

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145513.6 + phase: 0
(882 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
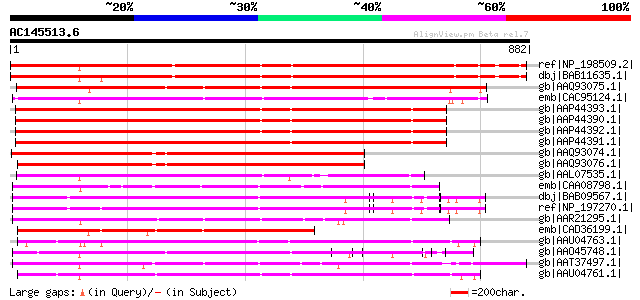
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_198509.2| disease resistance protein (TIR-NBS-LRR class),... 619 e-175
dbj|BAB11635.1| TMV resistance protein N [Arabidopsis thaliana] 617 e-175
gb|AAQ93075.1| TIR-NBS-LRR type R protein 7 [Malus baccata] 602 e-170
emb|CAC95124.1| TIR/NBS/LRR protein [Populus deltoides] 572 e-161
gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum] 543 e-152
gb|AAP44390.1| nematode resistance protein [Solanum tuberosum] 543 e-152
gb|AAP44392.1| nematode resistance-like protein [Solanum tuberosum] 541 e-152
gb|AAP44391.1| nematode resistance-like protein [Solanum tuberosum] 536 e-150
gb|AAQ93074.1| putative TIR-NBS type R protein 4 [Malus baccata] 516 e-145
gb|AAQ93076.1| putative TIR-NBS type R protein 4 [Malus baccata] 514 e-144
gb|AAL07535.1| resistance gene analog PU3 [Helianthus annuus] 487 e-136
emb|CAA08798.1| NL27 [Solanum tuberosum] 470 e-130
dbj|BAB09567.1| disease resistance protein-like [Arabidopsis tha... 469 e-130
ref|NP_197270.1| disease resistance protein (TIR-NBS-LRR class),... 465 e-129
gb|AAR21295.1| bacterial spot disease resistance protein 4 [Lyco... 464 e-129
emb|CAD36199.1| NLS-TIR-NBS disease resistance protein [Populus ... 459 e-127
gb|AAU04763.1| MRGH8 [Cucumis melo] 458 e-127
gb|AAO45748.1| MRGH5 [Cucumis melo] 456 e-126
gb|AAT37497.1| N-like protein [Nicotiana tabacum] 449 e-124
gb|AAU04761.1| MRGH13 [Cucumis melo] 447 e-124
>ref|NP_198509.2| disease resistance protein (TIR-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 1188
Score = 619 bits (1597), Expect = e-175
Identities = 373/892 (41%), Positives = 547/892 (60%), Gaps = 33/892 (3%)
Query: 1 MRNPRSKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPE 60
M R + +WTYDVF++FRGAD RK F+SHLY +L GI+TF+D+ LQ+G+ + PE
Sbjct: 1 MAQGRERIPERWTYDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPE 60
Query: 61 LIRAIQGSQIAIVVFSKNYVHSRWCLSELKQIMEC-KANDGQVVMPVFYCITPSNIR--- 116
L+ AI+ S+I IVV +K+Y S WCL EL IM+ K N +V P+F + PS+IR
Sbjct: 61 LLNAIETSKILIVVLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQ 120
Query: 117 -QYAVTRFSE--TTLFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLD 173
YA + FS+ + ++L + L + +SGWD+ N NE++ + +I ++LK L
Sbjct: 121 GSYAKS-FSKHKNSHPLNKLKDWREALTKVANISGWDIKN-RNEAECIADITREILKRLP 178
Query: 174 NKYLPLPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFE 233
+YL +P + VGL R + L + GV ++ I+GMGGIGK+T+AKV +N+ + FE
Sbjct: 179 CQYLHVPSYAVGLRSRLQHISSLLSIGSDGVRVIVIYGMGGIGKTTLAKVAFNEFSHLFE 238
Query: 234 NQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVL 293
SFL N RE +K GR LQ Q LSDIL+ I+ ++ +K++ R+KR+L V+
Sbjct: 239 GSSFLENFREYSKKPEGRTHLQHQLLSDILRRNDIEFKGLDHA---VKERFRSKRVLLVV 295
Query: 294 DDVSELEQFD-ALCQRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCK 352
DDV ++ Q + A R+ G GS IIITTR++ +L L + Y +EL+ ESLELF
Sbjct: 296 DDVDDVHQLNSAAIDRDCFGHGSRIIITTRNMHLLKQLRAEGSYSPKELDGDESLELFSW 355
Query: 353 HAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQI 412
HAFR + P ++FL S +VV YC G+PLA+EVLG++L +R +EW S L L++IPND I
Sbjct: 356 HAFRTSEPPKEFLQHSEEVVTYCAGLPLAVEVLGAFLIERSIREWESTLKLLKRIPNDNI 415
Query: 413 HEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIK 472
L+ISF+ L +K++FLD+ CFFIG D YV IL+GC L DI +++L+ER LI
Sbjct: 416 QAKLQISFNALTIE-QKDVFLDIACFFIGVDSYYVACILDGCNLYPDIVLSLLMERCLIT 474
Query: 473 VEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMK 532
+ N + MH LLRDMGR+IVRE SP++ + +RLW H DVV VL +GT AIEGL +K
Sbjct: 475 ISGNNIM-MHDLLRDMGRQIVREISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEGLSLK 533
Query: 533 LPKTNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQ 592
+ F+ AF KM LRLL+L V + G Y+ FPK LRWL W GF L+ P N
Sbjct: 534 ADVMDFQYFEVEAFAKMQELRLLELRYVDLNGSYEHFPKDLRWLCWHGFSLECFPINLSL 593
Query: 593 KNLVAMELKHSNLAQVWK---KPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDC 649
++L A++L++SNL + WK PQ +K L+LSHS L+ TPDFS PN+EKLI+ +C
Sbjct: 594 ESLAALDLQYSNLKRFWKAQSPPQPANMVKYLDLSHSVYLRETPDFSYFPNVEKLILINC 653
Query: 650 QSLLEVHPSIGDL-KNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIV 708
+SL+ VH SIG L K L++LNL C L LP EIY+L+ +E+L LS CSK+++L++ +
Sbjct: 654 KSLVLVHKSIGILDKKLVLLNLSSCIELDVLPEEIYKLKSLESLFLSNCSKLERLDDALG 713
Query: 709 QMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSHHVFPSLIRSWISPTMNSLP 768
++ESLTTL+A T +++ P +I + K + +SL G +GL +L S S +++ L
Sbjct: 714 ELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLY-SEKSHSVSLLR 772
Query: 769 RIPPFGGMSKSLFSLDIDSNNLALVSQSQILNSCSRLRSVSVQCDSEIQLKQEFGRFLDD 828
+ G + SL + + L+ + + S S LR + ++ +S L +F
Sbjct: 773 PVSLSGLTYMRILSLGYCNLSDELIPED--IGSLSFLRDLDLRGNSFCNLPTDFATL--- 827
Query: 829 LYDAGLTEMRTSHALQISNLTM--RSLLF-GIGSCHIVINTLRKSLSQVSSL 877
L E+ S ++ ++ RSLLF +G C ++ T +S+ S+L
Sbjct: 828 ---PNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRT--PDISKCSAL 874
Score = 37.7 bits (86), Expect = 1.7
Identities = 50/192 (26%), Positives = 79/192 (41%), Gaps = 31/192 (16%)
Query: 511 EDVVNVLADYTGTKAIEGLVMKLPKTNRVCF-----------DTIAFEKMIRLRLLQ--- 556
E + +LAD+T + I + +L K R+ D + EK + LL+
Sbjct: 716 ESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKSHSVSLLRPVS 775
Query: 557 ---LDNVQVIGDYKC------FPKH------LRWLSWQGFPLKYTPENFYQKNLVAMELK 601
L ++++ C P+ LR L +G P +F + EL
Sbjct: 776 LSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFATLPNLG-ELL 834
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S+ +++ L L L++ LKRTPD SK L KL + DC SL E+ P I +
Sbjct: 835 LSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISLFEI-PGIHN 893
Query: 662 LKNLLMLNLKDC 673
+ L + L C
Sbjct: 894 HEYLSFIVLDGC 905
Score = 37.7 bits (86), Expect = 1.7
Identities = 28/100 (28%), Positives = 41/100 (41%), Gaps = 21/100 (21%)
Query: 618 LKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSL--------------------LEVHP 657
L+ L+L + DF+ LPNL +L++ DC L L+ P
Sbjct: 807 LRDLDLRGNSFCNLPTDFATLPNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTP 866
Query: 658 SIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGC 697
I L L L DC SL +P I+ + ++L GC
Sbjct: 867 DISKCSALFKLQLNDCISLFEIP-GIHNHEYLSFIVLDGC 905
>dbj|BAB11635.1| TMV resistance protein N [Arabidopsis thaliana]
Length = 1130
Score = 617 bits (1592), Expect = e-175
Identities = 373/896 (41%), Positives = 547/896 (60%), Gaps = 36/896 (4%)
Query: 1 MRNPRSKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPE 60
M R + +WTYDVF++FRGAD RK F+SHLY +L GI+TF+D+ LQ+G+ + PE
Sbjct: 1 MAQGRERIPERWTYDVFVSFRGADVRKNFLSHLYDSLRRCGISTFMDDVELQRGEYISPE 60
Query: 61 LIRAIQGSQIAIVVFSKNYVHSRWCLSELKQIMEC-KANDGQVVMPVFYCITPSNIR--- 116
L+ AI+ S+I IVV +K+Y S WCL EL IM+ K N +V P+F + PS+IR
Sbjct: 61 LLNAIETSKILIVVLTKDYASSAWCLDELVHIMKSHKNNPSHMVFPIFLYVDPSDIRWQQ 120
Query: 117 -QYAVTRFSE--TTLFFDELVPFMNTLQDASYLSGWDLSNY----SNESKVVKEIVSQVL 169
YA + FS+ + ++L + L + +SGWD+ N NE++ + +I ++L
Sbjct: 121 GSYAKS-FSKHKNSHPLNKLKDWREALTKVANISGWDIKNRIYDSRNEAECIADITREIL 179
Query: 170 KNLDNKYLPLPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLC 229
K L +YL +P + VGL R + L + GV ++ I+GMGGIGK+T+AKV +N+
Sbjct: 180 KRLPCQYLHVPSYAVGLRSRLQHISSLLSIGSDGVRVIVIYGMGGIGKTTLAKVAFNEFS 239
Query: 230 YEFENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRI 289
+ FE SFL N RE +K GR LQ Q LSDIL+ I+ ++ +K++ R+KR+
Sbjct: 240 HLFEGSSFLENFREYSKKPEGRTHLQHQLLSDILRRNDIEFKGLDHA---VKERFRSKRV 296
Query: 290 LAVLDDVSELEQFD-ALCQRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLE 348
L V+DDV ++ Q + A R+ G GS IIITTR++ +L L + Y +EL+ ESLE
Sbjct: 297 LLVVDDVDDVHQLNSAAIDRDCFGHGSRIIITTRNMHLLKQLRAEGSYSPKELDGDESLE 356
Query: 349 LFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIP 408
LF HAFR + P ++FL S +VV YC G+PLA+EVLG++L +R +EW S L L++IP
Sbjct: 357 LFSWHAFRTSEPPKEFLQHSEEVVTYCAGLPLAVEVLGAFLIERSIREWESTLKLLKRIP 416
Query: 409 NDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIER 468
ND I L+ISF+ L +K++FLD+ CFFIG D YV IL+GC L DI +++L+ER
Sbjct: 417 NDNIQAKLQISFNALTIE-QKDVFLDIACFFIGVDSYYVACILDGCNLYPDIVLSLLMER 475
Query: 469 SLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEG 528
LI + N + MH LLRDMGR+IVRE SP++ + +RLW H DVV VL +GT AIEG
Sbjct: 476 CLITISGNNIM-MHDLLRDMGRQIVREISPKKCGERSRLWSHNDVVGVLKKKSGTNAIEG 534
Query: 529 LVMKLPKTNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPE 588
L +K + F+ AF KM LRLL+L V + G Y+ FPK LRWL W GF L+ P
Sbjct: 535 LSLKADVMDFQYFEVEAFAKMQELRLLELRYVDLNGSYEHFPKDLRWLCWHGFSLECFPI 594
Query: 589 NFYQKNLVAMELKHSNLAQVWK---KPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLI 645
N ++L A++L++SNL + WK PQ +K L+LSHS L+ TPDFS PN+EKLI
Sbjct: 595 NLSLESLAALDLQYSNLKRFWKAQSPPQPANMVKYLDLSHSVYLRETPDFSYFPNVEKLI 654
Query: 646 MKDCQSLLEVHPSIGDL-KNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLE 704
+ +C+SL+ VH SIG L K L++LNL C L LP EIY+L+ +E+L LS CSK+++L+
Sbjct: 655 LINCKSLVLVHKSIGILDKKLVLLNLSSCIELDVLPEEIYKLKSLESLFLSNCSKLERLD 714
Query: 705 EDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSHHVFPSLIRSWISPTM 764
+ + ++ESLTTL+A T +++ P +I + K + +SL G +GL +L S S ++
Sbjct: 715 DALGELESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLY-SEKSHSV 773
Query: 765 NSLPRIPPFGGMSKSLFSLDIDSNNLALVSQSQILNSCSRLRSVSVQCDSEIQLKQEFGR 824
+ L + G + SL + + L+ + + S S LR + ++ +S L +F
Sbjct: 774 SLLRPVSLSGLTYMRILSLGYCNLSDELIPED--IGSLSFLRDLDLRGNSFCNLPTDFAT 831
Query: 825 FLDDLYDAGLTEMRTSHALQISNLTM--RSLLF-GIGSCHIVINTLRKSLSQVSSL 877
L E+ S ++ ++ RSLLF +G C ++ T +S+ S+L
Sbjct: 832 L------PNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRT--PDISKCSAL 879
Score = 37.7 bits (86), Expect = 1.7
Identities = 50/192 (26%), Positives = 79/192 (41%), Gaps = 31/192 (16%)
Query: 511 EDVVNVLADYTGTKAIEGLVMKLPKTNRVCF-----------DTIAFEKMIRLRLLQ--- 556
E + +LAD+T + I + +L K R+ D + EK + LL+
Sbjct: 721 ESLTTLLADFTALREIPSTINQLKKLKRLSLNGCKGLLSDDIDNLYSEKSHSVSLLRPVS 780
Query: 557 ---LDNVQVIGDYKC------FPKH------LRWLSWQGFPLKYTPENFYQKNLVAMELK 601
L ++++ C P+ LR L +G P +F + EL
Sbjct: 781 LSGLTYMRILSLGYCNLSDELIPEDIGSLSFLRDLDLRGNSFCNLPTDFATLPNLG-ELL 839
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S+ +++ L L L++ LKRTPD SK L KL + DC SL E+ P I +
Sbjct: 840 LSDCSKLQSILSLPRSLLFLDVGKCIMLKRTPDISKCSALFKLQLNDCISLFEI-PGIHN 898
Query: 662 LKNLLMLNLKDC 673
+ L + L C
Sbjct: 899 HEYLSFIVLDGC 910
Score = 37.7 bits (86), Expect = 1.7
Identities = 28/100 (28%), Positives = 41/100 (41%), Gaps = 21/100 (21%)
Query: 618 LKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSL--------------------LEVHP 657
L+ L+L + DF+ LPNL +L++ DC L L+ P
Sbjct: 812 LRDLDLRGNSFCNLPTDFATLPNLGELLLSDCSKLQSILSLPRSLLFLDVGKCIMLKRTP 871
Query: 658 SIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGC 697
I L L L DC SL +P I+ + ++L GC
Sbjct: 872 DISKCSALFKLQLNDCISLFEIP-GIHNHEYLSFIVLDGC 910
>gb|AAQ93075.1| TIR-NBS-LRR type R protein 7 [Malus baccata]
Length = 1095
Score = 602 bits (1553), Expect = e-170
Identities = 352/834 (42%), Positives = 515/834 (61%), Gaps = 43/834 (5%)
Query: 12 WTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQIA 71
W YD+F++FRG DTR F HL+ AL + G ++D ++L +G+E+ EL RAI+GS+I+
Sbjct: 21 WNYDLFLSFRGEDTRNGFTGHLHAALKDRGYQAYMDQDDLNRGEEIKEELFRAIEGSRIS 80
Query: 72 IVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSETTLFFD 131
I+VFSK Y S WCL EL +IMEC++ G+ V+P+FY + PS++R+ +E L +
Sbjct: 81 IIVFSKRYADSSWCLDELVKIMECRSKLGRHVLPIFYHVDPSHVRKQDGD-LAEAFLKHE 139
Query: 132 ELV-----------------PFMNTLQDASYLSGWDL---SNYSNESKVVKEIVSQVLKN 171
E + + L +A+ LSG DL N + +EIV ++
Sbjct: 140 EGIGEGTDGKKREAKQERVKQWKKALTEAANLSGHDLRITDNGREANLCPREIVDNIITK 199
Query: 172 --LDNKYLPLPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLC 229
+ L + QVG+ R + I L V +VGIWGMGG+GK+T AK IYN +
Sbjct: 200 WLMSTNKLRVAKHQVGINSRIQDIISRLSSGGSNVIMVGIWGMGGLGKTTAAKAIYNQIH 259
Query: 230 YEFENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRI 289
+EF+ +SFL ++ K G + LQ++ + DILKT K K+ SV++G +I+ Q R +R+
Sbjct: 260 HEFQFKSFLPDVGNAASK-HGLVYLQKELIYDILKT-KSKISSVDEGIGLIEDQFRHRRV 317
Query: 290 LAVLDDVSELEQFDALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLE 348
L ++D++ E+ Q DA+ GPGS IIITTRD +L +VD Y A++L+ E+LE
Sbjct: 318 LVIMDNIDEVGQLDAIVGNPDWFGPGSRIIITTRDEHLLK--QVDKTYVAQKLDEREALE 375
Query: 349 LFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIP 408
LF HAF P +++L LS VV+YCGG+PLALEVLGS+LFKR EW+S L KL++ P
Sbjct: 376 LFSWHAFGNNWPNEEYLELSEKVVSYCGGLPLALEVLGSFLFKRPIAEWKSQLEKLKRTP 435
Query: 409 NDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIER 468
+I + L+ISF+GL D +K IFLD+ CFFIG+D+ YV K+L+GCG A IGI+VL ER
Sbjct: 436 EGKIIKSLRISFEGLDDA-QKAIFLDISCFFIGEDKDYVAKVLDGCGFYATIGISVLRER 494
Query: 469 SLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEG 528
L+ VE NK L MH LLR+M + I+ E SP +P K +RLW +V+NVL + +GT+ +EG
Sbjct: 495 CLVTVEHNK-LNMHDLLREMAKVIISEKSPGDPGKWSRLWDKREVINVLTNKSGTEEVEG 553
Query: 529 LVMKLPKTNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPE 588
L + + F T AF + +LRLLQL V++ G+YK PK L WL W PLK P+
Sbjct: 554 LALPWGYRHDTAFSTEAFANLKKLRLLQLCRVELNGEYKHLPKELIWLHWFECPLKSIPD 613
Query: 589 NFY-QKNLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMK 647
+F+ Q LV +E++ S L QVW+ + + LK L+LS S++L+++PDFS++PNLE+LI+
Sbjct: 614 DFFNQDKLVVLEMQWSKLVQVWEGSKSLHNLKTLDLSESRSLQKSPDFSQVPNLEELILY 673
Query: 648 DCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDI 707
+C+ L E+HPSIG LK L ++NL+ C L +LP + Y+ + VE L+L+GC + +L EDI
Sbjct: 674 NCKELSEIHPSIGHLKRLSLVNLEWCDKLISLPGDFYKSKSVEALLLNGCLILRELHEDI 733
Query: 708 VQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGL----SHHVFPSLIRSWISPT 763
+M SL TL A T +++ P SIVR K++ +SL E + S H SL +S
Sbjct: 734 GEMISLRTLEAEYTDIREVPPSIVRLKNLTRLSLSSVESIHLPHSLHGLNSLRELNLSSF 793
Query: 764 MNSLPRIPPFGGMSKSLFSLDIDSNNL-ALVSQSQI-------LNSCSRLRSVS 809
+ IP G SL L++ N+ L S S + L+ C +LR+++
Sbjct: 794 ELADDEIPKDLGSLISLQDLNLQRNDFHTLPSLSGLSKLETLRLHHCEQLRTIT 847
>emb|CAC95124.1| TIR/NBS/LRR protein [Populus deltoides]
Length = 1147
Score = 572 bits (1473), Expect = e-161
Identities = 346/841 (41%), Positives = 500/841 (59%), Gaps = 51/841 (6%)
Query: 5 RSKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRA 64
RS+P+ YDVF++FRG DTRKTF HLYTAL AGI+TF D++ L +G+E+ +RA
Sbjct: 32 RSRPEG--AYDVFLSFRGEDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHFLRA 89
Query: 65 IQGSQIAIVVFSKNYVHSRWCLSELKQIMEC-KANDGQVVMPVFYCITPSNIRQ----YA 119
IQ S+I+I VFSK Y SRWCL+EL +I++C K GQ+V+P+FY I PS++R+ +A
Sbjct: 90 IQESKISIAVFSKGYASSRWCLNELVEILKCKKRKTGQIVLPIFYDIDPSDVRKQNGSFA 149
Query: 120 VTRFSETTLFFDELV-PFMNTLQDASYLSGWDLSNYSN--ESKVVKEIVSQVLKNLDNKY 176
F ++LV + L++A LSGW+L++ +N E+K +KEI+ VL L+ KY
Sbjct: 150 EAFVKHEERFEEKLVKEWRKALEEAGNLSGWNLNDMANGHEAKFIKEIIKVVLNKLEPKY 209
Query: 177 LPLPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQS 236
L +P+ VG++ A FL T V +VGI GM GIGK+TIA+ ++N LCY FE
Sbjct: 210 LYVPEHLVGMDQLARNIFDFLSAATDDVRIVGIHGMPGIGKTTIAQAVFNQLCYGFEGSC 269
Query: 237 FLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDV 296
FL++I E ++ G + LQ+Q DILK ++GK +IK++LR KR+L V DDV
Sbjct: 270 FLSSINERSKQVNGLVPLQKQLHHDILKQDVANFDCADRGKVLIKERLRRKRVLVVADDV 329
Query: 297 SELEQFDALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAF 355
+ LEQ +AL R+ GPGS +IITTRD +L E D IY+ EEL ESL+LF +HAF
Sbjct: 330 AHLEQLNALMGDRSWFGPGSRVIITTRDSNLLR--EADQIYQIEELKPDESLQLFSRHAF 387
Query: 356 RKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEI 415
+ + P QD++ LS+ V YCGG+PLALEV+G+ L+++ + S + L +IPN I
Sbjct: 388 KDSKPAQDYIELSKKAVGYCGGLPLALEVIGALLYRKNRGRCVSEIDNLSRIPNQDIQGK 447
Query: 416 LKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNG-CGLNADIGITVLIERSLIKVE 474
L IS+ L +++ FLD+ CFFIG +R YVTK+L C N ++ + L ERSLI+V
Sbjct: 448 LLISYHALDGELQR-AFLDIACFFIGIEREYVTKVLGARCRPNPEVVLETLSERSLIQV- 505
Query: 475 KNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLAD--YTGTKAIEGLVMK 532
+ + MH LLRDMGRE+V ++SP++P K TR+W ED NVL GT ++GL +
Sbjct: 506 FGETVSMHDLLRDMGREVVCKASPKQPGKRTRIWNQEDAWNVLEQQKVRGTDVVKGLALD 565
Query: 533 LPKTNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQ 592
+ + +F +M L LLQ++ V + G K F K L W+ W PLKY P +F
Sbjct: 566 VRASEAKSLSAGSFAEMKCLNLLQINGVHLTGSLKLFSKELMWICWHECPLKYLPFDFTL 625
Query: 593 KNLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSL 652
NL +++++SNL ++WK G K+ N+ S + + + LEKL +K C SL
Sbjct: 626 DNLAVLDMQYSNLKELWK------GKKVRNMLQSPKFLQYVIYIYI--LEKLNLKGCSSL 677
Query: 653 LEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMES 712
+EVH SIG+L +L LNL+ C L NLP I ++ +ETL +SGCS+++KL E + MES
Sbjct: 678 VEVHQSIGNLTSLDFLNLEGCWRLKNLPESIGNVKSLETLNISGCSQLEKLPESMGDMES 737
Query: 713 LTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEG-------LSHHV------FPSLIRSW 759
L L+A +Q SI + K + +SL GY +S V P+ W
Sbjct: 738 LIELLADGIENEQFLSSIGQLKHVRRLSLRGYSSTPPSSSLISAGVLNLKRWLPTSFIQW 797
Query: 760 ISPTMNSLP---------RIPPFGGMSKSLFSLDIDSNNLALVSQSQILNSCSRLRSVSV 810
IS LP + F G+S +L LD+ N + + + S+L+ +SV
Sbjct: 798 ISVKRLELPHGGLSDRAAKCVDFSGLS-ALEVLDLIGNKFSSLPSG--IGFLSKLKFLSV 854
Query: 811 Q 811
+
Sbjct: 855 K 855
>gb|AAP44393.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 543 bits (1398), Expect = e-152
Identities = 307/740 (41%), Positives = 461/740 (61%), Gaps = 13/740 (1%)
Query: 11 KWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQI 70
+W+YDVF++FRG D RKTF+ HLY AL INTF D+E L+KGK + PEL+ +I+ S+I
Sbjct: 15 RWSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEESRI 74
Query: 71 AIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVT---RFSETT 127
A+++FSKNY +S WCL EL +IMECK GQ+V+PVFY + PS +R+ FS+
Sbjct: 75 ALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHE 134
Query: 128 LFF--DELVPFMNTLQDASYLSGWDLSNYSN--ESKVVKEIVSQVLKNLDN-KYLPLPDF 182
F D++ + L++A+ +SGWDL N SN E++V+++I ++ L + ++
Sbjct: 135 ARFQEDKVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHASNARN 194
Query: 183 QVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIR 242
VG+E + + L + GV +GI GM G+GK+T+A+VIY+++ +F+ FL +R
Sbjct: 195 LVGMESHMHQVYKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVR 254
Query: 243 EVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQF 302
+ K +G LQE LS+IL +K+++ +G M KQ+L+ K++L VLDDV ++Q
Sbjct: 255 DRSAK-QGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQL 313
Query: 303 DALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPT 361
+AL +R G GS IIITT+D +L E + IY + LN ESL+LF +HAF+K PT
Sbjct: 314 NALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPT 373
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
++F LS V+ + G+PLAL+VLGS+L+ R EW S + +L++IP ++I + L+ SF
Sbjct: 374 KEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFT 433
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL + E+ IFLD+ CFF GK + VT+IL IGI VL+E+ LI + + + +
Sbjct: 434 GLHNT-EQKIFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITTLQGR-ITI 491
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H L++DMG IVR + ++P +RLW ED+ VL GT IEG+ + L V F
Sbjct: 492 HQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKIEGMSLHLTNEEEVNF 551
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELK 601
AF +M RLR L+ N V + P LRWL W G+P K P +F LV+++LK
Sbjct: 552 GGKAFMQMTRLRFLKFQNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVSLKLK 611
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S + Q+WK + + LK +NLSHS+ L R PDFS PNLE+L++++C SL+E++ SI +
Sbjct: 612 KSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINFSIEN 671
Query: 662 LKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANT 721
L L++LNLK+C +L LP+ I +L ++E L+L+GCSK+ E +M L L T
Sbjct: 672 LGKLVLLNLKNCRNLKTLPKRI-RLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLDAT 730
Query: 722 GVKQPPFSIVRSKSIGYISL 741
+ + P S+ +G I+L
Sbjct: 731 SLSELPASVENLSGVGVINL 750
Score = 36.6 bits (83), Expect = 3.7
Identities = 40/160 (25%), Positives = 65/160 (40%), Gaps = 44/160 (27%)
Query: 614 LIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPS--------------I 659
L+ GL+ L+ +H+ S L NL++L + C +L S +
Sbjct: 789 LLVGLEQLHCTHTAIQTIPSSMSLLKNLKRLSLSGCNALSSQVSSSSHGQKSMGVNFQNL 848
Query: 660 GDLKNLLMLNLKDC--------TSLGNLP-----------------REIYQLRRVETLIL 694
L +L+ML+L DC ++LG LP I +L R++TL L
Sbjct: 849 SGLCSLIMLDLSDCNISDGGILSNLGFLPSLERLILDGNNFSNIPAASISRLTRLKTLKL 908
Query: 695 SGCSKIDKLEE-----DIVQMESLTTLMAANTGVKQPPFS 729
GC +++ L E + T+LM+ + K P S
Sbjct: 909 LGCGRLESLPELPPSIKGIYANECTSLMSIDQLTKYPMLS 948
>gb|AAP44390.1| nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 543 bits (1398), Expect = e-152
Identities = 306/740 (41%), Positives = 461/740 (61%), Gaps = 13/740 (1%)
Query: 11 KWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQI 70
+W+YDVF++FRG D RKTF+ HLY AL INTF D+E L+KGK + PEL+ +I+ S+I
Sbjct: 15 RWSYDVFLSFRGEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEESRI 74
Query: 71 AIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVT---RFSETT 127
A+++FSKNY +S WCL EL +IMECK GQ+V+PVFY + PS +R+ FS+
Sbjct: 75 ALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHE 134
Query: 128 LFF--DELVPFMNTLQDASYLSGWDLSNYSN--ESKVVKEIVSQVLKNLDN-KYLPLPDF 182
F D++ + L++A+ +SGWDL N +N E++V+++I ++ L + ++
Sbjct: 135 ARFQEDKVQKWRAALEEAANISGWDLPNTANGHEARVMEKIAEDIMARLGSQRHASNARN 194
Query: 183 QVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIR 242
VG+E K + L + GV +GI GM G+GK+T+A+VIY+++ +F+ FL +R
Sbjct: 195 LVGMESHMHKVYKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVR 254
Query: 243 EVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQF 302
+ K +G LQE LS+IL +K+++ +G M KQ+L+ K++L VLDDV ++Q
Sbjct: 255 DRSAK-QGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQL 313
Query: 303 DALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPT 361
+AL +R G GS IIITT+D +L E + IY + LN ESL+LF +HAF+K PT
Sbjct: 314 NALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPT 373
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
++F LS V+ + G+PLAL+VLGS+L+ R EW S + +L++IP ++I + L+ SF
Sbjct: 374 KEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFT 433
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL + E+ IFLD+ CFF GK + VT+IL IGI VL+E+ LI + + + + +
Sbjct: 434 GLHNT-EQKIFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITILQGR-ITI 491
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H L++DMG IVR + ++P +R+W ED+ VL GT EG+ + L V F
Sbjct: 492 HQLIQDMGWHIVRREATDDPRMCSRMWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNF 551
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELK 601
AF +M RLR L+ N V + P LRWL W G+P K P +F LV ++LK
Sbjct: 552 GGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLK 611
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S + Q+WK + + LK +NLSHS+ L RTPDFS PNLE+L++++C SL+E++ SI +
Sbjct: 612 KSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINFSIEN 671
Query: 662 LKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANT 721
L L++LNLK+C +L LP+ I +L ++E L+L+GCSK+ E +M L L T
Sbjct: 672 LGKLVLLNLKNCRNLKTLPKRI-RLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLGAT 730
Query: 722 GVKQPPFSIVRSKSIGYISL 741
+ + P S+ +G I+L
Sbjct: 731 SLSELPASVENLSGVGVINL 750
>gb|AAP44392.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 541 bits (1395), Expect = e-152
Identities = 308/740 (41%), Positives = 461/740 (61%), Gaps = 13/740 (1%)
Query: 11 KWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQI 70
+W+YDVF++FRG D RKTF+ HLY AL INTF D+E L+KGK + PEL+ +I+ S+I
Sbjct: 15 RWSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELMSSIEESRI 74
Query: 71 AIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVT---RFSETT 127
A+++FSKNY +S WCL EL +IMECK GQ+V+PVFY + PS +R+ FS+
Sbjct: 75 ALIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHE 134
Query: 128 LFF--DELVPFMNTLQDASYLSGWDLSNYSN--ESKVVKEIVSQVLKNLDN-KYLPLPDF 182
F D++ + L++A+ +SGWDL N SN E++V+++I ++ L + ++
Sbjct: 135 ARFQEDKVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHASNARN 194
Query: 183 QVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIR 242
VG+E K + L + GV +GI GM G+GK+T+A+VIY+++ +F+ FL +R
Sbjct: 195 LVGMESHMLKVYKMLGIGSGGVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACFLHEVR 254
Query: 243 EVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQF 302
+ K +G LQE LS+IL +K+++ + +G M KQ+L+ K++L VLDDV ++Q
Sbjct: 255 DRSAK-QGLERLQEILLSEILVVKKLRINNSFEGANMQKQRLQYKKVLLVLDDVDHIDQL 313
Query: 303 DALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPT 361
+AL +R G GS IIITT+D +L E + IY + LN ESL+LF +HAF+K PT
Sbjct: 314 NALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKKNRPT 373
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
++F LS V+ + G+PLAL+VLGS+L+ R EW S + +L++IP ++I + L+ SF
Sbjct: 374 KEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLEQSFT 433
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL + E+ IFLD+ CFF GK + VT+IL IGI VL+E+ LI + + + + +
Sbjct: 434 GLHNT-EQKIFLDIACFFSGKKKDSVTRILESFHFCPVIGIKVLMEKCLITILQGR-ITI 491
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H L++DMG IVR + ++P +RLW ED+ VL GT EG+ + L V F
Sbjct: 492 HQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKNEGMSLHLTNEEEVNF 551
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELK 601
AF +M RLR L+ N V + P LRWL W G+P K P +F LV ++LK
Sbjct: 552 GGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSFKGDQLVGLKLK 611
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S + Q+WK + + LK +NLSHS+ L RTPDFS PNLE+L++++C SL+E++ SI +
Sbjct: 612 KSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLERLVLEECTSLVEINFSIEN 671
Query: 662 LKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANT 721
L L++LNLK+C +L LP+ I +L ++E L+L+GCSK+ E +M L L T
Sbjct: 672 LGKLVLLNLKNCRNLKTLPKRI-RLEKLEILVLTGCSKLRTFPEIEEKMNCLAELYLGAT 730
Query: 722 GVKQPPFSIVRSKSIGYISL 741
+ P S+ +G I+L
Sbjct: 731 SLSGLPASVENLSGVGVINL 750
>gb|AAP44391.1| nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 536 bits (1381), Expect = e-150
Identities = 304/740 (41%), Positives = 459/740 (61%), Gaps = 13/740 (1%)
Query: 11 KWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQI 70
+W+YDVF++FRG + RKTF+ HLY AL INTF D+E L+KGK + PEL+ +I+ S+I
Sbjct: 15 RWSYDVFLSFRGENVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELMSSIEESRI 74
Query: 71 AIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVT---RFSETT 127
A+++FSKNY +S WCL EL +I+ECK GQ+V+PVFY + PS +R+ FS+
Sbjct: 75 ALIIFSKNYANSTWCLDELTKIIECKNVKGQIVVPVFYDVDPSTVRRQKNIFGEAFSKHE 134
Query: 128 LFFDE--LVPFMNTLQDASYLSGWDLSNYSN--ESKVVKEIVSQVLKNLDN-KYLPLPDF 182
F+E + + L++A+ +SGWDL N SN E++V+++I ++ L + ++
Sbjct: 135 ARFEEDKVKKWRAALEEAANISGWDLPNTSNGHEARVIEKITEDIMVRLGSQRHASNARN 194
Query: 183 QVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIR 242
VG+E + + L + GV +GI GM G+GK+T+A+VIY+++ +FE FL +R
Sbjct: 195 VVGMESHMHQVYKMLGIGSGGVRFLGILGMSGVGKTTLARVIYDNIQSQFEGACFLHEVR 254
Query: 243 EVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQF 302
+ K +G LQE LS+IL +K+++ +G M KQ+L+ K++L VLDDV ++Q
Sbjct: 255 DRSAK-QGLEHLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDHIDQL 313
Query: 303 DALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPT 361
+AL +R G GS IIITT+D +L E + IY L+ ESL+LF +HAF+K T
Sbjct: 314 NALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMGTLDKYESLQLFKQHAFKKNHST 373
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
++F LS V+ + GG+PLAL+VLGS+L+ R EW S + +L++IP ++I + L+ SF
Sbjct: 374 KEFEDLSAQVIEHTGGLPLALKVLGSFLYGRGLDEWISEVERLKQIPQNEILKKLEPSFT 433
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL + +E+ IFLD+ CFF GK + VT+IL + IGI VL+E+ LI + K + + +
Sbjct: 434 GLNN-IEQKIFLDIACFFSGKKKDSVTRILESFHFSPVIGIKVLMEKCLITILKGR-ITI 491
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H L+++MG IVR + P +RLW ED+ VL T IEG+ + L V F
Sbjct: 492 HQLIQEMGWHIVRREASYNPRICSRLWKREDICPVLEQNLCTDKIEGMSLHLTNEEEVNF 551
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELK 601
A +M LR L+ N V + P LRWL W G+P K P +F LV+++LK
Sbjct: 552 GGKALMQMTSLRFLKFRNAYVYQGPEFLPDELRWLDWHGYPSKNLPNSFKGDQLVSLKLK 611
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S + Q+WK + + LK +NLSHS+ L R PDFS PNLE+L++++C SL+E++ SIGD
Sbjct: 612 KSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLERLVLEECTSLVEINFSIGD 671
Query: 662 LKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANT 721
L L++LNLK+C +L +P+ I +L ++E L+LSGCSK+ E +M L L T
Sbjct: 672 LGKLVLLNLKNCRNLKTIPKRI-RLEKLEVLVLSGCSKLRTFPEIEEKMNRLAELYLGAT 730
Query: 722 GVKQPPFSIVRSKSIGYISL 741
+ + P S+ +G I+L
Sbjct: 731 SLSELPASVENFSGVGVINL 750
>gb|AAQ93074.1| putative TIR-NBS type R protein 4 [Malus baccata]
Length = 726
Score = 516 bits (1330), Expect = e-145
Identities = 276/606 (45%), Positives = 391/606 (63%), Gaps = 12/606 (1%)
Query: 3 NPRSKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELI 62
+P + Y+VFI+FRG DTRK F HL+ ALT AGIN F+D+E L++G+++ EL+
Sbjct: 114 SPSASSSKGSLYEVFISFRGEDTRKNFTGHLHEALTKAGINAFIDDEELRRGEDITTELV 173
Query: 63 RAIQGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTR 122
+AIQGS+I+I+VFS+ Y S WCL EL +IMEC+ GQ+V+P+FY + PSN+R+ +
Sbjct: 174 QAIQGSRISIIVFSRRYADSSWCLEELVKIMECRRTLGQLVLPIFYDVDPSNVRKLTGSF 233
Query: 123 FSETTLFFDE--LVPFMNTLQDASYLSGWDLSNY--SNESKVVKEIVSQVLKNLDNKYLP 178
DE + + L +AS LSGWDL N +E+K ++ I +QV L+N+Y
Sbjct: 234 AQSFLKHTDEKKVERWRAALTEASNLSGWDLKNTLDRHEAKFIRMITNQVTVKLNNRYFN 293
Query: 179 LPDFQVGLEPRAEKSIRFLR-QNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSF 237
+ +QVG++ R +L ++ V ++GI GMGGIGK+TI K IYN+ FE +SF
Sbjct: 294 VAPYQVGIDTRVLNISNYLGIGDSDDVRVIGISGMGGIGKTTIVKAIYNEFYERFEGKSF 353
Query: 238 LANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVS 297
L +RE + + LQ+Q L DIL+T K KV SV G ++ ++ R R+L ++DDV
Sbjct: 354 LEKVRE-----KKLVKLQKQLLFDILQT-KTKVSSVAVGTALVGERFRRLRVLVIVDDVD 407
Query: 298 ELEQFDALCQR-NSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFR 356
+++Q L +S GPGS IIITTR+ RVL VD IY ++ E+LEL HAF+
Sbjct: 408 DVKQLRELVGNCHSFGPGSRIIITTRNERVLKEFAVDEIYRENGMDQEEALELLSWHAFK 467
Query: 357 KAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEIL 416
+ +L+L+R+VV YCGG+PLALEVLGS +FKR EWRS+L +L+ IP +I L
Sbjct: 468 SSWCPSQYLVLTREVVNYCGGLPLALEVLGSTIFKRSVNEWRSILDELKMIPRGEIQAQL 527
Query: 417 KISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKN 476
KIS+DGL D ++ IFLD+ FFIG D+ V +IL+GCG A GI VL++R L+ + +
Sbjct: 528 KISYDGLNDHYKRQIFLDIAFFFIGMDKNDVMQILDGCGFYATTGIEVLLDRCLVTIGRK 587
Query: 477 KKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKT 536
K+ MH LLRDMGR+IV +P P + +RLW +DV +VL D +GT+ IEGL + LP
Sbjct: 588 NKIMMHDLLRDMGRDIVHAENPGFPRERSRLWHPKDVHDVLIDKSGTEKIEGLALNLPSL 647
Query: 537 NRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLV 596
F T AF M RLRLLQL+ V++ G Y+C K LRWL W GFPL++ P Q N+V
Sbjct: 648 EETSFSTDAFRNMKRLRLLQLNYVRLTGGYRCLSKKLRWLCWHGFPLEFIPIELCQPNIV 707
Query: 597 AMELKH 602
A+++++
Sbjct: 708 AIDMQY 713
>gb|AAQ93076.1| putative TIR-NBS type R protein 4 [Malus baccata]
Length = 726
Score = 514 bits (1323), Expect = e-144
Identities = 274/595 (46%), Positives = 387/595 (64%), Gaps = 12/595 (2%)
Query: 14 YDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQIAIV 73
Y+VFI+FRG DTRK F HL+ ALT AGIN F+D+E L++G+++ EL++AIQGS+I+I+
Sbjct: 125 YEVFISFRGEDTRKNFTGHLHEALTKAGINAFIDDEELRRGEDITTELVQAIQGSRISII 184
Query: 74 VFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSETTLFFDE- 132
VFS+ Y S WCL EL +IMEC+ GQ+V+P+FY + PSN+R+ + DE
Sbjct: 185 VFSRRYADSSWCLEELVKIMECRRTLGQLVLPIFYDVDPSNVRKLTGSFAQSFLKHTDEK 244
Query: 133 -LVPFMNTLQDASYLSGWDLSNY--SNESKVVKEIVSQVLKNLDNKYLPLPDFQVGLEPR 189
+ + L +AS LSGWDL N +E+K ++ I +QV L+N+Y + +QVG++ R
Sbjct: 245 KVERWRAALTEASNLSGWDLKNTLDRHEAKFIRMITNQVTVKLNNRYFNVAPYQVGIDTR 304
Query: 190 AEKSIRFLR-QNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIREVWEKD 248
+L ++ V ++GI G GGIGK+TI K IYN+ FE +SFL +RE
Sbjct: 305 VLNISNYLGIGDSDDVRVIGISGSGGIGKTTIVKAIYNEFYERFEGKSFLEKVRE----- 359
Query: 249 RGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFDALCQR 308
+ + LQ+Q L DIL+T K KV SV G ++ ++ R R+L ++DDV +++Q L
Sbjct: 360 KKLVKLQKQLLFDILQT-KTKVSSVAVGTALVGERFRRLRVLVIVDDVDDVKQLRELVGN 418
Query: 309 -NSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQDFLIL 367
+S GPGS IIITTR+ RVL VD IY ++ E+LEL HAF+ + +L+L
Sbjct: 419 CHSFGPGSRIIITTRNERVLKEFAVDEIYRENGMDQEEALELLSWHAFKSSWCPSQYLVL 478
Query: 368 SRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGLKDRM 427
+R+VV YCGG+PLALEVLGS +FKR EWRS+L +L+ IP +I LKIS+DGL D
Sbjct: 479 TREVVNYCGGLPLALEVLGSTIFKRSVNEWRSILDELKMIPRGEIQAQLKISYDGLNDHY 538
Query: 428 EKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGMHALLRD 487
++ IFLD+ FFIG D+ V +IL+GCG A GI VL++R L+ + + K+ MH LLRD
Sbjct: 539 KRQIFLDIAFFFIGMDKNDVMQILDGCGFYATTGIEVLLDRCLVTIGRKNKIMMHDLLRD 598
Query: 488 MGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFDTIAFE 547
MGR+IV +P P + +RLW +DV +VL D +GT+ IEGL + LP F T AF
Sbjct: 599 MGRDIVHAENPGFPRERSRLWHPKDVHDVLIDKSGTEKIEGLALNLPSLEETSFSTDAFR 658
Query: 548 KMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKH 602
M RLRLLQL+ V++ G Y+C K LRWL W GFPL++ P Q N+VA+++++
Sbjct: 659 NMKRLRLLQLNYVRLTGGYRCLSKKLRWLCWHGFPLEFIPIELCQPNIVAIDMQY 713
>gb|AAL07535.1| resistance gene analog PU3 [Helianthus annuus]
Length = 770
Score = 487 bits (1253), Expect = e-136
Identities = 278/704 (39%), Positives = 420/704 (59%), Gaps = 29/704 (4%)
Query: 12 WTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQIA 71
W +DVF++FRG DTR +F+ HLY AL GI T+ D++ L +G+ +GP L++AIQ S+IA
Sbjct: 81 WNHDVFLSFRGEDTRNSFVDHLYAALVQQGIQTYKDDQTLPRGERIGPALLKAIQESRIA 140
Query: 72 IVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIR----QYAVTRFSETT 127
+VVFS+NY S WCL EL IMEC GQ+V+P+FY + PS++R +Y
Sbjct: 141 VVVFSQNYADSSWCLDELAHIMECMDTRGQIVIPIFYFVDPSDVRKQKGKYGKAFRKHKR 200
Query: 128 LFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPLPDFQVGLE 187
++ + L+ A LSGW ++ S+E+K +KEIV+ + L + +G+E
Sbjct: 201 ENKQKVESWRKALEKAGNLSGWVINENSHEAKCIKEIVATISSRLPTLSTNVNKDLIGIE 260
Query: 188 PRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIREVWEK 247
R + L+ + V ++GIWG+GG GK+T+A Y ++ + FE L NIRE K
Sbjct: 261 TRLQDLKSKLKMESGDVRIIGIWGVGGGGKTTLASAAYAEISHRFEAHCLLQNIREESNK 320
Query: 248 DRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFDALCQ 307
G LQE+ LS +LKT+ + V S +G++MI+++LR K +L VLDDV +L+Q +AL
Sbjct: 321 -HGLEKLQEKILSLVLKTKDVVVGSEIEGRSMIERRLRNKSVLVVLDDVDDLKQLEALAG 379
Query: 308 RNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQDFLI 366
++ G GS IIITTRD +L D IYE L+ E++ELF KHA+R+ +D+ +
Sbjct: 380 SHAWFGKGSRIIITTRDEHLLT-RHADMIYEVSLLSDDEAMELFNKHAYREDELIEDYGM 438
Query: 367 LSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDGLKDR 426
LS+DVV+Y G+PLALE+LGS+L+ + K +W+S L+KL+ IPN ++ E LKIS+DGL+
Sbjct: 439 LSKDVVSYASGLPLALEILGSFLYDKNKDDWKSALAKLKCIPNVEVTERLKISYDGLEPE 498
Query: 427 MEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKV-----EKNKKLGM 481
+K +FLD+ CF+ +D +L+ C L+ IG+ VLI++SLIKV K K M
Sbjct: 499 HQK-LFLDIACFWRRRDMDEAMMVLDACNLHPRIGVKVLIQKSLIKVSDVRFSKQKVFDM 557
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H L+ +M IVR + P PEKH+R+W ED+ L D G A+
Sbjct: 558 HDLVEEMAHYIVRGAHPNHPEKHSRIWKMEDIA-YLCD-MGEDAVP-------------M 602
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELK 601
+T A + L N + D K L W+ + +P P NF+ L +EL+
Sbjct: 603 ETEALAFRCYIDDPGLSNAVGVSDVVANMKKLPWIRFDEYPASSFPSNFHPTELGCLELE 662
Query: 602 HSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGD 661
S ++W +L+ LKIL+L+ S NL TP+F LP LE+L ++ C+SL E+HPSIG
Sbjct: 663 RSRQKELWHGYKLLPNLKILDLAMSSNLITTPNFDGLPCLERLDLEGCESLEEIHPSIGY 722
Query: 662 LKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEE 705
K+L+ ++++ C++L I Q++ +ETLILS C ++ + +
Sbjct: 723 HKSLVYVDMRRCSTLKRF-SPIIQMQMLETLILSECRELQQFPD 765
>emb|CAA08798.1| NL27 [Solanum tuberosum]
Length = 821
Score = 470 bits (1209), Expect = e-130
Identities = 273/739 (36%), Positives = 442/739 (58%), Gaps = 30/739 (4%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S+ + +W YDVF++FRG DTR+TF SHLY L N GI TF D++ L+ G + EL++AI
Sbjct: 12 SQYRLRWKYDVFLSFRGVDTRRTFTSHLYEGLKNRGIFTFQDDKRLENGDSIPEELLKAI 71
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYA------ 119
+ SQ+A+++FSKNY SRWCL+EL +IMECK GQ+V+P+FY + PS +R+
Sbjct: 72 EESQVALIIFSKNYATSRWCLNELVKIMECKEEKGQIVIPIFYDVDPSEVRKQTKSFAEA 131
Query: 120 ----VTRFSETTLFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNK 175
++++ ++ + L DA+ L G+D+SN ES ++ IV + L
Sbjct: 132 FTEHESKYANDIEGMQKVKGWRTALSDAADLKGYDISN-RIESDYIQHIVDHI-SVLCKG 189
Query: 176 YLPLPDFQVGLEPRAEKSIRFLRQNTR--GVCLVGIWGMGGIGKSTIAKVIYNDLCYEFE 233
L VG++ K+IR L + GV +VGIWGM G+GK+TIA+ I++ L Y+FE
Sbjct: 190 SLSYIKNLVGIDTHF-KNIRSLLAELQMSGVLIVGIWGMPGVGKTTIARAIFDRLSYQFE 248
Query: 234 NQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVL 293
FLA+I+ E G LQ LS++LK + V + E G++++ +LR K++L VL
Sbjct: 249 AVCFLADIK---ENKCGMHSLQNILLSELLKEKDNCVNNKEDGRSLLAHRLRFKKVLVVL 305
Query: 294 DDVSELEQFDALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCK 352
DD+ ++Q D L + G GS II TTRD +++ + +YE L+ ++++LF +
Sbjct: 306 DDIDHIDQLDYLAGNLDWFGNGSRIIATTRD---KHLIGKNVVYELPTLHDHDAIKLFER 362
Query: 353 HAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQI 412
+AF++ + + F L+ +VV++ G+PLAL+V G + +R EWRS + +++ PN +I
Sbjct: 363 YAFKEQVSDKCFKELTLEVVSHAKGLPLALKVFGCFFHERDITEWRSAIKQIKNNPNSEI 422
Query: 413 HEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIK 472
E LKIS+DGL + ++++IFLD+ CF G+ + YV +IL C ADIG++VLI++SL+
Sbjct: 423 VEKLKISYDGL-ETIQQSIFLDIACFLRGRRKDYVMQILESCDFGADIGLSVLIDKSLVS 481
Query: 473 VEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMK 532
+ N + MH L++DMG+ +V++ ++P + +RLW +D V+ + TGTKA+E +
Sbjct: 482 ISGNNTIEMHDLIQDMGKYVVKKQ--KDPGERSRLWLTKDFEEVMINNTGTKAVEAI--W 537
Query: 533 LPKTNRVCFDTIAFEKMIRLRLLQL-DNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFY 591
+P NR F A M RLR+L + D+ + G + P LRW W +P + PENF
Sbjct: 538 VPNFNRPRFSKEAMTIMQRLRILCIHDSNCLDGSIEYLPNSLRWFVWNNYPCESLPENFE 597
Query: 592 QKNLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQS 651
+ LV ++L S+L +W + + L+ L+L S++L +TPDF+ +PNL+ L + C++
Sbjct: 598 PQKLVHLDLSLSSLHHLWTGKKHLPFLQKLDLRDSRSLMQTPDFTWMPNLKYLDLSYCRN 657
Query: 652 LLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQME 711
L EVH S+G + L+ LNL +C L P + ++ + L CS ++K M+
Sbjct: 658 LSEVHHSLGYSRELIELNLYNCGRLKRFP--CVNVESLDYMDLEFCSSLEKFPIIFGTMK 715
Query: 712 SLTTLMAANTGVKQPPFSI 730
+ +G+K+ P S+
Sbjct: 716 PELKIKMGLSGIKELPSSV 734
>dbj|BAB09567.1| disease resistance protein-like [Arabidopsis thaliana]
Length = 1295
Score = 469 bits (1207), Expect = e-130
Identities = 279/739 (37%), Positives = 428/739 (57%), Gaps = 22/739 (2%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S + W DVF++FRG D RKTF+SHL+ GI F D+ +LQ+GK + PELI AI
Sbjct: 10 SSSSTVWKTDVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQRGKSISPELIDAI 69
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSE 125
+GS+ AIVV S+NY S WCL EL +IMEC + ++P+FY + PS++R+ + +
Sbjct: 70 KGSRFAIVVVSRNYAASSWCLDELLKIMECNKD---TIVPIFYEVDPSDVRRQRGSFGED 126
Query: 126 TTLFFD--ELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPLPDFQ 183
D ++ + L+ + +SG D N+ ++SK++K+IV + L +
Sbjct: 127 VESHSDKEKVGKWKEALKKLAAISGEDSRNWRDDSKLIKKIVKDISDKLVSTSWDDSKGL 186
Query: 184 VGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIRE 243
+G+ + + + V ++GIWGMGG+GK+TIAK +YN L +F+ F+ N++E
Sbjct: 187 IGMSSHMDFLQSMISIVDKDVRMLGIWGMGGVGKTTIAKYLYNQLSGQFQVHCFMENVKE 246
Query: 244 VWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFD 303
V + G LQ +FL + + R + S +IK++ R K + VLDDV EQ +
Sbjct: 247 VCNR-YGVRRLQVEFLCRMFQERDKEAWSSVSCCNIIKERFRHKMVFIVLDDVDRSEQLN 305
Query: 304 ALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFR-KAIPT 361
L + GPGS II+TTRD +L ++ +Y+ + L E+L+LFC +AFR + I
Sbjct: 306 ELVKETGWFGPGSRIIVTTRDRHLLLSHGINLVYKVKCLPKKEALQLFCNYAFREEIILP 365
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
F LS V Y G+PLAL VLGS+L++R + EW S L++L+ P+ I E+L++S+D
Sbjct: 366 HGFEELSVQAVNYASGLPLALRVLGSFLYRRSQIEWESTLARLKTYPHSDIMEVLRVSYD 425
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL D EK IFL + CF+ K YV K+L+ CG A+IGIT+L E+SLI VE N + +
Sbjct: 426 GL-DEQEKAIFLYISCFYNMKQVDYVRKLLDLCGYAAEIGITILTEKSLI-VESNGCVKI 483
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H LL MGRE+VR+ + P + LW ED+ ++L++ +GT+ +EG+ + L + + V
Sbjct: 484 HDLLEQMGRELVRQQAVNNPAQRLLLWDPEDICHLLSENSGTQLVEGISLNLSEISEVFA 543
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKC--------FPKHLRWLSWQGFPLKYTPENFYQK 593
AFE + L+LL ++ G+ + P+ LR+L W G+PLK P F+ +
Sbjct: 544 SDRAFEGLSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYLRWDGYPLKTMPSRFFPE 603
Query: 594 NLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLL 653
LV + + +SNL ++W Q + LK ++LS K L PD SK NLE+L + CQSL+
Sbjct: 604 FLVELCMSNSNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLSKATNLEELNLSYCQSLV 663
Query: 654 EVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESL 713
EV PSI +LK L L +C L ++P I L+ +ET+ +SGCS + E +
Sbjct: 664 EVTPSIKNLKGLSCFYLTNCIQLKDIPIGII-LKSLETVGMSGCSSLKHFPE---ISWNT 719
Query: 714 TTLMAANTGVKQPPFSIVR 732
L ++T +++ P SI R
Sbjct: 720 RRLYLSSTKIEELPSSISR 738
Score = 61.6 bits (148), Expect = 1e-07
Identities = 66/253 (26%), Positives = 104/253 (41%), Gaps = 62/253 (24%)
Query: 618 LKILNLSHSKNLKRTPD-FSKLPNLEKLIMKDC--------------------QSLLEVH 656
LK LNL + L+ PD L +LE L + C S+ E+
Sbjct: 766 LKSLNLDGCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPRVSTSIEVLRISETSIEEIP 825
Query: 657 PSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCS------------------ 698
I +L L L++ + L +LP I +LR +E L LSGCS
Sbjct: 826 ARICNLSQLRSLDISENKRLASLPVSISELRSLEKLKLSGCSVLESFPLEICQTMSCLRW 885
Query: 699 ------KIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGY----EGLS 748
I +L E+I + +L L A+ T +++ P+SI R + +++ EGL
Sbjct: 886 FDLDRTSIKELPENIGNLVALEVLQASRTVIRRAPWSIARLTRLQVLAIGNSFFTPEGLL 945
Query: 749 HHVFPSLIR----SWISPTMNSLPRIPPFGGMSKSLFSLDIDSNNLALVSQS-------- 796
H + P L R +S + ++ IP G +L LD+ NN + S
Sbjct: 946 HSLCPPLSRFDDLRALSLSNMNMTEIPNSIGNLWNLLELDLSGNNFEFIPASIKRLTRLN 1005
Query: 797 -QILNSCSRLRSV 808
LN+C RL+++
Sbjct: 1006 RLNLNNCQRLQAL 1018
Score = 56.2 bits (134), Expect = 5e-06
Identities = 38/119 (31%), Positives = 58/119 (47%), Gaps = 3/119 (2%)
Query: 612 PQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLK 671
P++ + L LS +K + S+L L KL M DCQ L + +G L +L LNL
Sbjct: 713 PEISWNTRRLYLSSTKIEELPSSISRLSCLVKLDMSDCQRLRTLPSYLGHLVSLKSLNLD 772
Query: 672 DCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSI 730
C L NLP + L +ETL +SGC +++ S+ L + T +++ P I
Sbjct: 773 GCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPR---VSTSIEVLRISETSIEEIPARI 828
>ref|NP_197270.1| disease resistance protein (TIR-NBS-LRR class), putative
[Arabidopsis thaliana]
Length = 1294
Score = 465 bits (1196), Expect = e-129
Identities = 279/739 (37%), Positives = 428/739 (57%), Gaps = 23/739 (3%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S + W DVF++FRG D RKTF+SHL+ GI F D+ +LQ+GK + PELI AI
Sbjct: 10 SSSSTVWKTDVFVSFRGEDVRKTFVSHLFCEFDRMGIKAFRDDLDLQRGKSISPELIDAI 69
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQYAVTRFSE 125
+GS+ AIVV S+NY S WCL EL +IMEC + ++P+FY + PS++R+ + +
Sbjct: 70 KGSRFAIVVVSRNYAASSWCLDELLKIMECNKD---TIVPIFYEVDPSDVRRQRGSFGED 126
Query: 126 TTLFFD--ELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPLPDFQ 183
D ++ + L+ + +SG D N+ ++SK++K+IV + L +
Sbjct: 127 VESHSDKEKVGKWKEALKKLAAISGEDSRNW-DDSKLIKKIVKDISDKLVSTSWDDSKGL 185
Query: 184 VGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIRE 243
+G+ + + + V ++GIWGMGG+GK+TIAK +YN L +F+ F+ N++E
Sbjct: 186 IGMSSHMDFLQSMISIVDKDVRMLGIWGMGGVGKTTIAKYLYNQLSGQFQVHCFMENVKE 245
Query: 244 VWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFD 303
V + G LQ +FL + + R + S +IK++ R K + VLDDV EQ +
Sbjct: 246 VCNR-YGVRRLQVEFLCRMFQERDKEAWSSVSCCNIIKERFRHKMVFIVLDDVDRSEQLN 304
Query: 304 ALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFR-KAIPT 361
L + GPGS II+TTRD +L ++ +Y+ + L E+L+LFC +AFR + I
Sbjct: 305 ELVKETGWFGPGSRIIVTTRDRHLLLSHGINLVYKVKCLPKKEALQLFCNYAFREEIILP 364
Query: 362 QDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFD 421
F LS V Y G+PLAL VLGS+L++R + EW S L++L+ P+ I E+L++S+D
Sbjct: 365 HGFEELSVQAVNYASGLPLALRVLGSFLYRRSQIEWESTLARLKTYPHSDIMEVLRVSYD 424
Query: 422 GLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGM 481
GL D EK IFL + CF+ K YV K+L+ CG A+IGIT+L E+SLI VE N + +
Sbjct: 425 GL-DEQEKAIFLYISCFYNMKQVDYVRKLLDLCGYAAEIGITILTEKSLI-VESNGCVKI 482
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCF 541
H LL MGRE+VR+ + P + LW ED+ ++L++ +GT+ +EG+ + L + + V
Sbjct: 483 HDLLEQMGRELVRQQAVNNPAQRLLLWDPEDICHLLSENSGTQLVEGISLNLSEISEVFA 542
Query: 542 DTIAFEKMIRLRLLQLDNVQVIGDYKC--------FPKHLRWLSWQGFPLKYTPENFYQK 593
AFE + L+LL ++ G+ + P+ LR+L W G+PLK P F+ +
Sbjct: 543 SDRAFEGLSNLKLLNFYDLSFDGETRVHLPNGLSYLPRKLRYLRWDGYPLKTMPSRFFPE 602
Query: 594 NLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLL 653
LV + + +SNL ++W Q + LK ++LS K L PD SK NLE+L + CQSL+
Sbjct: 603 FLVELCMSNSNLEKLWDGIQPLRNLKKMDLSRCKYLVEVPDLSKATNLEELNLSYCQSLV 662
Query: 654 EVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESL 713
EV PSI +LK L L +C L ++P I L+ +ET+ +SGCS + E +
Sbjct: 663 EVTPSIKNLKGLSCFYLTNCIQLKDIPIGII-LKSLETVGMSGCSSLKHFPE---ISWNT 718
Query: 714 TTLMAANTGVKQPPFSIVR 732
L ++T +++ P SI R
Sbjct: 719 RRLYLSSTKIEELPSSISR 737
Score = 61.6 bits (148), Expect = 1e-07
Identities = 66/253 (26%), Positives = 104/253 (41%), Gaps = 62/253 (24%)
Query: 618 LKILNLSHSKNLKRTPD-FSKLPNLEKLIMKDC--------------------QSLLEVH 656
LK LNL + L+ PD L +LE L + C S+ E+
Sbjct: 765 LKSLNLDGCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPRVSTSIEVLRISETSIEEIP 824
Query: 657 PSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCS------------------ 698
I +L L L++ + L +LP I +LR +E L LSGCS
Sbjct: 825 ARICNLSQLRSLDISENKRLASLPVSISELRSLEKLKLSGCSVLESFPLEICQTMSCLRW 884
Query: 699 ------KIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGY----EGLS 748
I +L E+I + +L L A+ T +++ P+SI R + +++ EGL
Sbjct: 885 FDLDRTSIKELPENIGNLVALEVLQASRTVIRRAPWSIARLTRLQVLAIGNSFFTPEGLL 944
Query: 749 HHVFPSLIR----SWISPTMNSLPRIPPFGGMSKSLFSLDIDSNNLALVSQS-------- 796
H + P L R +S + ++ IP G +L LD+ NN + S
Sbjct: 945 HSLCPPLSRFDDLRALSLSNMNMTEIPNSIGNLWNLLELDLSGNNFEFIPASIKRLTRLN 1004
Query: 797 -QILNSCSRLRSV 808
LN+C RL+++
Sbjct: 1005 RLNLNNCQRLQAL 1017
Score = 56.2 bits (134), Expect = 5e-06
Identities = 38/119 (31%), Positives = 58/119 (47%), Gaps = 3/119 (2%)
Query: 612 PQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLK 671
P++ + L LS +K + S+L L KL M DCQ L + +G L +L LNL
Sbjct: 712 PEISWNTRRLYLSSTKIEELPSSISRLSCLVKLDMSDCQRLRTLPSYLGHLVSLKSLNLD 771
Query: 672 DCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSI 730
C L NLP + L +ETL +SGC +++ S+ L + T +++ P I
Sbjct: 772 GCRRLENLPDTLQNLTSLETLEVSGCLNVNEFPR---VSTSIEVLRISETSIEEIPARI 827
>gb|AAR21295.1| bacterial spot disease resistance protein 4 [Lycopersicon
esculentum]
Length = 1146
Score = 464 bits (1195), Expect = e-129
Identities = 284/784 (36%), Positives = 438/784 (55%), Gaps = 53/784 (6%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
SK +W Y VF++FRG DTRKTF HLY L N GINTF D++ L+ G + EL+RAI
Sbjct: 12 SKYYPRWKYVVFLSFRGEDTRKTFTGHLYEGLRNRGINTFQDDKRLEHGDSIPKELLRAI 71
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKAND-GQVVMPVFYCITPSNIRQYA----- 119
+ SQ+A+++FSKNY SRWCL+EL +IMECK + GQ V+P+FY + PS++R
Sbjct: 72 EDSQVALIIFSKNYATSRWCLNELVKIMECKEEENGQTVIPIFYNVDPSHVRYQTESFGA 131
Query: 120 -----VTRFSETTLFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDN 174
+++ + ++ + L A+ L G+D+ N ES+ +++IV +
Sbjct: 132 AFAKHESKYKDDVEGMQKVQRWRTALTAAANLKGYDIRN-GIESENIQQIVDCISSKFCT 190
Query: 175 KYLPLPDFQ--VGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEF 232
L Q VG+ EK L+ V ++GIWG+GG+GK+ IAK I++ L Y+F
Sbjct: 191 NAYSLSFLQDIVGINAHLEKLKSKLQIEINDVRILGIWGIGGVGKTRIAKAIFDTLSYQF 250
Query: 233 ENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAV 292
E FLA+++E +K++ LQ LS++L+ + V + GK MI +L + ++L V
Sbjct: 251 EASCFLADVKEFAKKNKLH-SLQNILLSELLRKKNDYVYNKYDGKCMIPNRLCSLKVLIV 309
Query: 293 LDDVSELEQFDALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFC 351
LDD+ +Q + L G GS +I+TTR+ ++ + D IYE L E+++LF
Sbjct: 310 LDDIDHGDQMEYLAGDICWFGNGSRVIVTTRNKHLIE--KDDAIYEVSTLPDHEAMQLFN 367
Query: 352 KHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQ 411
HAF+K +P +DF L+ ++V + G+PLAL+V G L K+ W+ + +++K N +
Sbjct: 368 MHAFKKEVPNEDFKELALEIVNHAKGLPLALKVWGCLLHKKNLSLWKITVEQIKKDSNSE 427
Query: 412 IHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLI 471
I E LKIS+DGL+ E+ IFLD+ CFF G+ R V +IL C A+ G+ VLI +SL+
Sbjct: 428 IVEQLKISYDGLESE-EQEIFLDIACFFRGEKRKEVMQILKSCDFGAEYGLDVLINKSLV 486
Query: 472 KVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVM 531
+ +N ++ MH L+RDMGR +V+ + +K +R+W ED V+ DYTGT +E +
Sbjct: 487 FISENDRIEMHDLIRDMGRYVVKMQKLQ--KKRSRIWDVEDFKEVMIDYTGTMTVEAIWF 544
Query: 532 KLPKTNRVCFDTIAFEKMIRLRLLQL-----------------------DNVQVIGDY-- 566
V F+ A +KM RLR+L + D+ ++ D+
Sbjct: 545 SC--FEEVRFNKEAMKKMKRLRILHIFDGFVKFFSSPPSSNSNDSEEEDDSYDLVVDHHD 602
Query: 567 ---KCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKHSNLAQVWKKPQLIEGLKILNL 623
+ +LRWL W + K PENF + LV +EL+ S+L +WKK + + L+ L+L
Sbjct: 603 DSIEYLSNNLRWLVWNHYSWKSLPENFKPEKLVHLELRWSSLHYLWKKTEHLPSLRKLDL 662
Query: 624 SHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREI 683
S SK+L +TPDF+ +PNLE L ++ C L EVH S+ + L+ LNL CT L P
Sbjct: 663 SLSKSLVQTPDFTGMPNLEYLNLEYCSKLEEVHYSLAYCEKLIELNLSWCTKLRRFP--Y 720
Query: 684 YQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCG 743
+ +E+L L C I E I M+ +++ANT + + P S+ + + L G
Sbjct: 721 INMESLESLDLQYCYGIMVFPEIIGTMKPELMILSANTMITELPSSLQYPTHLTELDLSG 780
Query: 744 YEGL 747
E L
Sbjct: 781 MENL 784
>emb|CAD36199.1| NLS-TIR-NBS disease resistance protein [Populus tremula]
Length = 516
Score = 459 bits (1181), Expect = e-127
Identities = 248/516 (48%), Positives = 342/516 (66%), Gaps = 18/516 (3%)
Query: 14 YDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQGSQIAIV 73
YDVF +FRG DTRKTF HLYTAL AGI+TF D++ L +G+E+ L++AI+ S+I IV
Sbjct: 1 YDVFFSFRGKDTRKTFTDHLYTALVQAGIHTFRDDDELPRGEEISDHLLKAIRESKICIV 60
Query: 74 VFSKNYVHSRWCLSELKQIMECK-ANDGQVVMPVFYCITPSNIRQYAVTRFSETTLFFDE 132
VFSK Y SRWCL EL +I++CK GQ+ +P+FY I PS +R+ + F+E + +E
Sbjct: 61 VFSKGYASSRWCLDELVEILKCKYRKTGQIALPIFYDIDPSYVRKQTGS-FAEAFVKHEE 119
Query: 133 -----LVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLPLPDFQVGLE 187
+ + L++A LSGW+L ++ E+K ++EI+ VL LD KYL +P VG++
Sbjct: 120 RSKEKVKEWREALEEAGNLSGWNLKDH--EAKFIQEIIKDVLTKLDPKYLHVPKHLVGID 177
Query: 188 PRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEF----ENQSFLANIRE 243
P A FL VC+VG+ GM GIGK+TIAKV++N LCY F E FL N++E
Sbjct: 178 PLAHNIFHFLSTAADDVCIVGLHGMPGIGKTTIAKVVFNQLCYGFGYGFEGNLFLLNVKE 237
Query: 244 VWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFD 303
E + + LQ+Q L DIL+ K+ +V++GK +IK++L KR+L V+DDV L+Q +
Sbjct: 238 KSEPN-DLVLLQQQLLHDILRQNTEKITNVDRGKVLIKERLCRKRVLVVVDDVDHLDQLN 296
Query: 304 ALC-QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQ 362
AL +R+ GPGS +IITTRD R+L LE D Y+ +E++ ESL+LFC+HAFR A P +
Sbjct: 297 ALMGERSWFGPGSRVIITTRDERLL--LEADQRYQVQEMDPYESLQLFCQHAFRDAKPAK 354
Query: 363 DFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDG 422
D++ LS DVV YCGG+PLALEVLGS L + + W SV+ +L +IP I E L+ISFD
Sbjct: 355 DYVELSNDVVEYCGGLPLALEVLGSCLIGKNQARWESVIDRLRRIPEHAIQERLRISFDS 414
Query: 423 LKDRMEKNIFLDVCCFFIGKDRAYVTKILNG-CGLNADIGITVLIERSLIKVEKNKKLGM 481
LK KN FLD+ CFFIG + YV ++L G G N + LIERS+IKV+ + + M
Sbjct: 415 LKAPNLKNTFLDISCFFIGGQKEYVAEVLEGRYGCNPEDDFGTLIERSVIKVDDSGTISM 474
Query: 482 HALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVL 517
H LLR+MGR IV++ SPE P + +R+WC ED VL
Sbjct: 475 HDLLREMGRGIVKDESPENPAQRSRIWCQEDAWKVL 510
>gb|AAU04763.1| MRGH8 [Cucumis melo]
Length = 1058
Score = 458 bits (1179), Expect = e-127
Identities = 292/826 (35%), Positives = 457/826 (54%), Gaps = 48/826 (5%)
Query: 14 YDVFINFRGADTR------KTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAIQG 67
YDVF++ R D R ++FIS L+ ALT+ GI F+D E+ + G + E ++A+
Sbjct: 33 YDVFLSHRAKDHRANNDTGRSFISDLHEALTSQGIVVFIDKEDEEDGGKPLTEKMKAVDE 92
Query: 68 SQIAIVVFSKNYVHSRW-CLSELKQIMECKANDGQVVMPVFYCITPSNIRQYA----VTR 122
S+ +IVVFS+NY W C+ E+++I C+ Q+V+P+FY + P ++R+ V
Sbjct: 93 SRSSIVVFSENY--GSWVCMKEIRKIRMCQKLRDQLVLPIFYKVDPGDVRKQEGESLVKF 150
Query: 123 FSE----TTLFFDELVPFMNTLQDASYLSGWDLSNY----------SNESKVVKEIVSQV 168
F+E + +E+ + ++ LSGW L + S+E +KEIV+ V
Sbjct: 151 FNEHEANPNISIEEVKKWRKSMNKVGNLSGWHLQDSQLNITFKQFCSSEEGAIKEIVNHV 210
Query: 169 LKNLDNKYLPLPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDL 228
L D VG+ R + L + VGIWGMGGIGK+T+A++IY +
Sbjct: 211 FNKLRPDLFRYDDKLVGISQRLHQINMLLGIGLDDIRFVGIWGMGGIGKTTLARIIYRSV 270
Query: 229 CYEFENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKR 288
+ F+ FL N++E +K +G LQE+ L+ L R I + + + G T+IK+++ +
Sbjct: 271 SHLFDGCYFLDNVKEALKK-QGIASLQEKLLTGALMKRNIDIPNAD-GATLIKRRISNIK 328
Query: 289 ILAVLDDVSELEQFDALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESL 347
L +LDDV L Q L + G GS II+TTR+ +L ++ Y+ E LN E+L
Sbjct: 329 ALIILDDVDHLSQLQQLAGSSDWFGSGSRIIVTTRNEHLLVSHGIEKRYKVEGLNVEEAL 388
Query: 348 ELFCKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKI 407
+LF + AF P +D+ LS VV Y G +PLA+EVLGS L + ++ W++ + KL++I
Sbjct: 389 QLFSQKAFGTNYPKKDYFDLSIQVVEYSGDLPLAIEVLGSSLRDKSREVWKNAVEKLKEI 448
Query: 408 PNDQIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIE 467
+ +I EIL++S+D L D+ EK IFLD+ CFF K + ++L G A IG+ +L E
Sbjct: 449 RDKKILEILRVSYD-LLDKSEKEIFLDLACFFKKKSKKQAIEVLQSFGFQAIIGLEILEE 507
Query: 468 RSLIKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIE 527
RSLI ++K+ MH L+++MG+E+VR P PEK TRLW EDV L+ G +AIE
Sbjct: 508 RSLITTP-HEKIQMHDLIQEMGQEVVRRMFPNNPEKRTRLWLREDVNLALSHDQGAEAIE 566
Query: 528 GLVMKLPKTNRVCFDTIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTP 587
G+VM + + F M LR+L+++NV + G+ LR+LSW G+P KY P
Sbjct: 567 GIVMDSSEEGESHLNAKVFSTMTNLRILKINNVSLCGELDYLSDQLRFLSWHGYPSKYLP 626
Query: 588 ENFYQKNLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMK 647
NF+ K+++ +EL +S + +WK + ++ LK +NLS S+ + +TPDFS +PNLE+LI+
Sbjct: 627 PNFHPKSILELELPNSFIHYLWKGSKRLDRLKTVNLSDSQFISKTPDFSGVPNLERLILS 686
Query: 648 DCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDI 707
C L ++H S+G LK L+ L+LK+C +L +P I L + L LS CS + +
Sbjct: 687 GCVRLTKLHQSLGSLKRLIQLDLKNCKALKAIPFSI-SLESLIVLSLSNCSSLKNFPNIV 745
Query: 708 VQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSHHVFPSLIRSWIS------ 761
M++LT L T +++ SI + ++L L P+ I S I
Sbjct: 746 GNMKNLTELHLDGTSIQELHPSIGHLTGLVLLNLENCTNLLE--LPNTIGSLICLKTLTL 803
Query: 762 PTMNSLPRIPPFGGMSKSLFSLDIDSN-------NLALVSQSQILN 800
+ L RIP G SL LD+ + +L L++ +IL+
Sbjct: 804 HGCSKLTRIPESLGFIASLEKLDVTNTCINQAPLSLQLLTNLEILD 849
>gb|AAO45748.1| MRGH5 [Cucumis melo]
Length = 1092
Score = 456 bits (1172), Expect = e-126
Identities = 270/721 (37%), Positives = 422/721 (58%), Gaps = 16/721 (2%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S P W+YDVF++FRG DTR F HLY L G+N F+D + L++G+++ L + I
Sbjct: 13 SSPIFNWSYDVFLSFRGEDTRSNFTGHLYMFLRQKGVNVFID-DGLERGEQISETLFKTI 71
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQ----YAVT 121
Q S I+IV+FS+NY S WCL EL +IMECK + GQ V+P+FY + PS++R+ +
Sbjct: 72 QNSLISIVIFSENYASSTWCLDELVEIMECKKSKGQKVLPIFYKVDPSDVRKQNGWFREG 131
Query: 122 RFSETTLFFDELVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDN-KYLPLP 180
F +++ + + L A+ LSGW L E+ ++++IV +VL L++ K L
Sbjct: 132 LAKHEANFMEKIPIWRDALTTAANLSGWHLGA-RKEAHLIQDIVKEVLSILNHTKPLNAN 190
Query: 181 DFQVGLEPRAEKSIRFLRQ-NTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLA 239
+ VG++ + E R + V ++GI+G+GGIGK+T+AK +Y+ + +FE +L
Sbjct: 191 EHLVGIDSKIEFLYRKEEMYKSECVNMLGIYGIGGIGKTTLAKALYDKMASQFEGCCYLR 250
Query: 240 NIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSEL 299
++RE + G LQ++ L ILK ++V+ ++ G +IK +LR+K++L +LDDV +L
Sbjct: 251 DVREASKLFDGLTQLQKKLLFQILKY-DLEVVDLDWGINIIKNRLRSKKVLILLDDVDKL 309
Query: 300 EQFDALCQRNS-VGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKA 358
EQ AL + G G+ II+TTR+ ++L D +YE + L+ E++ELF +HAF+
Sbjct: 310 EQLQALVGGHDWFGQGTKIIVTTRNKQLLVSHGFDKMYEVQGLSKHEAIELFRRHAFKNL 369
Query: 359 IPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKK-QEWRSVLSKLEKIPNDQIHEILK 417
P+ ++L LS YC G PLAL VLGS+L R EW +L E I +IL+
Sbjct: 370 QPSSNYLDLSERATRYCTGHPLALIVLGSFLCDRSDLAEWSGILDGFENSLRKDIKDILQ 429
Query: 418 ISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNK 477
+SFDGL+D + K IFLD+ C +GK +YV K+L+ C D GIT L + SLI+ E ++
Sbjct: 430 LSFDGLEDEV-KEIFLDISCLLVGKRVSYVKKMLSECHSILDFGITKLKDLSLIRFEDDR 488
Query: 478 KLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTN 537
+ MH L++ MG +IV + S ++P K +RLW +D++ V ++ +G+ A++ + + L
Sbjct: 489 -VQMHDLIKQMGHKIVHDESHDQPGKRSRLWLEKDILEVFSNNSGSDAVKAIKLVLTDPK 547
Query: 538 RVC-FDTIAFEKMIRLRLLQLD-NVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNL 595
RV D AF M LR+L +D NV+ K P L+W+ W F P F K+L
Sbjct: 548 RVIDLDPEAFRSMKNLRILMVDGNVRFCKKIKYLPNGLKWIKWHRFAHPSLPSCFITKDL 607
Query: 596 VAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEV 655
V ++L+HS + K Q LK+L+L HS LK+ + S PNLE+L + +C +L +
Sbjct: 608 VGLDLQHSFITNFGKGLQNCMRLKLLDLRHSVILKKISESSAAPNLEELYLSNCSNLKTI 667
Query: 656 HPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTT 715
S L+ L+ L+L C +L +PR +E L LS C K++K+ DI +L +
Sbjct: 668 PKSFLSLRKLVTLDLHHCVNLKKIPRSYISWEALEDLDLSHCKKLEKI-PDISSASNLRS 726
Query: 716 L 716
L
Sbjct: 727 L 727
Score = 86.7 bits (213), Expect = 3e-15
Identities = 55/143 (38%), Positives = 81/143 (56%), Gaps = 9/143 (6%)
Query: 600 LKHSNLAQVWKKPQLIEG--LKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHP 657
LK N + + K P+ I L+ LNLS K L+ PDFS NL+ L ++ C SL VH
Sbjct: 751 LKLQNCSNLKKLPRYISWNFLQDLNLSWCKKLEEIPDFSSTSNLKHLSLEQCTSLRVVHD 810
Query: 658 SIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLM 717
SIG L L+ LNL+ C++L LP + +L+ ++ L LSGC K++ E M+SL L
Sbjct: 811 SIGSLSKLVSLNLEKCSNLEKLPSYL-KLKSLQNLTLSGCCKLETFPEIDENMKSLYILR 869
Query: 718 AANTGVKQPPFSIVRSKSIGYIS 740
+T +++ P SIGY++
Sbjct: 870 LDSTAIRELP------PSIGYLT 886
Score = 76.6 bits (187), Expect = 3e-12
Identities = 60/214 (28%), Positives = 103/214 (48%), Gaps = 14/214 (6%)
Query: 583 LKYTPENFYQ-KNLVAMELKHS-NLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPN 640
LK P++F + LV ++L H NL ++ + E L+ L+LSH K L++ PD S N
Sbjct: 664 LKTIPKSFLSLRKLVTLDLHHCVNLKKIPRSYISWEALEDLDLSHCKKLEKIPDISSASN 723
Query: 641 LEKLIMKDCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKI 700
L L + C +L+ +H SIG L L+ L L++C++L LPR I ++ L LS C K+
Sbjct: 724 LRSLSFEQCTNLVMIHDSIGSLTKLVTLKLQNCSNLKKLPRYI-SWNFLQDLNLSWCKKL 782
Query: 701 DKLEE-------DIVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSHHVFP 753
+++ + + +E T+L + + S+ + S+ E L ++
Sbjct: 783 EEIPDFSSTSNLKHLSLEQCTSLRVVHDSIG----SLSKLVSLNLEKCSNLEKLPSYLKL 838
Query: 754 SLIRSWISPTMNSLPRIPPFGGMSKSLFSLDIDS 787
+++ L P KSL+ L +DS
Sbjct: 839 KSLQNLTLSGCCKLETFPEIDENMKSLYILRLDS 872
Score = 55.1 bits (131), Expect = 1e-05
Identities = 50/185 (27%), Positives = 82/185 (44%), Gaps = 38/185 (20%)
Query: 548 KMIRLRLLQLDNVQVIGDYKCFPKHLRW-------LSWQGFPLKYTPENFYQKNLVAMEL 600
K++ L+L N+ K P+++ W LSW L+ P+ NL + L
Sbjct: 747 KLVTLKLQNCSNL------KKLPRYISWNFLQDLNLSWCK-KLEEIPDFSSTSNLKHLSL 799
Query: 601 KH-SNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDC---------- 649
+ ++L V + L LNL NL++ P + KL +L+ L + C
Sbjct: 800 EQCTSLRVVHDSIGSLSKLVSLNLEKCSNLEKLPSYLKLKSLQNLTLSGCCKLETFPEID 859
Query: 650 -------------QSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSG 696
++ E+ PSIG L +L M +LK CT+L +LP + L+ + L LSG
Sbjct: 860 ENMKSLYILRLDSTAIRELPPSIGYLTHLYMFDLKGCTNLISLPCTTHLLKSLGELHLSG 919
Query: 697 CSKID 701
S+ +
Sbjct: 920 SSRFE 924
>gb|AAT37497.1| N-like protein [Nicotiana tabacum]
Length = 941
Score = 449 bits (1154), Expect = e-124
Identities = 298/893 (33%), Positives = 473/893 (52%), Gaps = 54/893 (6%)
Query: 6 SKPQSKWTYDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKGKELGPELIRAI 65
S S+W+YDVF++FRG DTRKTF SHLY L + GI TF D + L+ G + EL +AI
Sbjct: 4 SSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLKDRGIKTFQDEKRLEYGATIPEELCKAI 63
Query: 66 QGSQIAIVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIR----QYAVT 121
+ SQ AIVVFS+NY SRWCL+EL +IMECK Q ++P+FY + PS++R +A
Sbjct: 64 EESQFAIVVFSENYATSRWCLNELVKIMECKTQFRQTIIPIFYDVDPSHVRNQKESFAKA 123
Query: 122 RFSETTLFFDE---LVPFMNTLQDASYLSGWDLSNYSNESKVVKEIVSQVLKNLDNKYLP 178
T + D+ + + L A+ L G + ++ +++IV Q+ L L
Sbjct: 124 FEEHETKYKDDVEGIQRWRTALNAAANLKGSCDNRDKTDADCIRQIVDQISSKLSKISLS 183
Query: 179 LPDFQVGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYN------DLCYEF 232
VG++ E+ L V +VGIWGMGG+GK+TIA+ +++ D Y+F
Sbjct: 184 YLQNIVGIDTHLEEIESLLGIGINDVRIVGIWGMGGVGKTTIARAMFDTLLGRRDSSYQF 243
Query: 233 ENQSFLANIREVWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAV 292
+ FL +I+ E RG LQ L ++L+ + + GK + +LR+K++L V
Sbjct: 244 DGACFLKDIK---ENKRGMHSLQNTLLFELLR-ENANYNNEDDGKHQMASRLRSKKVLIV 299
Query: 293 LDDVSELEQFDALC--QRNSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELF 350
LDD+ + + + + G GS II+TTRD ++ + D IYE L E+++LF
Sbjct: 300 LDDIDDKDHYLEYLAGDLDWFGNGSRIIVTTRDKHLIG--KNDIIYEVTALPDHEAIQLF 357
Query: 351 CKHAFRKAIPTQDFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPND 410
+HAF+K +P + F LS +VV + G+PLAL+V GS L KR W+S + +++ PN
Sbjct: 358 YQHAFKKEVPDECFKELSLEVVNHAKGLPLALKVWGSSLHKRDITVWKSAIEQMKINPNS 417
Query: 411 QIHEILKISFDGLKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSL 470
+I E LKIS+DGL + M++ +FLD+ CFF G+ + Y+ ++L C A+ G+ VLIE+SL
Sbjct: 418 KIVEKLKISYDGL-ESMQQEMFLDIACFFRGRQKDYIMQVLKSCHFGAEYGLDVLIEKSL 476
Query: 471 IKVEKNKKLGMHALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLV 530
+ + + ++ MH L++DMG+ IV + ++P + +RLW EDV V+ + GT ++E V
Sbjct: 477 VFISEYNQVEMHDLIQDMGKYIV--NFKKDPGERSRLWLAEDVEEVMNNNAGTMSVE--V 532
Query: 531 MKLPKTNRVCFDTIAFEKMIRLRLLQ----LDNVQVIGDYKCFPKHLRWLSWQGFPLKYT 586
+ + + F A + M RLR+L L + G + P +LRW +P +
Sbjct: 533 IWVHYDFGLYFSNDAMKNMKRLRILHIKGYLSSTSHDGSIEYLPSNLRWFVLDDYPWESL 592
Query: 587 PENFYQKNLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIM 646
P F K LV +EL S+L +W + + + L+ ++LS S+ L+RTPDF+ +PNLE L M
Sbjct: 593 PSTFDLKMLVHLELSRSSLHYLWTETKHLPSLRRIDLSSSRRLRRTPDFTGMPNLEYLNM 652
Query: 647 KDCQSLLEVHPSIGDLKNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEED 706
C++L EVH S+ L+ LNL +C SL P + +E L L CS ++K E
Sbjct: 653 LYCRNLEEVHHSLRCCSKLIRLNLNNCKSLKRFP--CVNVESLEYLSLEYCSSLEKFPEI 710
Query: 707 IVQMESLTTLMAANTGVKQPPFSIVRSKSIGYISLCGYEGLSHHVFPSLIRSWISPTMNS 766
+M+ + +G+++ P SI + ++ H+ +R M
Sbjct: 711 HGRMKPEIQIHMQGSGIRELPSSITQYQT--------------HITKLDLRG-----MEK 751
Query: 767 LPRIPPFGGMSKSLFSLDIDSNNLALVSQSQILNSCSRLRSVSVQCDSEIQLKQEFGRFL 826
L +P KSL SL + S L S + + L + C + R
Sbjct: 752 LVALPSSICRLKSLVSLSV-SGCFKLESLPEEVGDLENLEELDASCTLISRPPSSIIRLS 810
Query: 827 D-DLYDAGLTEMRTSHALQISNLTMRSL-LFGIGSCHIVINTLRKSLSQVSSL 877
++D G ++ R L RSL + +C+++ L + + +SSL
Sbjct: 811 KLKIFDFGSSKDRVHFELPPVVEGFRSLETLSLRNCNLIDGGLPEDMGSLSSL 863
Score = 36.6 bits (83), Expect = 3.7
Identities = 22/70 (31%), Positives = 37/70 (52%)
Query: 594 NLVAMELKHSNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLL 653
+L + L +N + + + L+IL L + K L + P+F+ + NLE L ++ C L
Sbjct: 862 SLKKLYLSGNNFEHLPRSIAQLGALRILELRNCKRLTQLPEFTGMLNLEYLDLEGCSYLE 921
Query: 654 EVHPSIGDLK 663
EVH G L+
Sbjct: 922 EVHHFPGVLQ 931
>gb|AAU04761.1| MRGH13 [Cucumis melo]
Length = 1024
Score = 447 bits (1151), Expect = e-124
Identities = 287/811 (35%), Positives = 452/811 (55%), Gaps = 32/811 (3%)
Query: 14 YDVFINFRGADTRKTFISHLYTALTNAGINTFLDNENLQKG-KELG-PELIRAIQGSQIA 71
YDVF++ R DT ++F + L+ ALT+ GI F D+ + + G K G E ++A++ S+ +
Sbjct: 38 YDVFLSHRAKDTGQSFAADLHEALTSQGIVVFRDDVDEEDGEKPYGVEEKMKAVEESRSS 97
Query: 72 IVVFSKNYVHSRWCLSELKQIMECKANDGQVVMPVFYCITPSNIRQ-------YAVTRFS 124
IVVFS+NY S C+ E+ +I CK Q+V+P+FY I P N+R+ Y +
Sbjct: 98 IVVFSENY-GSFVCMKEVGKIAMCKELMDQLVLPIFYKIDPGNVRKQEGNFEKYFNEHEA 156
Query: 125 ETTLFFDELVPFMNTLQDASYLSGWDLSN-YSNESKVVKEIVSQVLKNLDNKYLPLPDFQ 183
+ +E+ + ++ +LSGW + + S E ++ E+V + L D
Sbjct: 157 NPKIDIEEVENWRYSMNQVGHLSGWHVQDSQSEEGSIIDEVVKHIFNKLRPDLFRYDDKL 216
Query: 184 VGLEPRAEKSIRFLRQNTRGVCLVGIWGMGGIGKSTIAKVIYNDLCYEFENQSFLANIRE 243
VG+ PR + L V VGIWGMGGIGK+T+A++IY + + F+ FL N++E
Sbjct: 217 VGITPRLHQINMLLGIGLDDVRFVGIWGMGGIGKTTLARIIYKSVSHLFDGCYFLDNVKE 276
Query: 244 VWEKDRGRIDLQEQFLSDILKTRKIKVLSVEQGKTMIKQQLRAKRILAVLDDVSELEQFD 303
+K+ LQ++ ++ L R I + + + G T+IK+++ + L +LDDV+ L Q
Sbjct: 277 ALKKE-DIASLQQKLITGTLMKRNIDIPNAD-GATLIKRRISKIKALIILDDVNHLSQLQ 334
Query: 304 ALCQR-NSVGPGSIIIITTRDLRVLNILEVDFIYEAEELNASESLELFCKHAFRKAIPTQ 362
L + G GS +I+TTRD +L ++ Y E L E L+LF + AF + P +
Sbjct: 335 KLAGGLDWFGSGSRVIVTTRDEHLLISHGIERRYNVEVLKIEEGLQLFSQKAFGEEHPKE 394
Query: 363 DFLILSRDVVAYCGGIPLALEVLGSYLFKRKKQEWRSVLSKLEKIPNDQIHEILKISFDG 422
++ L VV Y GG+PLA+EVLGS L + ++W + + KL ++ + +I E LKIS+
Sbjct: 395 EYFDLCSQVVNYAGGLPLAIEVLGSSLHNKPMEDWINAVEKLWEVRDKEIIEKLKISYYM 454
Query: 423 LKDRMEKNIFLDVCCFFIGKDRAYVTKILNGCGLNADIGITVLIERSLIKVEKNKKLGMH 482
L++ E+ IFLD+ CFF K + +IL G A +G+ +L E+ LI +K L +H
Sbjct: 455 LEES-EQKIFLDIACFFKRKSKNQAIEILESFGFPAVLGLEILEEKCLITAPHDK-LQIH 512
Query: 483 ALLRDMGREIVRESSPEEPEKHTRLWCHEDVVNVLADYTGTKAIEGLVMKLPKTNRVCFD 542
L+++MG+EIVR + P EPEK TRLW ED+ L+ GT+AIEG++M + +
Sbjct: 513 DLIQEMGQEIVRHTFPNEPEKRTRLWLREDINLALSRDQGTEAIEGIMMDFDEEGESHLN 572
Query: 543 TIAFEKMIRLRLLQLDNVQVIGDYKCFPKHLRWLSWQGFPLKYTPENFYQKNLVAMELKH 602
AF M LR+L+L+NV + + + LR+L+W G+PLK P NF NL+ +EL +
Sbjct: 573 AKAFSSMTNLRVLKLNNVHLCEEIEYLSDQLRFLNWHGYPLKTLPSNFNPTNLLELELPN 632
Query: 603 SNLAQVWKKPQLIEGLKILNLSHSKNLKRTPDFSKLPNLEKLIMKDCQSLLEVHPSIGDL 662
S++ +W + +E LK++NLS S+ L +TPDFS +PNLE+L++ C L ++H S+G+L
Sbjct: 633 SSIHLLWTTSKSMETLKVINLSDSQFLSKTPDFSVVPNLERLVLSGCVELHQLHHSLGNL 692
Query: 663 KNLLMLNLKDCTSLGNLPREIYQLRRVETLILSGCSKIDKLEEDIVQMESLTTLMAANTG 722
K+L+ L+L++C L N+P I L ++ L+LSGCS + + M L L T
Sbjct: 693 KHLIQLDLRNCKKLTNIPFNIC-LESLKILVLSGCSSLTHFPKISSNMNYLLELHLEETS 751
Query: 723 VKQPPFSIVRSKSIGYISLCGYEGLSHHVFPSLIRSWIS-PTMN-----SLPRIPPFGGM 776
+K SI S+ ++L L PS I S S T+N L +P G
Sbjct: 752 IKVLHSSIGHLTSLVVLNLKNCTNLLK--LPSTIGSLTSLKTLNLNGCSELDSLPESLGN 809
Query: 777 SKSLFSLDIDSN-------NLALVSQSQILN 800
SL LDI S + L+++ +ILN
Sbjct: 810 ISSLEKLDITSTCVNQAPMSFQLLTKLEILN 840
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.138 0.404
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,425,049,103
Number of Sequences: 2540612
Number of extensions: 59177979
Number of successful extensions: 172587
Number of sequences better than 10.0: 3229
Number of HSP's better than 10.0 without gapping: 1441
Number of HSP's successfully gapped in prelim test: 1801
Number of HSP's that attempted gapping in prelim test: 160842
Number of HSP's gapped (non-prelim): 7271
length of query: 882
length of database: 863,360,394
effective HSP length: 137
effective length of query: 745
effective length of database: 515,296,550
effective search space: 383895929750
effective search space used: 383895929750
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC145513.6


