

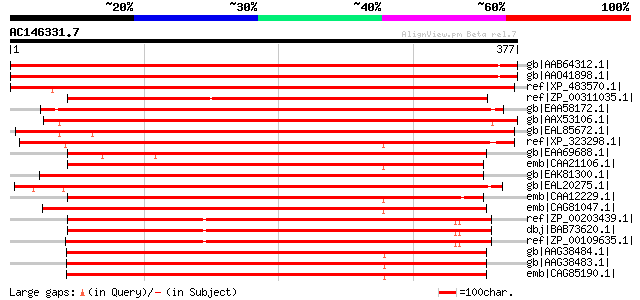
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146331.7 - phase: 0
(377 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAB64312.1| biotin synthase (Bio B) [Arabidopsis thaliana] gi... 615 e-175
gb|AAO41898.1| putative biotin synthase (Bio B) [Arabidopsis tha... 614 e-174
ref|XP_483570.1| putative biotin synthase [Oryza sativa (japonic... 600 e-170
ref|ZP_00311035.1| COG0502: Biotin synthase and related enzymes ... 426 e-118
gb|EAA58172.1| hypothetical protein AN6643.2 [Aspergillus nidula... 406 e-112
gb|AAX53106.1| biotin synthase [Aspergillus niger] 402 e-111
gb|EAL85672.1| biotin synthase, putative [Aspergillus fumigatus ... 397 e-109
ref|XP_323298.1| hypothetical protein [Neurospora crassa] gi|289... 394 e-108
gb|EAA69688.1| hypothetical protein FG00278.1 [Gibberella zeae P... 390 e-107
emb|CAA21106.1| bio2 [Schizosaccharomyces pombe] gi|2995363|emb|... 387 e-106
gb|EAK81300.1| hypothetical protein UM00315.1 [Ustilago maydis 5... 385 e-105
gb|EAL20275.1| hypothetical protein CNBF0870 [Cryptococcus neofo... 382 e-105
emb|CAA12229.1| Biotin synthase [Schizosaccharomyces pombe] 377 e-103
emb|CAG81047.1| unnamed protein product [Yarrowia lipolytica CLI... 376 e-103
ref|ZP_00203439.1| COG0502: Biotin synthase and related enzymes ... 372 e-101
dbj|BAB73620.1| biotin synthase [Nostoc sp. PCC 7120] gi|1722941... 371 e-101
ref|ZP_00109635.1| COG0502: Biotin synthase and related enzymes ... 368 e-100
gb|AAG38484.1| biotin synthase short form [Pichia jadinii] 366 e-100
gb|AAG38483.1| biotin synthase long form [Pichia jadinii] 366 e-100
emb|CAG85190.1| unnamed protein product [Debaryomyces hansenii C... 365 1e-99
>gb|AAB64312.1| biotin synthase (Bio B) [Arabidopsis thaliana]
gi|1705463|sp|P54967|BIOB_ARATH Biotin synthase (Biotin
synthetase) gi|1403662|gb|AAC49445.1| BIO2 protein
gi|15224273|ref|NP_181864.1| biotin synthase (BioB)
(BIO2) [Arabidopsis thaliana] gi|1769457|gb|AAB39953.1|
biotin synthase [Arabidopsis thaliana]
gi|1045316|gb|AAA80226.1| biotin sythase
gi|1589016|prf||2209438A biotin synthase
Length = 378
Score = 615 bits (1587), Expect = e-175
Identities = 297/378 (78%), Positives = 342/378 (89%), Gaps = 2/378 (0%)
Query: 1 MFWLRPILRSQSRSSI-WVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSP 59
M +R + RSQ R S+ LQ + +S+ SAA+ +AE+TI+ GPRNDW++DE+KS+YDSP
Sbjct: 1 MMLVRSVFRSQLRPSVSGGLQSASCYSSLSAASAEAERTIREGPRNDWSRDEIKSVYDSP 60
Query: 60 ILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKD 119
+LDLLFHGAQVHRH HNFREVQQCTLLS+KTGGCSEDCSYCPQSSRY TG+K Q+L++KD
Sbjct: 61 LLDLLFHGAQVHRHVHNFREVQQCTLLSIKTGGCSEDCSYCPQSSRYSTGVKAQRLMSKD 120
Query: 120 AVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKD 179
AV+ AA KAKEAGSTRFCMGAAWRDTIGRKTNF+QILEY+KEI+GMGMEVCCTLGM++K
Sbjct: 121 AVIDAAKKAKEAGSTRFCMGAAWRDTIGRKTNFSQILEYIKEIRGMGMEVCCTLGMIEKQ 180
Query: 180 QAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGE 239
QA ELKKAGLTAYNHNLDTSREYYPN+ITTR+YD+RL+TL VRDAGINVCSGGIIGLGE
Sbjct: 181 QALELKKAGLTAYNHNLDTSREYYPNVITTRSYDDRLETLSHVRDAGINVCSGGIIGLGE 240
Query: 240 AEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKA 299
AE+DR+GLLHTL+TLP+HPESVPINAL+AVKGTPL+DQKPVEIWEMIRMI TARI MPKA
Sbjct: 241 AEEDRIGLLHTLATLPSHPESVPINALLAVKGTPLEDQKPVEIWEMIRMIGTARIVMPKA 300
Query: 300 MVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSL 359
MVRLSAGRVRFS+ EQALCFLAGANSIF GEKLLTT NNDFDADQLMFK LGL+PK PS
Sbjct: 301 MVRLSAGRVRFSMSEQALCFLAGANSIFTGEKLLTTPNNDFDADQLMFKTLGLIPKPPSF 360
Query: 360 DDDETNEAENYKEAASSS 377
+D+ +E+EN ++ AS+S
Sbjct: 361 SEDD-SESENCEKVASAS 377
>gb|AAO41898.1| putative biotin synthase (Bio B) [Arabidopsis thaliana]
Length = 378
Score = 614 bits (1583), Expect = e-174
Identities = 296/378 (78%), Positives = 342/378 (90%), Gaps = 2/378 (0%)
Query: 1 MFWLRPILRSQSRSSI-WVLQHCNSFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSP 59
M +R + RSQ R S+ LQ + +S+ SAA+ +AE+TI+ GPRNDW++DE+KS+YDSP
Sbjct: 1 MMLVRSVFRSQLRPSVSGGLQSASCYSSLSAASAEAERTIREGPRNDWSRDEIKSVYDSP 60
Query: 60 ILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKD 119
+LDLLFHGAQVHRH HNFREVQQCTLLS+KTGGCSEDCSYCPQSSRY TG+K Q+L++KD
Sbjct: 61 LLDLLFHGAQVHRHVHNFREVQQCTLLSIKTGGCSEDCSYCPQSSRYSTGVKAQRLMSKD 120
Query: 120 AVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKD 179
AV+ AA KAKEAGSTRFCMGAAWRDTIGRKTNF+QILEY+KEI+GMGMEVCCTLGM+++
Sbjct: 121 AVIDAAKKAKEAGSTRFCMGAAWRDTIGRKTNFSQILEYIKEIRGMGMEVCCTLGMIEEQ 180
Query: 180 QAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGE 239
QA ELKKAGLTAYNHNLDTSREYYPN+ITTR+YD+RL+TL VRDAGINVCSGGIIGLGE
Sbjct: 181 QALELKKAGLTAYNHNLDTSREYYPNVITTRSYDDRLETLSHVRDAGINVCSGGIIGLGE 240
Query: 240 AEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKA 299
AE+DR+GLLHTL+TLP+HPESVPINAL+AVKGTPL+DQKPVEIWEMIRMI TARI MPKA
Sbjct: 241 AEEDRIGLLHTLATLPSHPESVPINALLAVKGTPLEDQKPVEIWEMIRMIGTARIVMPKA 300
Query: 300 MVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSL 359
MVRLSAGRVRFS+ EQALCFLAGANSIF GEKLLTT NNDFDADQLMFK LGL+PK PS
Sbjct: 301 MVRLSAGRVRFSMSEQALCFLAGANSIFTGEKLLTTPNNDFDADQLMFKTLGLIPKPPSF 360
Query: 360 DDDETNEAENYKEAASSS 377
+D+ +E+EN ++ AS+S
Sbjct: 361 SEDD-SESENCEKVASAS 377
>ref|XP_483570.1| putative biotin synthase [Oryza sativa (japonica cultivar-group)]
gi|50725683|dbj|BAD33149.1| putative biotin synthase
[Oryza sativa (japonica cultivar-group)]
Length = 381
Score = 600 bits (1547), Expect = e-170
Identities = 295/379 (77%), Positives = 335/379 (87%), Gaps = 4/379 (1%)
Query: 1 MFWLRPILRSQSRSSIWVLQHCNSFSTSSA---AAIQAEKTIQNGPRNDWTKDEVKSIYD 57
M R +LRS+ R ++ + SSA AA +AE+ +++GPRNDW++ E++++YD
Sbjct: 2 MLLARAVLRSRLRCPPLSAAAASTAAFSSAPSPAAAEAERAVRDGPRNDWSRPEIQAVYD 61
Query: 58 SPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLN 117
SP+LDLLFHGAQVHR H FREVQQCTLLS+KTGGCSEDCSYCPQSSRY TGLK QKL+N
Sbjct: 62 SPLLDLLFHGAQVHRSVHKFREVQQCTLLSIKTGGCSEDCSYCPQSSRYSTGLKAQKLMN 121
Query: 118 KDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLD 177
KDAVL+AA KAKEAGSTRFCMGAAWR+TIGRKTNFNQILEYVK+I+GMGMEVCCTLGML+
Sbjct: 122 KDAVLEAAKKAKEAGSTRFCMGAAWRETIGRKTNFNQILEYVKDIRGMGMEVCCTLGMLE 181
Query: 178 KDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGL 237
K QA ELKKAGLTAYNHNLDTSREYYPNII+TR+YD+RL+TLE VR+AGI++CSGGIIGL
Sbjct: 182 KQQAEELKKAGLTAYNHNLDTSREYYPNIISTRSYDDRLQTLEHVREAGISICSGGIIGL 241
Query: 238 GEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMP 297
GEAE+DRVGLLHTL+TLP HPESVPINAL+AVKGTPLQDQKPVEIWEMIRMIATARI MP
Sbjct: 242 GEAEEDRVGLLHTLATLPAHPESVPINALVAVKGTPLQDQKPVEIWEMIRMIATARIVMP 301
Query: 298 KAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAP 357
KAMVRLSAGRVRFS+PEQALCFLAGANSIFAGEKLLTT NNDFDADQ MFK+LGL+PKAP
Sbjct: 302 KAMVRLSAGRVRFSMPEQALCFLAGANSIFAGEKLLTTTNNDFDADQAMFKILGLIPKAP 361
Query: 358 SLDDD-ETNEAENYKEAAS 375
S D+ + E +EAAS
Sbjct: 362 SFGDEAPAADTERCEEAAS 380
>ref|ZP_00311035.1| COG0502: Biotin synthase and related enzymes [Cytophaga
hutchinsonii]
Length = 337
Score = 426 bits (1095), Expect = e-118
Identities = 204/312 (65%), Positives = 259/312 (82%), Gaps = 1/312 (0%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQS 103
RN+WTK+E+ +IY+SPILDL++ GA VHR H+ +EVQ CTLLS+KTGGC EDCSYCPQ+
Sbjct: 5 RNNWTKEEISAIYNSPILDLMYRGATVHREFHDPQEVQVCTLLSIKTGGCPEDCSYCPQA 64
Query: 104 SRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIK 163
+RY T +K +KL++ VL +A++AKE+GSTRFCMGAAWR+ K +F+++++ VK +
Sbjct: 65 ARYHTDVKVEKLMDVKDVLNSALEAKESGSTRFCMGAAWREVRDNK-DFDKVIDMVKGVS 123
Query: 164 GMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVR 223
MGMEVCCTLGML +QA +LK AGL AYNHNLDTS E+Y +ITTRTYD+RL+TL+ VR
Sbjct: 124 TMGMEVCCTLGMLTPEQADKLKDAGLYAYNHNLDTSAEHYDKVITTRTYDDRLETLDNVR 183
Query: 224 DAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIW 283
+A I+VCSGGIIG+GE+ DRVG+LHTL+ + HPESVP+NAL+ V+GTPL+DQ V +W
Sbjct: 184 NAKISVCSGGIIGMGESHGDRVGMLHTLANMVEHPESVPVNALVPVEGTPLEDQPRVSVW 243
Query: 284 EMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDAD 343
EM+RMIATARI MPKAMVRLSAGRVR + EQALCFLAGANSIFAG+KLLTT N + +AD
Sbjct: 244 EMVRMIATARIIMPKAMVRLSAGRVRMNTEEQALCFLAGANSIFAGDKLLTTPNPEVNAD 303
Query: 344 QLMFKVLGLLPK 355
+ MF+VL L P+
Sbjct: 304 KEMFQVLNLKPR 315
>gb|EAA58172.1| hypothetical protein AN6643.2 [Aspergillus nidulans FGSC A4]
gi|67540946|ref|XP_664247.1| hypothetical protein
AN6643_2 [Aspergillus nidulans FGSC A4]
gi|49099578|ref|XP_410780.1| hypothetical protein
AN6643.2 [Aspergillus nidulans FGSC A4]
Length = 393
Score = 406 bits (1043), Expect = e-112
Identities = 198/344 (57%), Positives = 252/344 (72%), Gaps = 3/344 (0%)
Query: 24 SFSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQC 83
SF++ A+Q E PR +WT+DEV+ IY++P+ L + A VHR H+ +Q C
Sbjct: 28 SFASRIPPALQ-EAVAATAPRTNWTRDEVQQIYETPLNQLTYAAAAVHRRFHDPSAIQMC 86
Query: 84 TLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWR 143
TL+++KTGGCSEDCSYC QSSRY TGLK K+ D VL+ A AK GSTRFCMGAAWR
Sbjct: 87 TLMNIKTGGCSEDCSYCAQSSRYSTGLKATKMSPVDDVLEKARIAKANGSTRFCMGAAWR 146
Query: 144 DTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYY 203
D GRKT+ + + V ++ MGMEVC TLGM+D DQA ELK AGLTAYNHNLDTSRE+Y
Sbjct: 147 DMRGRKTSLKNVKQMVSGVREMGMEVCVTLGMIDADQAKELKDAGLTAYNHNLDTSREFY 206
Query: 204 PNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPI 263
P IITTR+YDERLKTL VRDAGINVCSGGI+GLGEA+ DR+GL+HT+S+LP+HPES P+
Sbjct: 207 PTIITTRSYDERLKTLSHVRDAGINVCSGGILGLGEADSDRIGLIHTVSSLPSHPESFPV 266
Query: 264 NALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGA 323
NAL+ +KGTPL D+K + +++R +ATARI +P +VRL+AGR+ + +Q CF+AGA
Sbjct: 267 NALVPIKGTPLGDRKMISFDKLLRTVATARIVLPATIVRLAAGRISLTEEQQVACFMAGA 326
Query: 324 NSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSLDDDETNEA 367
N++F GEK+LTT N +D D+ MF G P + ETN A
Sbjct: 327 NAVFTGEKMLTTDCNGWDEDRAMFDRWGFYPMRSF--EKETNAA 368
>gb|AAX53106.1| biotin synthase [Aspergillus niger]
Length = 384
Score = 402 bits (1034), Expect = e-111
Identities = 201/364 (55%), Positives = 258/364 (70%), Gaps = 12/364 (3%)
Query: 26 STSSAAAIQA-------EKTIQNGPRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFR 78
+T SAAAI + E T PR +WT++E++ IY++P+ L + A VHR H+
Sbjct: 21 TTPSAAAIASRIPPALREATAATAPRTNWTREEIQQIYETPLNQLTYAAAAVHRRFHDPS 80
Query: 79 EVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCM 138
+Q CTL+++KTGGCSEDCSYC QSSRY TGLK K+ D VL+AA AK GSTRFCM
Sbjct: 81 AIQMCTLMNIKTGGCSEDCSYCAQSSRYQTGLKATKMSPVDEVLEAARIAKSNGSTRFCM 140
Query: 139 GAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDT 198
GAAWRD GRKT+ I V ++GMGMEVC TLGM+D+ QA ELK AGLTAYNHNLDT
Sbjct: 141 GAAWRDMRGRKTSLKNIKSMVSGVRGMGMEVCVTLGMIDEAQAKELKDAGLTAYNHNLDT 200
Query: 199 SREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHP 258
SRE+YP IITTR+YDERLKTLE VRDAGINVCSGGI+GLGE ++DRVGL++T+S L HP
Sbjct: 201 SREFYPEIITTRSYDERLKTLENVRDAGINVCSGGILGLGEKDEDRVGLIYTMSGLEKHP 260
Query: 259 ESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALC 318
ES P+NAL+ + GTPL ++K + +++R +ATAR+ MP +VRL+AGR+ S +Q C
Sbjct: 261 ESFPVNALVPIPGTPLGERKMISFDKILRTVATARVVMPATIVRLAAGRISLSEEQQVAC 320
Query: 319 FLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAP-----SLDDDETNEAENYKEA 373
F+AGAN++F GEK+LTT N +D D+ MF+ G P D+ + E E +A
Sbjct: 321 FMAGANAVFTGEKMLTTECNGWDEDRAMFEKWGFYPMKSFEGEHVKDEVKVKEVEGTAQA 380
Query: 374 ASSS 377
A S+
Sbjct: 381 AVSA 384
>gb|EAL85672.1| biotin synthase, putative [Aspergillus fumigatus Af293]
Length = 415
Score = 397 bits (1021), Expect = e-109
Identities = 204/391 (52%), Positives = 267/391 (68%), Gaps = 20/391 (5%)
Query: 5 RPILRSQSRSSIWVLQHCNSFSTSSAAAIQA---EKTIQNGPRNDWTKDEVKSIYDSPI- 60
R + R+ R S +Q S ++ +A I E T PR +W+++EV+ IY++P+
Sbjct: 7 RSLHRALLRRSYGTVQAPPSAASLAANRIPPALQEATAATAPRTNWSREEVQQIYETPLS 66
Query: 61 ----------------LDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSS 104
+D F A VHR H+ +Q CTL+++KTGGCSEDCSYC QSS
Sbjct: 67 QLTYAAVGRSLISYFSMDTDFCKATVHRRFHDPAAIQMCTLMNIKTGGCSEDCSYCAQSS 126
Query: 105 RYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKG 164
RY+TGLK K+ D VL+AA AK+ GSTRFCMGAAWRD GRKT+ I E V ++
Sbjct: 127 RYNTGLKATKMSPVDEVLKAARIAKDNGSTRFCMGAAWRDMRGRKTSLKNIKEMVSGVRA 186
Query: 165 MGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRD 224
MGMEVC TLGM+D++QA ELK AGLTAYNHNLDTSRE+YP+IITTR+YDERL+TL VRD
Sbjct: 187 MGMEVCVTLGMIDENQAKELKDAGLTAYNHNLDTSREFYPSIITTRSYDERLRTLSHVRD 246
Query: 225 AGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWE 284
AGINVCSGGI+GLGEA+ DR+GL+HT++ LP+HPES P+NAL+ +KGTPL D+K + +
Sbjct: 247 AGINVCSGGILGLGEADSDRIGLIHTVANLPSHPESFPVNALVPIKGTPLGDRKMIPFDK 306
Query: 285 MIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQ 344
++R IATARI +P +VRL+AGR+ S +Q CF+AGAN++F GEK+LTT N +D D+
Sbjct: 307 LLRTIATARIVLPATIVRLAAGRISLSEEQQVACFMAGANAVFTGEKMLTTDCNGWDEDR 366
Query: 345 LMFKVLGLLPKAPSLDDDETNEAENYKEAAS 375
MF+ G P + T A AA+
Sbjct: 367 EMFERWGFYPMRSFEKEGSTQPAVEAPSAAA 397
>ref|XP_323298.1| hypothetical protein [Neurospora crassa] gi|28917633|gb|EAA27328.1|
hypothetical protein [Neurospora crassa]
Length = 439
Score = 394 bits (1013), Expect = e-108
Identities = 203/397 (51%), Positives = 266/397 (66%), Gaps = 33/397 (8%)
Query: 8 LRSQSRSSIWVLQHCNSFSTSSAAAIQAEKTIQ------------------NGPRNDWTK 49
LR QSR S + +S + S A I A Q + PRNDWTK
Sbjct: 28 LRFQSRLSHAQFRSLSSAAGSQVANIPATNKYQKLDSFAGRNSLLQATIAADAPRNDWTK 87
Query: 50 DEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTG 109
+E++ IY +P+++L++ + +HR + VQ CTL+++KTGGCSEDCSYC QSSRY TG
Sbjct: 88 EEIREIYQTPLMELVYSASTLHRRFFDPASVQLCTLMNIKTGGCSEDCSYCAQSSRYKTG 147
Query: 110 LKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEV 169
+K K+ + D VLQAA AKE GSTRFCMGAAWRD GRKTN I V ++ +GME
Sbjct: 148 VKATKISSVDEVLQAAKIAKENGSTRFCMGAAWRDMRGRKTNLKNIKAMVSGVRELGMEA 207
Query: 170 CCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINV 229
C TLGM+D+ QA EL+ AGLTAYNHN+DTSREYYPN+ITTRTYDERLKT+ VR AGI+V
Sbjct: 208 CVTLGMIDESQAAELRDAGLTAYNHNVDTSREYYPNVITTRTYDERLKTINNVRQAGIHV 267
Query: 230 CSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQD-----------QK 278
CSGGI+GLGE + DRV L+T++T+ +HPES PINAL+ +KGTPL D +
Sbjct: 268 CSGGILGLGEDDTDRVSFLYTVATMESHPESFPINALVPIKGTPLGDASAAKSETGEPES 327
Query: 279 PVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANN 338
V + +RM+ATAR+ +P +++RL+AGR+ + EQALCF+AGAN+IF GEK+LTT +
Sbjct: 328 TVTFHDTVRMVATARLVLPGSVIRLAAGRINLTESEQALCFMAGANAIFTGEKMLTTPCS 387
Query: 339 DFDADQLMFKVLGLLPKAPSLDDDETNEAENYKEAAS 375
+D D+ MF GL P P DE + +E + A +
Sbjct: 388 GWDEDKAMFDRWGLRPMLP----DELHGSEKRQHAGT 420
>gb|EAA69688.1| hypothetical protein FG00278.1 [Gibberella zeae PH-1]
gi|46105300|ref|XP_380454.1| hypothetical protein
FG00278.1 [Gibberella zeae PH-1]
Length = 436
Score = 390 bits (1003), Expect = e-107
Identities = 196/339 (57%), Positives = 244/339 (71%), Gaps = 28/339 (8%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHG--------------------------AQVHRHAHNF 77
R+DWTKDE+ +IY P+L+L + + VHR HN
Sbjct: 81 RHDWTKDEIAAIYYQPVLELAYQATADLIPDSNHSIAEQYHRLPRLTLNQSTVHRRWHNP 140
Query: 78 REVQQCTLLSVKTGGCSEDCSYCPQSSRYD--TGLKGQKLLNKDAVLQAAVKAKEAGSTR 135
EVQ CTL+++KTGGC+EDCSYC QS+RY TG+ +++ + ++VL AA AKE GSTR
Sbjct: 141 SEVQLCTLMNIKTGGCTEDCSYCAQSTRYQEGTGVPAKRVESVESVLAAARIAKEKGSTR 200
Query: 136 FCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHN 195
FCMGAAWRD GRK + I V+ +KGMGMEVC TLGM+D +QA ELK AGLTAYNHN
Sbjct: 201 FCMGAAWRDMRGRKNSLKNIKAMVEGVKGMGMEVCVTLGMIDAEQAKELKAAGLTAYNHN 260
Query: 196 LDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLP 255
+DTSRE+YPN+ITTR+YDERL+TL VRDAGINVCSGGI+GLGE+ +DRVGLLHT+STLP
Sbjct: 261 VDTSREFYPNVITTRSYDERLQTLSHVRDAGINVCSGGILGLGESSEDRVGLLHTVSTLP 320
Query: 256 THPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQ 315
+HPES P+NAL+ +KGTPL D+ VE M+R IATARI MP ++R++AGR S +Q
Sbjct: 321 SHPESFPVNALVPIKGTPLGDRPMVEFTSMLRTIATARIIMPSTIIRIAAGRKTMSEEKQ 380
Query: 316 ALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLP 354
ALCF+AGAN+IF GEK+LTT N +D D MF GL P
Sbjct: 381 ALCFMAGANAIFTGEKMLTTECNGWDEDAAMFGRWGLEP 419
>emb|CAA21106.1| bio2 [Schizosaccharomyces pombe] gi|2995363|emb|CAA18303.1| bio2
[Schizosaccharomyces pombe] gi|19075228|ref|NP_587728.1|
hypothetical protein SPCC1235.02 [Schizosaccharomyces
pombe 972h-] gi|12643615|sp|O59778|BIOB_SCHPO Biotin
synthase (Biotin synthetase)
Length = 363
Score = 387 bits (993), Expect = e-106
Identities = 185/311 (59%), Positives = 238/311 (76%), Gaps = 2/311 (0%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQS 103
RN+WT++E++ IYD+P++DL+F A +HR H+ ++VQQCTLLS+KTGGC+EDC YC QS
Sbjct: 20 RNNWTREEIQKIYDTPLIDLIFRAASIHRKFHDPKKVQQCTLLSIKTGGCTEDCKYCAQS 79
Query: 104 SRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIK 163
SRY+TG+K KL+ D VL+ A AK GSTRFCMG+AWRD GR F ILE +KE++
Sbjct: 80 SRYNTGVKATKLMKIDEVLEKAKIAKAKGSTRFCMGSAWRDLNGRNRTFKNILEIIKEVR 139
Query: 164 GMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVR 223
M MEVC TLGML++ QA ELK AGLTAYNHNLDTSREYY II+TRTYDERL T++ +R
Sbjct: 140 SMDMEVCVTLGMLNEQQAKELKDAGLTAYNHNLDTSREYYSKIISTRTYDERLNTIDNLR 199
Query: 224 DAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQD--QKPVE 281
AG+ VCSGGI+GLGE + DRVGL+H+L+T+PTHPESVP N L+ + GTP+ D ++ +
Sbjct: 200 KAGLKVCSGGILGLGEKKHDRVGLIHSLATMPTHPESVPFNLLVPIPGTPVGDAVKERLP 259
Query: 282 IWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFD 341
I +R IATARI MPK ++R +AGR S EQAL F+AGAN++F GEK+LTT +D
Sbjct: 260 IHPFLRSIATARICMPKTIIRFAAGRNTCSESEQALAFMAGANAVFTGEKMLTTPAVSWD 319
Query: 342 ADQLMFKVLGL 352
+D +F GL
Sbjct: 320 SDSQLFYNWGL 330
>gb|EAK81300.1| hypothetical protein UM00315.1 [Ustilago maydis 521]
gi|49067280|ref|XP_397930.1| hypothetical protein
UM00315.1 [Ustilago maydis 521]
Length = 436
Score = 385 bits (989), Expect = e-105
Identities = 186/331 (56%), Positives = 245/331 (73%), Gaps = 1/331 (0%)
Query: 23 NSFSTSSAAAIQAEKTIQNGP-RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQ 81
+S S +SA+ G R++W++ EV+ IYDSP+L+L+F A VHR H VQ
Sbjct: 61 SSLSVASASTAATSLPSNEGAIRHNWSRKEVQRIYDSPLLELVFRSANVHRMHHPVNRVQ 120
Query: 82 QCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAA 141
CTL+++K GGCSEDC YC QSS+Y+T KL + + VL A KAK GSTRFCMGAA
Sbjct: 121 LCTLMNIKEGGCSEDCGYCSQSSKYETPSNASKLEDLETVLTEARKAKANGSTRFCMGAA 180
Query: 142 WRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSRE 201
WR+ GRK FN+IL+ V+EI+GMGMEVC TLGML +QA +LK+AGLTAYNHNLDTSRE
Sbjct: 181 WREVGGRKRGFNRILDMVREIRGMGMEVCTTLGMLTPEQAKQLKEAGLTAYNHNLDTSRE 240
Query: 202 YYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESV 261
YY ++TTR+YD+R+ T+ VR+AGI+VCSGGI+GLGE +DRVGL+ +STLP HPES
Sbjct: 241 YYGKVVTTRSYDDRIGTIANVREAGISVCSGGILGLGEEHEDRVGLIWEMSTLPEHPESF 300
Query: 262 PINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLA 321
P+NAL+A+ GTP++ KP+ EM+R +ATARI +P +++RL+AGR F+ EQA+ FLA
Sbjct: 301 PVNALVAIPGTPMEQNKPITFQEMLRTVATARIVLPGSIIRLAAGRHLFTESEQAMAFLA 360
Query: 322 GANSIFAGEKLLTTANNDFDADQLMFKVLGL 352
GAN+IF GE++LTT + +D D+ M GL
Sbjct: 361 GANAIFTGERMLTTPTSSWDEDKAMLGRWGL 391
>gb|EAL20275.1| hypothetical protein CNBF0870 [Cryptococcus neoformans var.
neoformans B-3501A] gi|57227917|gb|AAW44375.1| biotin
synthase, putative [Cryptococcus neoformans var.
neoformans JEC21] gi|58269052|ref|XP_571682.1| biotin
synthase, putative [Cryptococcus neoformans var.
neoformans JEC21]
Length = 388
Score = 382 bits (982), Expect = e-105
Identities = 186/367 (50%), Positives = 265/367 (71%), Gaps = 5/367 (1%)
Query: 4 LRPILRSQSRSSI--WVLQHCNSFSTSSAAAIQAEKT--IQNGPRNDWTKDEVKSIYDSP 59
LRP+LRS + ++ + F S + Q+EK + R+DW + E++ ++D+P
Sbjct: 7 LRPVLRSTAPTASRGHAVAVDAPFLPPSPSVQQSEKRRYVDGDVRHDWRRSEIQKVFDAP 66
Query: 60 ILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKD 119
++++++ A VHR + +Q CTL+++KTGGC+E+C YC QSS Y T K +L++ +
Sbjct: 67 LMEIIYRAATVHRLHQDASRIQLCTLMNIKTGGCTENCKYCSQSSSYKTPTKASRLVSIE 126
Query: 120 AVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKD 179
VL+AA +AK GSTRFCMGAAWRD GRK+ F +ILE VKE++GMGMEVC TLGML +
Sbjct: 127 PVLEAARQAKANGSTRFCMGAAWRDLAGRKSGFEKILEMVKEVRGMGMEVCTTLGMLSPE 186
Query: 180 QAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGE 239
QA LK+AGL+AYNHNLDTSRE+YP ++T+R+YD+RL T+ VR+AGI+VCSGGI+GLGE
Sbjct: 187 QAIRLKEAGLSAYNHNLDTSREFYPEVVTSRSYDDRLSTIAAVREAGISVCSGGILGLGE 246
Query: 240 AEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIWEMIRMIATARITMPKA 299
++DRVGL+H +S +P HPES P+N L+ + GTPL+ +PV++ ++R IATARI +PK
Sbjct: 247 RDEDRVGLIHEVSRMPEHPESFPVNTLVPIPGTPLEGNEPVKVHTVLRTIATARIVLPKT 306
Query: 300 MVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLPKAPSL 359
++RL+AGR FS EQA+ F+AGAN+IF GEK+LTT + +D D+ M GL + S
Sbjct: 307 IIRLAAGRHEFSETEQAMAFMAGANAIFTGEKMLTTPCSGWDEDKAMLDRWGLRGQR-SF 365
Query: 360 DDDETNE 366
+D E+ E
Sbjct: 366 EDKESLE 372
>emb|CAA12229.1| Biotin synthase [Schizosaccharomyces pombe]
Length = 362
Score = 377 bits (967), Expect = e-103
Identities = 183/311 (58%), Positives = 235/311 (74%), Gaps = 3/311 (0%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQS 103
RN+WT++E++ IYD+P++DL+F A +HR H+ ++VQQCTLLS+KTGGC+EDC YC QS
Sbjct: 20 RNNWTREEIQKIYDTPLIDLIFRAASIHRKFHDPKKVQQCTLLSIKTGGCTEDCKYCAQS 79
Query: 104 SRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIK 163
SRY+TG+K KL+ D VL+ A AK GSTRFCMG+AWRD GR F ILE +KE++
Sbjct: 80 SRYNTGVKATKLMKIDEVLEKAKIAKAKGSTRFCMGSAWRDLNGRNRTFKNILEIIKEVR 139
Query: 164 GMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVR 223
M MEVC TLGML++ QA ELK AGLTAYNHNLDTSREYY II+TRTYDERL T++ +R
Sbjct: 140 SMDMEVCVTLGMLNEQQAKELKDAGLTAYNHNLDTSREYYSKIISTRTYDERLNTIDNLR 199
Query: 224 DAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQD--QKPVE 281
AG+ VCSGGI+GLGE + DRVGL+H+L+T+PTHPESVP N L+ + GTP+ D ++ +
Sbjct: 200 KAGLKVCSGGILGLGEKKHDRVGLIHSLATMPTHPESVPFNLLVPIPGTPVGDAVKERLP 259
Query: 282 IWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANNDFD 341
I +R IATARI MPK ++R +AGR S EQAL F+AGAN++F GEK+L D
Sbjct: 260 IHPFLRSIATARICMPKTIIRFAAGRNTCSESEQALAFMAGANAVFTGEKMLLLL-LFLD 318
Query: 342 ADQLMFKVLGL 352
+D +F GL
Sbjct: 319 SDSQLFYNWGL 329
>emb|CAG81047.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50550773|ref|XP_502859.1| hypothetical protein
[Yarrowia lipolytica]
Length = 384
Score = 376 bits (965), Expect = e-103
Identities = 180/335 (53%), Positives = 240/335 (70%), Gaps = 5/335 (1%)
Query: 25 FSTSSAAAIQAEKTIQNGPRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCT 84
F + A+ + T + R +WT+DEV IY+ P++DL+F A VHR HN EVQ CT
Sbjct: 14 FVRNLASHASFQPTTSSAIRTNWTRDEVSKIYNMPLMDLVFKAATVHRQHHNPAEVQLCT 73
Query: 85 LLSVKTGGCSEDCSYCPQSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRD 144
L ++K+GGC+EDC YC QSSRY TG++ +K + V+ AA +AK GSTRFCMGAAWRD
Sbjct: 74 LYNIKSGGCTEDCKYCAQSSRYTTGVEAEKAVKPAEVIAAAKEAKANGSTRFCMGAAWRD 133
Query: 145 TIGRKTNFNQILEYVKEIKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYP 204
GRK + E + + +GME C TLGM+DK QA EL++AGLTAYNHN+DTSRE+YP
Sbjct: 134 MKGRKNALKNVKEMIGAVHELGMESCVTLGMVDKAQAEELRRAGLTAYNHNIDTSREHYP 193
Query: 205 NIITTRTYDERLKTLEFVRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPIN 264
+ITTRT+DERL+T+E VR+AG++VCSGGI+GLGE DD VG L L+T+P+HPES+PIN
Sbjct: 194 KVITTRTFDERLQTIENVREAGVSVCSGGILGLGETADDHVGFLWALATMPSHPESLPIN 253
Query: 265 ALIAVKGTPLQD-----QKPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCF 319
L+ +KGTPL D +K + ++R +ATAR+ MP + +RL+AGR PEQA+CF
Sbjct: 254 RLVPIKGTPLGDDPAMKEKQLTYDTVVRTVATARVVMPGSKIRLAAGRYTMKEPEQAMCF 313
Query: 320 LAGANSIFAGEKLLTTANNDFDADQLMFKVLGLLP 354
+AGAN+IF G+K+LTT N +D D+ M + GL P
Sbjct: 314 MAGANAIFTGKKMLTTMCNGWDEDKAMLEKWGLKP 348
>ref|ZP_00203439.1| COG0502: Biotin synthase and related enzymes [Anabaena variabilis
ATCC 29413]
Length = 335
Score = 372 bits (955), Expect = e-101
Identities = 184/319 (57%), Positives = 236/319 (73%), Gaps = 5/319 (1%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQS 103
R DW E++++YD P+L+L++ A VHR HN +++Q C L+S+KTGGC EDCSYC QS
Sbjct: 7 RYDWHTAEIRAVYDMPLLELIYQAASVHRQFHNPKQIQVCKLISIKTGGCPEDCSYCAQS 66
Query: 104 SRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIK 163
SRY T +K Q LL+K V++ A AK+ G +R CMGAAWR+ + + F+++LE VK++
Sbjct: 67 SRYQTEVKPQALLDKQTVVEIAQNAKQKGVSRVCMGAAWRE-VRDNSQFDRVLEMVKDVT 125
Query: 164 GMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVR 223
MG+EVCCTLGML DQA +L+ AGL AYNHNLDTS +YY IITTRTY +RL T+E VR
Sbjct: 126 DMGLEVCCTLGMLTTDQAKKLETAGLYAYNHNLDTSSDYYSTIITTRTYSDRLNTIENVR 185
Query: 224 DAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIW 283
+ VCSGGI+GLGE+ DDRV +L TL+TL HPESVPIN L V+GTPL+DQ V +W
Sbjct: 186 QTNVTVCSGGILGLGESIDDRVAMLQTLATLNPHPESVPINILSQVEGTPLEDQPDVPVW 245
Query: 284 EMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGE--KLL--TTANND 339
+++RMIATARI MP + VRLSAGR R S EQA CF+AGANSIF+ + K+L TT D
Sbjct: 246 DVVRMIATARIVMPTSDVRLSAGRARLSQVEQAFCFMAGANSIFSSDDNKMLTVTTPCPD 305
Query: 340 FDADQLMFKVLGLLPKAPS 358
+DADQ M +LGL + PS
Sbjct: 306 YDADQEMLNLLGLEMRPPS 324
>dbj|BAB73620.1| biotin synthase [Nostoc sp. PCC 7120] gi|17229413|ref|NP_485961.1|
biotin synthase [Nostoc sp. PCC 7120]
gi|25294063|pir||AC2046 biotin synthase [imported] -
Nostoc sp. (strain PCC 7120)
Length = 335
Score = 371 bits (953), Expect = e-101
Identities = 183/319 (57%), Positives = 237/319 (73%), Gaps = 5/319 (1%)
Query: 44 RNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQS 103
R DW K E++++YD+P+L+L++ A VHR HN +++Q C L+S+KTG C EDCSYC QS
Sbjct: 7 RYDWHKAEIRAVYDTPLLELIYQAASVHRQFHNPKQIQVCKLISIKTGACPEDCSYCAQS 66
Query: 104 SRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEIK 163
SRY T +K Q LL+K V++ A AK+ G +R CMGAAWR+ + + F+++LE VK++
Sbjct: 67 SRYQTEVKPQALLDKQTVVEIAQNAKQKGVSRVCMGAAWRE-VRDNSQFDRVLEMVKDVT 125
Query: 164 GMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFVR 223
MG+EVCCTLGML +QA +L+ AGL AYNHNLDTS +YY IITTRTY +RL T+E VR
Sbjct: 126 DMGLEVCCTLGMLTSEQAKKLETAGLYAYNHNLDTSSDYYSTIITTRTYGDRLNTIENVR 185
Query: 224 DAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVEIW 283
+ VCSGGI+GLGE+ DDRV +L TL+TL HPESVPIN L V+GTPL+DQ V +W
Sbjct: 186 QTNVTVCSGGILGLGESIDDRVAMLQTLATLNPHPESVPINILSQVEGTPLEDQPDVPVW 245
Query: 284 EMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGE--KLL--TTANND 339
+++RMIATARI MP + VRLSAGR R S EQA CF+AGANSIF+ + K+L TT D
Sbjct: 246 DVVRMIATARIVMPTSDVRLSAGRARLSQVEQAFCFMAGANSIFSSDDNKMLTVTTPCPD 305
Query: 340 FDADQLMFKVLGLLPKAPS 358
+DADQ M +LGL + PS
Sbjct: 306 YDADQEMLNLLGLEMRPPS 324
>ref|ZP_00109635.1| COG0502: Biotin synthase and related enzymes [Nostoc punctiforme
PCC 73102]
Length = 335
Score = 368 bits (944), Expect = e-100
Identities = 184/321 (57%), Positives = 236/321 (73%), Gaps = 5/321 (1%)
Query: 42 GPRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCP 101
G R DW + E+++IY++P+L+L++ A VHR H+ ++Q C L+S+KTGGC EDCSYC
Sbjct: 3 GIRYDWQELEIRAIYNTPLLELIYQAASVHRQYHDPTKIQVCKLISIKTGGCPEDCSYCA 62
Query: 102 QSSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKE 161
QSSRY T +K + LL K+ V+ A KAKE G +R CMGAAWR+ + + F ++LE VK+
Sbjct: 63 QSSRYKTEVKAEALLEKETVVNIAQKAKETGVSRICMGAAWRE-VRDNSQFEEVLEMVKD 121
Query: 162 IKGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEF 221
I MG+EVCCTLGML +QA +L++AGL AYNHNLDTS+EYY IITTRTY +RL T+E
Sbjct: 122 ITAMGLEVCCTLGMLTANQARKLEEAGLYAYNHNLDTSQEYYSTIITTRTYSDRLNTIEN 181
Query: 222 VRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQKPVE 281
VR + VCSGGI+GLGE DDRVG+L TL+ L HPESVPIN L V GTPL++Q V
Sbjct: 182 VRQTNVTVCSGGILGLGETVDDRVGMLQTLANLHPHPESVPINILSQVPGTPLENQPDVP 241
Query: 282 IWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGE--KLL--TTAN 337
IW+++RMIATARI MP + VRLSAGR R S EQA CF+AGANSIF+ + K+L TT
Sbjct: 242 IWDIVRMIATARILMPASDVRLSAGRARLSQVEQAFCFMAGANSIFSSDDNKMLTVTTPC 301
Query: 338 NDFDADQLMFKVLGLLPKAPS 358
D+D D+ M +LGL + PS
Sbjct: 302 PDYDTDREMLNLLGLGMRPPS 322
>gb|AAG38484.1| biotin synthase short form [Pichia jadinii]
Length = 352
Score = 366 bits (940), Expect = e-100
Identities = 167/315 (53%), Positives = 235/315 (74%), Gaps = 3/315 (0%)
Query: 43 PRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQ 102
P N WTK+E+K+IYD+P++DL+ + HR EVQ CTL+++KTGGC+EDC YC Q
Sbjct: 23 PVNTWTKEEIKAIYDTPLMDLMHYAQVQHRRFQQPSEVQLCTLMNIKTGGCTEDCKYCAQ 82
Query: 103 SSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEI 162
S RY+TG+K ++++ D V++AA +AK GSTRFCMGAAWR+ GRK+N +I E + +
Sbjct: 83 SQRYNTGVKAERIIQVDEVIEAAKEAKANGSTRFCMGAAWREMKGRKSNLKKIKEMITAV 142
Query: 163 KGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFV 222
+GME C TLGM+DKDQA ELK AGLTAYNHN+DT +E+YP +I+TR++D+RLKT + V
Sbjct: 143 HDLGMESCVTLGMVDKDQATELKSAGLTAYNHNIDTYKEHYPKVISTRSFDDRLKTFKNV 202
Query: 223 RDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQ---KP 279
+ +G+ C+GGI+GLGE ++DRV L+TL+T+ HPES+PIN L+ +KGTP+ ++ K
Sbjct: 203 QGSGLKACTGGILGLGETQEDRVSFLYTLATMDQHPESLPINRLVPIKGTPMYEEVKNKQ 262
Query: 280 VEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANND 339
VE+ E++R IATAR+ MP +++RL+AGR EQ +CF+AG N+IF G+K+LTT N
Sbjct: 263 VEVDEIVRTIATARLVMPTSIIRLAAGRYTMKEAEQVMCFMAGCNAIFTGKKMLTTMCNG 322
Query: 340 FDADQLMFKVLGLLP 354
+D D+ M GL P
Sbjct: 323 WDEDKAMLAKWGLKP 337
>gb|AAG38483.1| biotin synthase long form [Pichia jadinii]
Length = 395
Score = 366 bits (940), Expect = e-100
Identities = 167/315 (53%), Positives = 235/315 (74%), Gaps = 3/315 (0%)
Query: 43 PRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQ 102
P N WTK+E+K+IYD+P++DL+ + HR EVQ CTL+++KTGGC+EDC YC Q
Sbjct: 66 PVNTWTKEEIKAIYDTPLMDLMHYAQVQHRRFQQPSEVQLCTLMNIKTGGCTEDCKYCAQ 125
Query: 103 SSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEI 162
S RY+TG+K ++++ D V++AA +AK GSTRFCMGAAWR+ GRK+N +I E + +
Sbjct: 126 SQRYNTGVKAERIIQVDEVIEAAKEAKANGSTRFCMGAAWREMKGRKSNLKKIKEMITAV 185
Query: 163 KGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEFV 222
+GME C TLGM+DKDQA ELK AGLTAYNHN+DT +E+YP +I+TR++D+RLKT + V
Sbjct: 186 HDLGMESCVTLGMVDKDQATELKSAGLTAYNHNIDTYKEHYPKVISTRSFDDRLKTFKNV 245
Query: 223 RDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQ---KP 279
+ +G+ C+GGI+GLGE ++DRV L+TL+T+ HPES+PIN L+ +KGTP+ ++ K
Sbjct: 246 QGSGLKACTGGILGLGETQEDRVSFLYTLATMDQHPESLPINRLVPIKGTPMYEEVKNKQ 305
Query: 280 VEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKLLTTANND 339
VE+ E++R IATAR+ MP +++RL+AGR EQ +CF+AG N+IF G+K+LTT N
Sbjct: 306 VEVDEIVRTIATARLVMPTSIIRLAAGRYTMKEAEQVMCFMAGCNAIFTGKKMLTTMCNG 365
Query: 340 FDADQLMFKVLGLLP 354
+D D+ M GL P
Sbjct: 366 WDEDKAMLAKWGLKP 380
>emb|CAG85190.1| unnamed protein product [Debaryomyces hansenii CBS767]
gi|50413016|ref|XP_457195.1| unnamed protein product
[Debaryomyces hansenii]
Length = 426
Score = 365 bits (937), Expect = 1e-99
Identities = 172/322 (53%), Positives = 237/322 (73%), Gaps = 10/322 (3%)
Query: 43 PRNDWTKDEVKSIYDSPILDLLFHGAQVHRHAHNFREVQQCTLLSVKTGGCSEDCSYCPQ 102
P N WTKD+++ IY++P+++L+F HR HN +EVQ CTLLS+KTGGC+EDC YC Q
Sbjct: 69 PLNSWTKDQIREIYNTPLMELVFQSQIQHRKYHNPQEVQLCTLLSIKTGGCTEDCKYCAQ 128
Query: 103 SSRYDTGLKGQKLLNKDAVLQAAVKAKEAGSTRFCMGAAWRDTIGRKTNFNQILEYVKEI 162
S+RYDTG K +++++ D V++AA AK GSTRFCMGAAWRD GRK+ +I + V +I
Sbjct: 129 STRYDTGTKAERMISVDEVMKAARTAKANGSTRFCMGAAWRDMTGRKSGLKRISQMVTQI 188
Query: 163 -KGMGMEVCCTLGMLDKDQAGELKKAGLTAYNHNLDTSREYYPNIITTRTYDERLKTLEF 221
+ +GME C TLGM+DK QA +L AGLTAYNHN+DTSRE+YP +I+TR+YDERL+T+
Sbjct: 189 NQELGMETCVTLGMVDKKQAADLADAGLTAYNHNIDTSREHYPKVISTRSYDERLQTIRN 248
Query: 222 VRDAGINVCSGGIIGLGEAEDDRVGLLHTLSTLPTHPESVPINALIAVKGTPLQDQ---- 277
V+DAGI C+GGI+GLGE E+D +G L+TLST+ HPES+PIN L+ +KGTP++ +
Sbjct: 249 VQDAGIKACTGGILGLGETEEDHIGFLYTLSTMEKHPESLPINRLVPIKGTPMEKEISSL 308
Query: 278 -----KPVEIWEMIRMIATARITMPKAMVRLSAGRVRFSVPEQALCFLAGANSIFAGEKL 332
+ ++ +++ IATAR+ MP++++RL+AGR EQ LCF++G N+IF GEK+
Sbjct: 309 PRESGRRLKFEHILKTIATARLIMPESIIRLAAGRYTMKEHEQFLCFMSGCNAIFTGEKM 368
Query: 333 LTTANNDFDADQLMFKVLGLLP 354
LTT N +D D M K GL P
Sbjct: 369 LTTMCNGWDEDIQMLKKWGLRP 390
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 627,285,802
Number of Sequences: 2540612
Number of extensions: 25560989
Number of successful extensions: 56844
Number of sequences better than 10.0: 388
Number of HSP's better than 10.0 without gapping: 249
Number of HSP's successfully gapped in prelim test: 139
Number of HSP's that attempted gapping in prelim test: 56175
Number of HSP's gapped (non-prelim): 395
length of query: 377
length of database: 863,360,394
effective HSP length: 130
effective length of query: 247
effective length of database: 533,080,834
effective search space: 131670965998
effective search space used: 131670965998
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146331.7


