbjct: 74 IKKIENLSMLKTLVELELYDNQITKVENLEELTKLEMLDLSFNRLTKIENLDALVNLEKL 133
Query: 69 SVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQM 128
V N++ +E + LT LT+L G NKLK + I +L +R L L N+I I NLD +
Sbjct: 134 YFVANRITVIENLGMLTSLTMLELGDNKLKKIQNIDTLVNLRQLFLGKNKIAKIENLDTL 193
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
L L L N I KI E L+K+ ++ +L +S +E I+ +L+ L L LA N +K
Sbjct: 194 VNLEILSLQANRIVKI-ENLEKLTNLKELYISENGIEVIE-NLEENKNLETLDLAKNRLK 251
Query: 189 SLPE-ELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKN 247
++ E + N L L L +N I W +I++LK L+ + L+ NP+A + + K+++
Sbjct: 252 AVGNLETLEN--LEELWLNHNGIDDWKNIELLKGNKALQTIYLESNPLAKDVRYRSKLRD 309
Query: 248 ALPKLQVFNA 257
LP+LQ +A
Sbjct: 310 ILPQLQKIDA 319
Score = 64.3 bits (155), Expect = 8e-09
Identities = 46/173 (26%), Positives = 79/173 (45%), Gaps = 7/173 (4%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L ++ K N ++ L L ++ + L + LE L L+ N + +E L
Sbjct: 154 MLELGDNKLKKIQNIDTLVNLRQLFLGKNKIAKIENLDTLVNLEILSLQANRIVKIENLE 213
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
LK L + EN +E +E ++ L L+ KN+LK++ + +L + L LN N I
Sbjct: 214 KLTNLKELYISENGIEVIENLEENKNLETLDLAKNRLKAVGNLETLENLEELWLNHNGID 273
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKVKSI----TKLSLSHCQLEG 166
N++ + K L T+ L NP+ K K++ I K+ + C + G
Sbjct: 274 DWKNIELLKGNKALQTIYLESNPLAKDVRYRSKLRDILPQLQKIDATMCVMPG 326
>emb|CAA90626.1| yeast sds22 homolog [Homo sapiens] gi|54697128|gb|AAV38936.1|
protein phosphatase 1, regulatory subunit 7 [Homo
sapiens] gi|61355994|gb|AAX41197.1| protein phosphatase
1 regulatory subunit 7 [synthetic construct]
gi|12654185|gb|AAH00910.1| Protein phosphatase 1,
regulatory subunit 7 [Homo sapiens]
gi|4633067|gb|AAD26611.1| protein phosphatase-1
regulatory subunit 7 alpha1 [Homo sapiens]
gi|2136139|pir||S68209 sds22 protein homolog - human
gi|4506013|ref|NP_002703.1| protein phosphatase 1,
regulatory subunit 7 [Homo sapiens]
gi|1585165|prf||2124310A sds22 gene
Length = 360
Score = 124 bits (310), Expect = 8e-27
Identities = 74/238 (31%), Positives = 133/238 (55%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 121 SLRELDLYDNQIKKIENLEALTELEILDISFNLLRNIEGVDKLTRLKKLFLVNNKISKIE 180
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
+ L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 181 NLSNLHQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSN 240
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 241 RLTKI-EGLQNLVNLRELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 297
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 298 LQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 355
Score = 80.1 bits (196), Expect = 1e-13
Identities = 46/154 (29%), Positives = 85/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 190 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSNRLTKIEGLQ 249
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ ++ +NDN +
Sbjct: 250 NLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTELQEFWMNDNLLE 309
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 310 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 343
>ref|XP_516206.1| PREDICTED: similar to protein phosphatase 1, regulatory subunit 7
[Pan troglodytes]
Length = 360
Score = 124 bits (310), Expect = 8e-27
Identities = 74/238 (31%), Positives = 133/238 (55%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 121 SLRELDLYDNQIKKIENLEALTELEILDISFNLLRNIEGVDKLTRLKKLFLVNNKISKIE 180
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
+ L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 181 NLSNLHQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSN 240
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 241 RLTKI-EGLQNLVNLRELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 297
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 298 LQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 355
Score = 80.1 bits (196), Expect = 1e-13
Identities = 46/154 (29%), Positives = 85/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 190 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSNRLTKIEGLQ 249
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ ++ +NDN +
Sbjct: 250 NLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTELQEFWMNDNLLE 309
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 310 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 343
>gb|AAD26610.1| protein phosphatase-1 regulatory subunit 7 alpha2 [Homo sapiens]
Length = 317
Score = 124 bits (310), Expect = 8e-27
Identities = 74/238 (31%), Positives = 133/238 (55%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 78 SLRELDLYDNQIKKIENLEALTELEILDISFNLLRNIEGVDKLTRLKKLFLVNNKISKIE 137
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
+ L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 138 NLSNLHQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSN 197
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 198 RLTKI-EGLQNLVNLRELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 254
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 255 LQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 312
Score = 80.1 bits (196), Expect = 1e-13
Identities = 46/154 (29%), Positives = 85/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 147 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSNRLTKIEGLQ 206
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ ++ +NDN +
Sbjct: 207 NLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTELQEFWMNDNLLE 266
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 267 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 300
>ref|NP_075689.1| protein phosphatase-1 regulatory subunit 7 [Mus musculus]
gi|15488779|gb|AAH13524.1| Protein phosphatase-1
regulatory subunit 7 [Mus musculus]
gi|12831470|gb|AAK08624.1| protein phosphatase-1
regulatory subunit 7 [Mus musculus]
gi|12655852|gb|AAK00624.1| protein phosphatase-1
regulatory subunit 7 [Mus musculus]
gi|63485042|ref|XP_622874.1| PREDICTED: similar to
protein phosphatase-1 regulatory subunit 7 [Mus
musculus]
Length = 361
Score = 122 bits (307), Expect = 2e-26
Identities = 74/238 (31%), Positives = 133/238 (55%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 122 SLRELDLYDNQIKKIENLEALTELEVLDISFNMLRNIEGIDKLTQLKKLFLVNNKINKIE 181
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
I L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 182 NISNLHQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSVQSN 241
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LS+ +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 242 RLAKI-EGLQSLVNLRELYLSNNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 298
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 299 LQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 356
Score = 79.0 bits (193), Expect = 3e-13
Identities = 45/154 (29%), Positives = 85/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + L L ++ N L +EGL+
Sbjct: 191 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSVQSNRLAKIEGLQ 250
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
+ V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ ++ +NDN +
Sbjct: 251 SLVNLRELYLSNNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTELQEFWMNDNLLE 310
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 311 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 344
>ref|XP_534627.1| PREDICTED: similar to protein phosphatase 1, regulatory subunit 7
[Canis familiaris]
Length = 376
Score = 122 bits (305), Expect = 3e-26
Identities = 74/238 (31%), Positives = 134/238 (56%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + LE LD+ FN L ++EG+ L+ L +V NK+ +E
Sbjct: 137 SLRELDLYDNQIKKIENLEALTHLEILDISFNLLRNIEGVDKLTRLRKLFLVNNKISKIE 196
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
I L +L +L G N++++++ I +L+++ +L L N+I + NLD + L L + N
Sbjct: 197 NISNLHQLQMLELGSNRIRAIENIDTLTSLESLFLGKNKITKLQNLDALTNLTVLSMQSN 256
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ K+ E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 257 RLTKM-EGLQSLVNLRELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENVSHLTE 313
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD+ LK+ L + L+ NP+ + + RKI ALP ++ +A
Sbjct: 314 LQEFWMNDNLLESWSDLDELKAAKSLETVYLERNPLQKDPQYRRKIMLALPTVRQIDA 371
Score = 81.6 bits (200), Expect = 5e-14
Identities = 45/154 (29%), Positives = 86/154 (55%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +S+ SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 206 MLELGSNRIRAIENIDTLTSLESLFLGKNKITKLQNLDALTNLTVLSMQSNRLTKMEGLQ 265
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
+ V L+ L + N +E +EG++ KLT+L+ N++K ++ + L+ ++ +NDN +
Sbjct: 266 SLVNLRELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENVSHLTELQEFWMNDNLLE 325
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKV 151
S +LD++ K L T+ L +NP++K + +K+
Sbjct: 326 SWSDLDELKAAKSLETVYLERNPLQKDPQYRRKI 359
Score = 65.9 bits (159), Expect = 3e-09
Identities = 51/166 (30%), Positives = 90/166 (53%), Gaps = 6/166 (3%)
Query: 74 KLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNT 133
++ +EG + L K+ L +N +K ++ +G L ++R L L DN+I I NL+ + L
Sbjct: 103 RIGKIEGFEVLKKVKTLCLRQNLIKCIENLGELQSLRELDLYDNQIKKIENLEALTHLEI 162
Query: 134 LVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEE 193
L +S N +R I E + K+ + KL L + ++ I+ ++ +L L L N I+++ E
Sbjct: 163 LDISFNLLRNI-EGVDKLTRLRKLFLVNNKISKIE-NISNLHQLQMLELGSNRIRAI-EN 219
Query: 194 LMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNE 239
+ + L +L LG N I K ++ L +LT L L++Q N + E
Sbjct: 220 IDTLTSLESLFLGKNKITK---LQNLDALTNLTVLSMQSNRLTKME 262
>emb|CAH89380.1| hypothetical protein [Pongo pygmaeus]
Length = 360
Score = 121 bits (303), Expect = 5e-26
Identities = 73/238 (30%), Positives = 132/238 (54%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
S+ L L + + L + +LE LD+ FN L ++EG+ LK L +V NK+ +E
Sbjct: 121 SLRELDLYDNQIKKIENLEALTELEILDISFNLLRNIEGVDKVTQLKKLFLVNNKISKIE 180
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
+ L +L +L G N++++++ I +L+ + +L L N+I + NLD + L L + N
Sbjct: 181 NLSNLHQLQMLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSN 240
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LSH +E I+ L+ +LT L +A N IK + E + H ++
Sbjct: 241 RLTKI-EGLQNLVNLQELYLSHNGIEVIE-GLENNNKLTMLDIASNRIKKI-ENISHLTE 297
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
+ + +N++ WSD+ LK L + L+ NP+ + + RK+ ALP ++ +A
Sbjct: 298 PQEFWMNDNLLESWSDLDELKGARSLETVYLERNPLQKDPQYRRKVMLALPSVRQIDA 355
Score = 77.8 bits (190), Expect = 7e-13
Identities = 46/154 (29%), Positives = 84/154 (53%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +++ SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 190 MLELGSNRIRAIENIDTLTNLESLFLGKNKITKLQNLDALTNLTVLSMQSNRLTKIEGLQ 249
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + N +E +EG++ KLT+L+ N++K ++ I L+ + +NDN +
Sbjct: 250 NLVNLQELYLSHNGIEVIEGLENNNKLTMLDIASNRIKKIENISHLTEPQEFWMNDNLLE 309
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
S +LD++K L T+ L +NP++K + +KV
Sbjct: 310 SWSDLDELKGARSLETVYLERNPLQKDPQYRRKV 343
>gb|AAF23505.1| putative mitotic protein phosphatase 1 regulator [Drosophila
melanogaster]
Length = 326
Score = 119 bits (297), Expect = 3e-25
Identities = 83/249 (33%), Positives = 131/249 (52%), Gaps = 3/249 (1%)
Query: 9 VLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWL 68
+ K N + ++ L L ++ + L LE LD+ FN LT +E L V L+ +
Sbjct: 73 IKKIENLSSLKTLIELELYDNQITKIENLDDLPHLEVLDISFNRLTKIENLDKLVKLEKV 132
Query: 69 SVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQM 128
V N++ +E + LT LT+L G NKLK ++ I L +R L L N+I I NLD +
Sbjct: 133 YFVSNRITQIENLDMLTNLTMLELGDNKLKKIENIEMLVNLRQLFLGKNKIAKIENLDTL 192
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
L L L N I KI E L+K+ ++ +L +S +E I+ +L +L L LA N +K
Sbjct: 193 VNLEILSLQANRIVKI-ETLEKLANLRELYVSENGVETIE-NLSENTKLETLDLAKNRLK 250
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNA 248
+ L L L L +N + W DI++LK L+ + L+ NP+A + + K+++
Sbjct: 251 GI-ANLEKLELLEELWLNHNGVDDWKDIELLKVNKALQTIYLEYNPLAKDVRYRSKLRDI 309
Query: 249 LPKLQVFNA 257
LP+LQ +A
Sbjct: 310 LPQLQKIDA 318
Score = 62.8 bits (151), Expect = 2e-08
Identities = 48/174 (27%), Positives = 80/174 (45%), Gaps = 9/174 (5%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L ++ K N ++ L L ++ + L + LE L L+ N + +E L
Sbjct: 153 MLELGDNKLKKIENIEMLVNLRQLFLGKNKIAKIENLDTLVNLEILSLQANRIVKIETLE 212
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
L+ L V EN +E++E + TKL L+ KN+LK + + L + L LN N +
Sbjct: 213 KLANLRELYVSENGVETIENLSENTKLETLDLAKNRLKGIANLEKLELLEELWLNHNGVD 272
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSL 171
+++ + K L T+ L NP+ K K++ I QL+ ID +L
Sbjct: 273 DWKDIELLKVNKALQTIYLEYNPLAKDVRYRSKLRDILP------QLQKIDATL 320
>gb|AAM50611.1| GH07711p [Drosophila melanogaster] gi|7300250|gb|AAF55413.1|
CG5851-PA [Drosophila melanogaster]
gi|21358617|ref|NP_650619.1| CG5851-PA [Drosophila
melanogaster] gi|17944820|gb|AAL48476.1| GM06266p
[Drosophila melanogaster]
Length = 326
Score = 118 bits (295), Expect = 5e-25
Identities = 83/249 (33%), Positives = 131/249 (52%), Gaps = 3/249 (1%)
Query: 9 VLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWL 68
+ K N + ++ L L ++ + L LE LD+ FN LT +E L V L+ +
Sbjct: 73 IKKIENLSSLKTLIELELYDNQITKIENLDDLPHLEVLDISFNRLTKIENLDKLVKLEKV 132
Query: 69 SVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQM 128
V N++ +E + LT LT+L G NKLK ++ I L +R L L N+I I NLD +
Sbjct: 133 YFVSNRITQIENLDMLTNLTMLELGDNKLKKIENIEMLVNLRQLFLGKNKIAKIENLDTL 192
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
L L L N I KI E L+K+ ++ +L +S +E I+ +L +L L LA N +K
Sbjct: 193 VNLEILSLQANRIVKI-ENLEKLANLRELYVSENGVETIE-NLSENTKLETLDLAKNRLK 250
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNA 248
+ L L L L +N + W DI++LK L+ + L+ NP+A + + K+++
Sbjct: 251 GI-ANLEKLELLEELWLNHNGVDDWKDIELLKVNKALQTIYLEYNPLAKDVRYRSKLRDI 309
Query: 249 LPKLQVFNA 257
LP+LQ +A
Sbjct: 310 LPQLQKIDA 318
Score = 63.5 bits (153), Expect = 1e-08
Identities = 48/174 (27%), Positives = 80/174 (45%), Gaps = 9/174 (5%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L ++ K N ++ L L ++ + L + LE L L+ N + +E L
Sbjct: 153 MLELGDNKLKKIENIEMLVNLRQLFLGKNKIAKIENLDTLVNLEILSLQANRIVKIENLE 212
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
L+ L V EN +E++E + TKL L+ KN+LK + + L + L LN N +
Sbjct: 213 KLANLRELYVSENGVETIENLSENTKLETLDLAKNRLKGIANLEKLELLEELWLNHNGVD 272
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSL 171
+++ + K L T+ L NP+ K K++ I QL+ ID +L
Sbjct: 273 DWKDIELLKVNKALQTIYLEYNPLAKDVRYRSKLRDILP------QLQKIDATL 320
>emb|CAG07637.1| unnamed protein product [Tetraodon nigroviridis]
Length = 281
Score = 117 bits (293), Expect = 8e-25
Identities = 77/247 (31%), Positives = 134/247 (54%), Gaps = 3/247 (1%)
Query: 11 KDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSV 70
K N + +S+ L L + + L +LE+LD+ FN L +EGL LK L +
Sbjct: 33 KIENLDSLTSLRELDLYDNQIRKLENLHHLPELEQLDVSFNILRKVEGLEQLTRLKKLFL 92
Query: 71 VENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKE 130
+ NK+ S+ + L +L G N+++ ++ + L+++++L L N+I + NLD +
Sbjct: 93 LHNKISSIANLDHFKCLEMLELGSNRIRVIENLDGLTSLQSLFLGTNKITKLQNLDGLHN 152
Query: 131 LNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSL 190
L L + N I K+ E L+ + S+ +L LSH +E I+ L+ +LT L +A N IK +
Sbjct: 153 LTILSIQSNRITKL-EGLQNLISLKELYLSHNGIEVIE-GLENNKKLTTLDIAANRIKKI 210
Query: 191 PEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALP 250
E + H ++L+ + +N I WSD+ LK+ L + L+ NP+ + + RKI ALP
Sbjct: 211 -ENISHLTELQEFWMNDNQIDNWSDLDELKNAKSLETVYLERNPLQKDPQYRRKIMLALP 269
Query: 251 KLQVFNA 257
++ +A
Sbjct: 270 TVRQIDA 276
Score = 80.9 bits (198), Expect = 8e-14
Identities = 46/154 (29%), Positives = 86/154 (54%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L S ++ N +S+ SL L ++ + L + L L ++ N +T LEGL+
Sbjct: 111 MLELGSNRIRVIENLDGLTSLQSLFLGTNKITKLQNLDGLHNLTILSIQSNRITKLEGLQ 170
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
++LK L + N +E +EG++ KLT L+ N++K ++ I L+ ++ +NDN+I
Sbjct: 171 NLISLKELYLSHNGIEVIEGLENNKKLTTLDIAANRIKKIENISHLTELQEFWMNDNQID 230
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKV 151
+ +LD++ K L T+ L +NP++K + +K+
Sbjct: 231 NWSDLDELKNAKSLETVYLERNPLQKDPQYRRKI 264
Score = 72.0 bits (175), Expect = 4e-11
Identities = 56/173 (32%), Positives = 96/173 (55%), Gaps = 8/173 (4%)
Query: 68 LSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQ 127
+ +V ++ +EG++ L K L+ +N LK ++ + SL+++R L L DN+I + NL
Sbjct: 2 VDLVHCRIGKIEGLEVLRKAKTLSLRQNLLKKIENLDSLTSLRELDLYDNQIRKLENLHH 61
Query: 128 MKELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKF-CVELTELRLAHND 186
+ EL L +S N +RK+ E L+++ + KL L H ++ I F C+E+ L L N
Sbjct: 62 LPELEQLDVSFNILRKV-EGLEQLTRLKKLFLLHNKISSIANLDHFKCLEM--LELGSNR 118
Query: 187 IKSLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNE 239
I+ + E L + L++L LG N I K ++ L +LT L++Q N + E
Sbjct: 119 IRVI-ENLDGLTSLQSLFLGTNKITKLQNLDGLHNLT---ILSIQSNRITKLE 167
>emb|CAE60033.1| Hypothetical protein CBG03541 [Caenorhabditis briggsae]
Length = 1201
Score = 117 bits (292), Expect = 1e-24
Identities = 84/290 (28%), Positives = 144/290 (48%), Gaps = 9/290 (3%)
Query: 19 SSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESL 78
S+++ L+L + + L + + LE LD+ +N +T +EGL LK L +V NK+ +
Sbjct: 19 SALTHLNLNDNQIEKIENLETLSNLEFLDVSYNRITKIEGLSGLAKLKELHLVHNKIVVI 78
Query: 79 EGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSK 138
EG++ T L L G N+++ ++ +G LS +R L L N+I I NLD++ L L L
Sbjct: 79 EGLEENTCLEYLELGDNRIRKIENLGHLSKLRRLFLGANQIRKIENLDELSTLRELSLPG 138
Query: 139 NPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNS 198
N I+ I E L K+ + LS++ + ID L L L L N I+ L E +
Sbjct: 139 NAIQVI-EGLDKLSGLRSLSVAQNGIRKID-GLSGLTSLVSLDLNDNIIEKL-ENVEQFK 195
Query: 199 KLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIR-KIKNALPKLQVFNA 257
+ NL L N + W D+ L + +L L L+ NP+ +++ R ++K LP++++ +
Sbjct: 196 GVANLMLRKNKLDSWHDLYQLLEMKELTALTLEMNPIYSSDYTYRNRMKQILPEIKILDG 255
Query: 258 KPIDKDTKNEKGHMTDDAHDFSFDHVDQ-----NEDDHLEAADKRKSNKK 302
P + + D +D +V + ++ + A D S KK
Sbjct: 256 FPTTWRQGDAYQPLDSDFYDMGDSYVTEPPFVHHDTGPILAIDCHPSGKK 305
>ref|XP_318952.2| ENSANGP00000015763 [Anopheles gambiae str. PEST]
gi|55236061|gb|EAA14303.3| ENSANGP00000015763 [Anopheles
gambiae str. PEST]
Length = 321
Score = 116 bits (290), Expect = 2e-24
Identities = 78/249 (31%), Positives = 137/249 (54%), Gaps = 3/249 (1%)
Query: 9 VLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWL 68
+ K N + +S+ L L ++++ L + LE LD+ FN L ++ L A L+ L
Sbjct: 72 IKKIENLDHLTSLLELELYDNQITELENLDNLVNLEMLDVSFNRLHQIKNLSALTNLRKL 131
Query: 69 SVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQM 128
+ N++ +E + + LT+L G NK++ ++ + +LS++ L L N+I I NLD++
Sbjct: 132 FLCANRISLIENLDHFSSLTMLELGDNKIRKIENLDNLSSLTHLYLGKNKITKIENLDKL 191
Query: 129 KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIK 188
+L L L N + KI E L ++ ++T+L LS +E I+ +L +L L LA N +K
Sbjct: 192 VKLECLSLQCNRLTKI-ENLDQLVNLTELYLSENGIETIE-NLDQNKQLETLDLAKNRVK 249
Query: 189 SLPEELMHNSKLRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNA 248
+ E + H L + +N +++W+ + L S KL + L+ NPVA++ RK+K A
Sbjct: 250 RI-ENIEHLEMLEEFWMNDNGVSEWTCVDKLASNKKLATVYLERNPVASDVNYRRKLKLA 308
Query: 249 LPKLQVFNA 257
+P LQ +A
Sbjct: 309 VPWLQKIDA 317
Score = 72.8 bits (177), Expect = 2e-11
Identities = 52/174 (29%), Positives = 86/174 (48%), Gaps = 9/174 (5%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
M L ++ K N N SS++ L+L ++ + L KLE L L+ N LT +E L
Sbjct: 152 MLELGDNKIRKIENLDNLSSLTHLYLGKNKITKIENLDKLVKLECLSLQCNRLTKIENLD 211
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L L + EN +E++E + +L L+ KN++K ++ I L + +NDN +
Sbjct: 212 QLVNLTELYLSENGIETIENLDQNKQLETLDLAKNRVKRIENIEHLEMLEEFWMNDNGVS 271
Query: 121 SICNLDQM---KELNTLVLSKNPIRKIGEALKKVKSITKLSLSHCQLEGIDTSL 171
+D++ K+L T+ L +NP+ V KL L+ L+ ID +L
Sbjct: 272 EWTCVDKLASNKKLATVYLERNPV------ASDVNYRRKLKLAVPWLQKIDATL 319
>ref|XP_667696.1| leucine rich repeat protein [Cryptosporidium hominis]
gi|54658852|gb|EAL37464.1| leucine rich repeat protein
[Cryptosporidium hominis]
Length = 310
Score = 114 bits (285), Expect = 7e-24
Identities = 81/259 (31%), Positives = 130/259 (49%), Gaps = 18/259 (6%)
Query: 24 LHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQG 83
L L ++ + L L K+D+ N L S+ L+ C L +++ N L S++ I
Sbjct: 25 LKLRELEITGIDELTQAKNLRKIDVSKNKLESISFLKYCWNLTDINLSNNNLTSVDEICN 84
Query: 84 LTKLTVLNAGKNKLKSMDEI----GSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
L + +LN N +KS++ + G +++ LI N N+I I +L KEL T++LS N
Sbjct: 85 LENVKILNISNNSIKSIENLCRSPGLQKSLKVLIANHNKIRYIPDLSHFKELETVILSHN 144
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+I + K+SLS+ L+ SL F + + ELRL +N I SLP ++ +
Sbjct: 145 EAIEIENPKNHCPKLKKMSLSNNCLKQFPFSLNFNM-VQELRLNNNKILSLPSDITYMGN 203
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNE----------KVIRKIKNAL 249
++ L+LG+N I ++I L SLTKL++LN+ NP +E K + IK L
Sbjct: 204 IKILELGHNFI---TEIDPLLSLTKLKSLNISKNPCIISEGTEGDTSNSIKTLNLIKEKL 260
Query: 250 PKLQVFNAKPIDKDTKNEK 268
P L N I K ++
Sbjct: 261 PSLDSLNGSFISKSNDKKR 279
>gb|AAH75482.1| Protein phosphatase 1, regulatory subunit 7 [Xenopus tropicalis]
gi|55742306|ref|NP_001006731.1| protein phosphatase 1,
regulatory subunit 7 [Xenopus tropicalis]
Length = 346
Score = 114 bits (284), Expect = 9e-24
Identities = 74/238 (31%), Positives = 131/238 (54%), Gaps = 3/238 (1%)
Query: 20 SISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLE 79
+++ L L + + L + L+ LDL FN L +EGL + L+ L +V NK+ +E
Sbjct: 107 TLTELDLYDNQIRKIGNLETLRDLQILDLSFNLLRRIEGLESLSHLQRLYLVNNKISRIE 166
Query: 80 GIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
LT+L +L G N+L+ ++ + SL + +L L N+I + NL+ + L L + N
Sbjct: 167 NFGTLTQLRLLELGSNRLRVIENLDSLRELDSLFLGKNKITKLQNLETLTNLTVLSVQSN 226
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+ KI E L+ + ++ +L LS ++ I+ L+ +LT L LA N IK + E + H S+
Sbjct: 227 RLTKI-EGLQNLVNLRELYLSDNGIQVIE-GLENNNKLTTLDLASNRIKRI-ENIKHLSE 283
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNEKVIRKIKNALPKLQVFNA 257
L+ + +N++ WSD++ L L+ + L+ NP+ + + RKI ALP ++ +A
Sbjct: 284 LQEFWMNDNLVENWSDLEELSGAPGLQTVYLERNPLQKDAQYRRKIMLALPSVRQIDA 341
Score = 71.6 bits (174), Expect = 5e-11
Identities = 41/154 (26%), Positives = 83/154 (53%), Gaps = 3/154 (1%)
Query: 1 MTRLSSEQVLKDNNAVNPSSISSLHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLR 60
+ L S ++ N + + SL L ++ + L + L L ++ N LT +EGL+
Sbjct: 176 LLELGSNRLRVIENLDSLRELDSLFLGKNKITKLQNLETLTNLTVLSVQSNRLTKIEGLQ 235
Query: 61 ACVTLKWLSVVENKLESLEGIQGLTKLTVLNAGKNKLKSMDEIGSLSTIRALILNDNEIV 120
V L+ L + +N ++ +EG++ KLT L+ N++K ++ I LS ++ +NDN +
Sbjct: 236 NLVNLRELYLSDNGIQVIEGLENNNKLTTLDLASNRIKRIENIKHLSELQEFWMNDNLVE 295
Query: 121 SICNLDQMK---ELNTLVLSKNPIRKIGEALKKV 151
+ +L+++ L T+ L +NP++K + +K+
Sbjct: 296 NWSDLEELSGAPGLQTVYLERNPLQKDAQYRRKI 329
>emb|CAD98612.1| conserved hypothetical Leucine Rich Repeat protein [Cryptosporidium
parvum]
Length = 310
Score = 112 bits (281), Expect = 2e-23
Identities = 81/259 (31%), Positives = 129/259 (49%), Gaps = 18/259 (6%)
Query: 24 LHLTHKALSDVSCLASFNKLEKLDLKFNNLTSLEGLRACVTLKWLSVVENKLESLEGIQG 83
L L ++ + L L K+D+ N L S+ L+ C L +++ N L S++ I
Sbjct: 25 LKLRELEITGIDELTQAKNLRKIDVSKNKLESVSFLKYCWNLTDINLSNNNLTSVDEICN 84
Query: 84 LTKLTVLNAGKNKLKSMDEI----GSLSTIRALILNDNEIVSICNLDQMKELNTLVLSKN 139
L + +LN N +KS++ + G +++ LI N N+I I +L KEL T++LS N
Sbjct: 85 LENVKILNISNNSIKSIENLCRSPGLQKSLKVLIANHNKIRYIPDLSHFKELETVILSHN 144
Query: 140 PIRKIGEALKKVKSITKLSLSHCQLEGIDTSLKFCVELTELRLAHNDIKSLPEELMHNSK 199
+I + K+SLS+ L+ SL F + + ELRL +N I SLP ++ +
Sbjct: 145 EAIEIENPKNHCPKLKKMSLSNNCLKQFPFSLNFNM-VQELRLNNNKILSLPSDITYMGN 203
Query: 200 LRNLDLGNNVIAKWSDIKVLKSLTKLRNLNLQGNPVATNE----------KVIRKIKNAL 249
++ L+LG+N I ++I L SLTKL++LN+ NP E K + IK L
Sbjct: 204 IKILELGHNFI---TEIDPLLSLTKLKSLNISKNPCIIAEGTEGDNSNSIKTLNLIKEKL 260
Query: 250 PKLQVFNAKPIDKDTKNEK 268
P L N I K ++
Sbjct: 261 PSLDSLNGSFISKSNDKKR 279
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.308 0.128 0.345
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 758,243,522
Number of Sequences: 2540612
Number of extensions: 32557227
Number of successful extensions: 256481
Number of sequences better than 10.0: 10645
Number of HSP's better than 10.0 without gapping: 2232
Number of HSP's successfully gapped in prelim test: 8907
Number of HSP's that attempted gapping in prelim test: 176871
Number of HSP's gapped (non-prelim): 42335
length of query: 468
length of database: 863,360,394
effective HSP length: 132
effective length of query: 336
effective length of database: 527,999,610
effective search space: 177407868960
effective search space used: 177407868960
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC146332.10


