

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145331.13 + phase: 0 /partial
(365 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
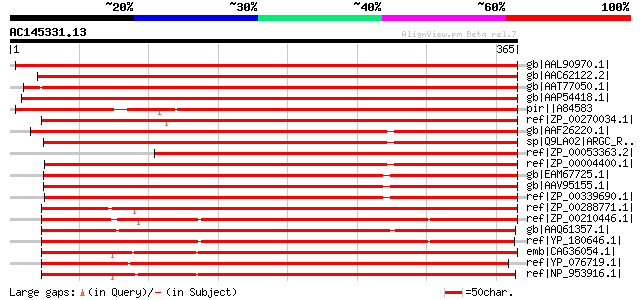
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL90970.1| At2g19940/F6F22.3 [Arabidopsis thaliana] gi|16604... 547 e-154
gb|AAC62122.2| putative N-acetyl-gamma-glutamyl-phosphate reduct... 547 e-154
gb|AAT77050.1| putative Semialdehyde dehydrogenase [Oryza sativa... 530 e-149
gb|AAP54418.1| putative N-acetyl-gamma-glutamyl-phosphate reduct... 526 e-148
pir||A84583 hypothetical protein At2g19940 [imported] - Arabidop... 499 e-140
ref|ZP_00270034.1| COG0002: Acetylglutamate semialdehyde dehydro... 394 e-108
gb|AAF26220.1| N-acetyl-gamma-glutamyl-phosphate reductase [Rhod... 349 7e-95
sp|Q9LA02|ARGC_RHOCA N-acetyl-gamma-glutamyl-phosphate reductase... 348 1e-94
ref|ZP_00053363.2| COG0002: Acetylglutamate semialdehyde dehydro... 345 1e-93
ref|ZP_00004400.1| COG0002: Acetylglutamate semialdehyde dehydro... 345 2e-93
gb|EAM67725.1| N-acetyl-gamma-glutamyl-phosphate reductase [Jann... 334 2e-90
gb|AAV95155.1| N-acetyl-gamma-glutamyl-phosphate reductase [Sili... 331 2e-89
ref|ZP_00339690.1| COG0002: Acetylglutamate semialdehyde dehydro... 330 5e-89
ref|ZP_00288771.1| COG0002: Acetylglutamate semialdehyde dehydro... 326 6e-88
ref|ZP_00210446.1| COG0002: Acetylglutamate semialdehyde dehydro... 323 5e-87
gb|AAQ61357.1| N-acetyl-gamma-glutamyl-phosphate reductase [Chro... 311 2e-83
ref|YP_180646.1| N-acetyl-gamma-glutamyl-phosphate reductase [Eh... 309 8e-83
emb|CAG36054.1| probable N-acetyl-gamma-glutamyl-phosphate reduc... 306 5e-82
ref|YP_076719.1| putative N-acetyl-gamma-glutamyl-phosphate redu... 304 2e-81
ref|NP_953916.1| N-acetyl-gamma-glutamyl-phosphate reductase [Ge... 304 2e-81
>gb|AAL90970.1| At2g19940/F6F22.3 [Arabidopsis thaliana] gi|16604368|gb|AAL24190.1|
At2g19940/F6F22.3 [Arabidopsis thaliana]
Length = 401
Score = 547 bits (1410), Expect = e-154
Identities = 264/361 (73%), Positives = 311/361 (86%)
Query: 5 PQNVKLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAI 64
P ++ S + ++R+G+LGASGYTG+EI+RLLANHP F V LMTADRKAGQ++
Sbjct: 40 PSSMSFRVSASSSVKPEKDIRIGLLGASGYTGAEIVRLLANHPHFQVTLMTADRKAGQSM 99
Query: 65 SSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRL 124
SVFPHL Q LP L+++KDA+FS VDAVFCCLPHGTTQEIIK LP LKIVDLSADFRL
Sbjct: 100 ESVFPHLRVQKLPTLVSVKDADFSTVDAVFCCLPHGTTQEIIKELPTALKIVDLSADFRL 159
Query: 125 RDVSEYEEWYGQPHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIK 184
R+++EYEEWYGQPH+A +LQKE +YGLTE+LRE+IK ARLVANPGCYPT+IQLPLVPL+K
Sbjct: 160 RNIAEYEEWYGQPHKAVELQKEVVYGLTEILREDIKKARLVANPGCYPTTIQLPLVPLLK 219
Query: 185 ANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAG 244
ANLI+ NII+DAKSGVSGAGR AKE L++E+ EG+SSYGVTRHRH PEIEQGL+D A
Sbjct: 220 ANLIKHENIIIDAKSGVSGAGRGAKEANLYSEIAEGISSYGVTRHRHVPEIEQGLSDVAQ 279
Query: 245 SKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRT 304
SKVT+SFTPHL+PM RGMQSTIYVEMAPGVR +DL QL+ SYEDEEFV +L+ GV+PRT
Sbjct: 280 SKVTVSFTPHLMPMIRGMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDEGVVPRT 339
Query: 305 HSVKGSNYCLFNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
H+V+GSNYC +VFPDRIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE GL + PL
Sbjct: 340 HNVRGSNYCHMSVFPDRIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTGLLHQPL 399
Query: 365 F 365
F
Sbjct: 400 F 400
>gb|AAC62122.2| putative N-acetyl-gamma-glutamyl-phosphate reductase [Arabidopsis
thaliana] gi|30680834|ref|NP_849993.1| semialdehyde
dehydrogenase family protein [Arabidopsis thaliana]
gi|18399202|ref|NP_565461.1| semialdehyde dehydrogenase
family protein [Arabidopsis thaliana]
gi|56967313|pdb|1XYG|D Chain D, X-Ray Structure Of Gene
Product From Arabidopsis Thaliana At2g19940
gi|56967312|pdb|1XYG|C Chain C, X-Ray Structure Of Gene
Product From Arabidopsis Thaliana At2g19940
gi|56967311|pdb|1XYG|B Chain B, X-Ray Structure Of Gene
Product From Arabidopsis Thaliana At2g19940
gi|56967310|pdb|1XYG|A Chain A, X-Ray Structure Of Gene
Product From Arabidopsis Thaliana At2g19940
Length = 359
Score = 547 bits (1409), Expect = e-154
Identities = 262/345 (75%), Positives = 307/345 (88%)
Query: 21 QNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLI 80
+ ++R+G+LGASGYTG+EI+RLLANHP F V LMTADRKAGQ++ SVFPHL Q LP L+
Sbjct: 14 EKDIRIGLLGASGYTGAEIVRLLANHPHFQVTLMTADRKAGQSMESVFPHLRAQKLPTLV 73
Query: 81 AIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRA 140
++KDA+FS VDAVFCCLPHGTTQEIIK LP LKIVDLSADFRLR+++EYEEWYGQPH+A
Sbjct: 74 SVKDADFSTVDAVFCCLPHGTTQEIIKELPTALKIVDLSADFRLRNIAEYEEWYGQPHKA 133
Query: 141 PDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSG 200
+LQKE +YGLTE+LRE+IK ARLVANPGCYPT+IQLPLVPL+KANLI+ NII+DAKSG
Sbjct: 134 VELQKEVVYGLTEILREDIKKARLVANPGCYPTTIQLPLVPLLKANLIKHENIIIDAKSG 193
Query: 201 VSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSR 260
VSGAGR AKE L++E+ EG+SSYGVTRHRH PEIEQGL+D A SKVT+SFTPHL+PM R
Sbjct: 194 VSGAGRGAKEANLYSEIAEGISSYGVTRHRHVPEIEQGLSDVAQSKVTVSFTPHLMPMIR 253
Query: 261 GMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
GMQSTIYVEMAPGVR +DL QL+ SYEDEEFV +L+ GV+PRTH+V+GSNYC +VFPD
Sbjct: 254 GMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDEGVVPRTHNVRGSNYCHMSVFPD 313
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
RIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE GL + PLF
Sbjct: 314 RIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTGLLHQPLF 358
>gb|AAT77050.1| putative Semialdehyde dehydrogenase [Oryza sativa (japonica
cultivar-group)]
Length = 415
Score = 530 bits (1364), Expect = e-149
Identities = 262/355 (73%), Positives = 301/355 (83%), Gaps = 1/355 (0%)
Query: 11 SPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPH 70
SP +KS + VR+ VLGASGYTG+EI+RLLANHPQF + +MTADRKAG+ SVFPH
Sbjct: 61 SPKTSGVKSGEE-VRIAVLGASGYTGAEIVRLLANHPQFRIKVMTADRKAGEQFGSVFPH 119
Query: 71 LGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEY 130
L TQDLP+L+A+KDA+FS+VDAVFCCLPHGTTQEIIKGLP+ LKIVDLSADFRLRD++EY
Sbjct: 120 LITQDLPNLVAVKDADFSNVDAVFCCLPHGTTQEIIKGLPQELKIVDLSADFRLRDINEY 179
Query: 131 EEWYGQPHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQT 190
EWYG HRAP+LQ+EA+YGLTEVLR EI+NARLVANPGCYPTSIQLPLVPLIKA LI+
Sbjct: 180 AEWYGHSHRAPELQQEAVYGLTEVLRNEIRNARLVANPGCYPTSIQLPLVPLIKAKLIKV 239
Query: 191 TNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTIS 250
+NII+DAKSGVSGAGR AKE L+TE+ EG+ +YG+ HRH PEIEQGL++AA SKVTIS
Sbjct: 240 SNIIIDAKSGVSGAGRGAKEANLYTEIAEGIHAYGIKGHRHVPEIEQGLSEAAESKVTIS 299
Query: 251 FTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGS 310
FTP+LI M RGMQST++VEMAPGV +DL L+ +YE EEFV LL +P T V GS
Sbjct: 300 FTPNLICMKRGMQSTMFVEMAPGVTANDLYQHLKSTYEGEEFVKLLNGSSVPHTRHVVGS 359
Query: 311 NYCLFNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
NYC NVF DRIPGRAIIISVIDNLVKGASGQA+QNLNL+MG PEN GLQY PLF
Sbjct: 360 NYCFMNVFEDRIPGRAIIISVIDNLVKGASGQAVQNLNLMMGLPENTGLQYQPLF 414
>gb|AAP54418.1| putative N-acetyl-gamma-glutamyl-phosphate reductase [Oryza sativa
(japonica cultivar-group)] gi|37535658|ref|NP_922131.1|
putative N-acetyl-gamma-glutamyl-phosphate reductase
[Oryza sativa (japonica cultivar-group)]
gi|22128709|gb|AAM92821.1| putative
N-acetyl-gamma-glutamyl-phosphate reductase [Oryza
sativa (japonica cultivar-group)]
Length = 416
Score = 526 bits (1356), Expect = e-148
Identities = 262/357 (73%), Positives = 299/357 (83%)
Query: 9 KLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVF 68
KL K + S V++ VLGASGYTG+EI+RLLANHPQF + +MTADRKA + SVF
Sbjct: 59 KLYSSKTTQVKSGEEVQIAVLGASGYTGAEIVRLLANHPQFHIKVMTADRKASEQFGSVF 118
Query: 69 PHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVS 128
PHL TQDLP+L+AIKDA+FS+VDAVFCCLPHGTTQEIIKGLPK LKIVDLSADFRLRD++
Sbjct: 119 PHLITQDLPNLVAIKDADFSNVDAVFCCLPHGTTQEIIKGLPKQLKIVDLSADFRLRDIN 178
Query: 129 EYEEWYGQPHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLI 188
EY EWYG HRAP+LQ+EA+YGLTEVLR+EI+NARLVANPGCYPTSIQLPLVPLIKA LI
Sbjct: 179 EYAEWYGHAHRAPELQQEAVYGLTEVLRDEIRNARLVANPGCYPTSIQLPLVPLIKAKLI 238
Query: 189 QTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVT 248
+ +NII+DAKSGV GAGR AKE L+TE+ EG+ +YG+ HRHAPEIEQGL++AA SKVT
Sbjct: 239 KLSNIIIDAKSGVCGAGRGAKEANLYTEIAEGIHAYGIKGHRHAPEIEQGLSEAAKSKVT 298
Query: 249 ISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVK 308
ISFTP+LI M RGMQST++VEMAPGV DL L+ +YE EEFV LL +P T V
Sbjct: 299 ISFTPNLICMKRGMQSTMFVEMAPGVTAGDLYQHLKSTYEGEEFVKLLHGSTVPHTRHVV 358
Query: 309 GSNYCLFNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GSNYC NVF DRIPGRAIIISVIDNLVKGASGQA+QNLNL+MG PEN GLQY PLF
Sbjct: 359 GSNYCFMNVFEDRIPGRAIIISVIDNLVKGASGQAVQNLNLMMGLPENRGLQYQPLF 415
>pir||A84583 hypothetical protein At2g19940 [imported] - Arabidopsis thaliana
Length = 389
Score = 499 bits (1285), Expect = e-140
Identities = 249/367 (67%), Positives = 297/367 (80%), Gaps = 16/367 (4%)
Query: 5 PQNVKLSPIKCSIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAI 64
P ++ S + ++R+G+LGASGYTG+EI+RLLANHP F V LMTADRKAGQ++
Sbjct: 32 PSSMSFRVSASSSVKPEKDIRIGLLGASGYTGAEIVRLLANHPHFQVTLMTADRKAGQSM 91
Query: 65 SSVFPHLGTQDLPDLIAIKDANFSDVDAVFCCLPHGTTQEII------KGLPKHLKIVDL 118
SVFPHL QD A+FS VDAVFCCLPHGTTQ + L L + ++
Sbjct: 92 ESVFPHLRAQD---------ADFSTVDAVFCCLPHGTTQVHFPFILPFQFLKAFLSLTEM 142
Query: 119 SADFRLRDVSEYEEWYGQPHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLP 178
S DFRLR+++EYEEWYGQPH+A +LQKE +YGLTE+LRE+IK ARLVANPGCYPT+IQLP
Sbjct: 143 S-DFRLRNIAEYEEWYGQPHKAVELQKEVVYGLTEILREDIKKARLVANPGCYPTTIQLP 201
Query: 179 LVPLIKANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQG 238
LVPL+KANLI+ NII+DAKSGVSGAGR AKE L++E+ EG+SSYGVTRHRH PEIEQG
Sbjct: 202 LVPLLKANLIKHENIIIDAKSGVSGAGRGAKEANLYSEIAEGISSYGVTRHRHVPEIEQG 261
Query: 239 LADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLEN 298
L+D A SKVT+SFTPHL+PM RGMQSTIYVEMAPGVR +DL QL+ SYEDEEFV +L+
Sbjct: 262 LSDVAQSKVTVSFTPHLMPMIRGMQSTIYVEMAPGVRTEDLHQQLKTSYEDEEFVKVLDE 321
Query: 299 GVIPRTHSVKGSNYCLFNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLG 358
GV+PRTH+V+GSNYC +VFPDRIPGRAIIISVIDNLVKGASGQALQNLN+++G+PE G
Sbjct: 322 GVVPRTHNVRGSNYCHMSVFPDRIPGRAIIISVIDNLVKGASGQALQNLNIMLGYPETTG 381
Query: 359 LQYLPLF 365
L + PLF
Sbjct: 382 LLHQPLF 388
>ref|ZP_00270034.1| COG0002: Acetylglutamate semialdehyde dehydrogenase [Rhodospirillum
rubrum]
Length = 352
Score = 394 bits (1011), Expect = e-108
Identities = 201/347 (57%), Positives = 246/347 (69%), Gaps = 5/347 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
V V +LGASGYTG+E++RLLA HP+ +A +TA+RKAGQA +SVFPHLG DLP L I+
Sbjct: 5 VNVAILGASGYTGAELVRLLARHPRVTLAALTANRKAGQAFASVFPHLGGLDLPVLSTIE 64
Query: 84 DANFSDVDAVFCCLPHGTTQEIIKGLPK-----HLKIVDLSADFRLRDVSEYEEWYGQPH 138
++S +D VFC LPHGTTQ II L L+I DLSADFRL D Y+ WYG H
Sbjct: 65 AVDWSAIDFVFCALPHGTTQTIIGDLLNGPHGGRLRIADLSADFRLADPMVYQTWYGHAH 124
Query: 139 RAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAK 198
A +LQKEA+YGLTE+ R I ARLVA PGCYPTS QLPL+PL++A LI II+DAK
Sbjct: 125 EAVELQKEAVYGLTEINRAAIATARLVAVPGCYPTSAQLPLIPLLRAGLIDPDAIIIDAK 184
Query: 199 SGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPM 258
SG SGAGR AKE L EV+EG+ +YGV HRH PEIEQGL+ A G V ++FTPHL+PM
Sbjct: 185 SGASGAGRDAKEGSLHCEVSEGIHAYGVGTHRHGPEIEQGLSLAVGRPVAVTFTPHLMPM 244
Query: 259 SRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVF 318
+RG+ STIY+ G LR L +Y DE FV ++ GV P T V+GSN+ L V
Sbjct: 245 NRGILSTIYLRATAGNDATTLRQALSAAYADEAFVRVVPEGVSPHTRHVRGSNFVLIGVH 304
Query: 319 PDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
DR+PGR I+ V DNLVKGASGQA+Q++N+++GFPE LGL PLF
Sbjct: 305 ADRVPGRVIVTCVEDNLVKGASGQAIQDMNVMLGFPETLGLDQQPLF 351
>gb|AAF26220.1| N-acetyl-gamma-glutamyl-phosphate reductase [Rhodobacter
capsulatus]
Length = 418
Score = 349 bits (896), Expect = 7e-95
Identities = 171/351 (48%), Positives = 233/351 (65%), Gaps = 5/351 (1%)
Query: 16 SIKSSQNNVRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQD 75
+I ++ ++ +LGASGYTG+E+ R++A HP+ + ++ DRKAG + VFPHL
Sbjct: 71 TIAETKMTQKIAILGASGYTGAELARIIATHPEMEITALSGDRKAGMRMGEVFPHLRHIG 130
Query: 76 LPDLIAIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYG 135
LPDL+ I++ +FS +D FC LPH T+Q +I LP+ LK+VDLSADFRLRD +EYE+WYG
Sbjct: 131 LPDLVKIEEIDFSGIDLAFCALPHATSQAVIAELPRDLKVVDLSADFRLRDAAEYEKWYG 190
Query: 136 QPHRAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIV 195
+PH A DLQ EA+YGLTE REE+K ARL A GC + Q + PLI+A +I I++
Sbjct: 191 KPHAAMDLQAEAVYGLTEFYREELKTARLCAGTGCNAAAGQYAIRPLIEAGVIDLDEIVI 250
Query: 196 DAKSGVSGAGRSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPH 254
D K+GVSGAGRS KENLL E+ G Y +HRH E +Q + AG V + FTPH
Sbjct: 251 DLKAGVSGAGRSLKENLLHAELAGGTMPYSAGGKHRHLGEFDQEFSKVAGRPVRVQFTPH 310
Query: 255 LIPMSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCL 314
L+P +RG+ +T+YV P +D+ L YE+E F+ +L G + T SV GSN+
Sbjct: 311 LMPFNRGILATVYVRGTP----EDIHAALASRYENEVFLEVLPFGQLASTRSVAGSNFVH 366
Query: 315 FNVFPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
V DR+PGRA++ +DNL KG+SGQA+QN NL++G PE LGL P+F
Sbjct: 367 LGVIGDRLPGRAVVTVALDNLTKGSSGQAIQNANLMLGLPETLGLMLAPVF 417
>sp|Q9LA02|ARGC_RHOCA N-acetyl-gamma-glutamyl-phosphate reductase (AGPR)
(N-acetyl-glutamate semialdehyde dehydrogenase) (NAGSA
dehydrogenase)
Length = 342
Score = 348 bits (894), Expect = 1e-94
Identities = 170/342 (49%), Positives = 229/342 (66%), Gaps = 5/342 (1%)
Query: 25 RVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKD 84
++ +LGASGYTG+E+ R++A HP+ + ++ DRKAG + VFPHL LPDL+ I++
Sbjct: 4 KIAILGASGYTGAELARIIATHPEMEITALSGDRKAGMRMGEVFPHLRHIGLPDLVKIEE 63
Query: 85 ANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQ 144
+FS +D FC LPH T+Q +I LP+ LK+VDLSADFRLRD +EYE+WYG+PH A DLQ
Sbjct: 64 IDFSGIDLAFCALPHATSQAVIAELPRDLKVVDLSADFRLRDAAEYEKWYGKPHAAMDLQ 123
Query: 145 KEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGA 204
EA+YGLTE REE+K ARL A GC + Q + PLI+A +I I++D K+GVSGA
Sbjct: 124 AEAVYGLTEFYREELKTARLCAGTGCNAAAGQYAIRPLIEAGVIDLDEIVIDLKAGVSGA 183
Query: 205 GRSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQ 263
GRS KENLL E+ G Y +HRH E +Q + AG V + FTPHL+P +RG+
Sbjct: 184 GRSLKENLLHAELAGGTMPYSAGGKHRHLGEFDQEFSKVAGRPVRVQFTPHLMPFNRGIL 243
Query: 264 STIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIP 323
+T+YV P +D+ L YE+E F+ +L G + T SV GSN+ V DR+P
Sbjct: 244 ATVYVRGTP----EDIHAALASRYENEVFLEVLPFGQLASTRSVAGSNFVHLGVIGDRLP 299
Query: 324 GRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GRA++ +DNL KG+SGQA+QN NL++G PE LGL P+F
Sbjct: 300 GRAVVTVALDNLTKGSSGQAIQNANLMLGLPETLGLMLAPVF 341
>ref|ZP_00053363.2| COG0002: Acetylglutamate semialdehyde dehydrogenase
[Magnetospirillum magnetotacticum MS-1]
Length = 262
Score = 345 bits (885), Expect = 1e-93
Identities = 166/261 (63%), Positives = 203/261 (77%)
Query: 105 IIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQKEAIYGLTEVLREEIKNARL 164
+I LP +K+VDLSADFRL DV Y +WYG HRAP+LQK+A+YGLTE+ R+ + ARL
Sbjct: 1 MIAALPASVKVVDLSADFRLFDVDTYAQWYGHEHRAPELQKQAVYGLTELARDSVAKARL 60
Query: 165 VANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAGRSAKENLLFTEVTEGMSSY 224
VANPGCYPTS+QLPL+PL++A LI+ ++I++DAKSGVSGAGR+AKE L+TEVTEGM +Y
Sbjct: 61 VANPGCYPTSVQLPLIPLLRAGLIEASDIVIDAKSGVSGAGRAAKEANLYTEVTEGMHAY 120
Query: 225 GVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQSTIYVEMAPGVRIDDLRHQLQ 284
GV HRHAPEIEQGL+ AAG V +SFTPHLIPMSRGM STIYV+ G + DLR L
Sbjct: 121 GVASHRHAPEIEQGLSQAAGKPVLVSFTPHLIPMSRGMLSTIYVKTKNGAKAADLRAHLS 180
Query: 285 LSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIPGRAIIISVIDNLVKGASGQAL 344
++ E FV ++ GV+P T V+GSN+CL N F DR+PGR II SVIDNLVKGASGQAL
Sbjct: 181 KTFGGEPFVRVVPEGVVPHTRHVRGSNHCLINAFDDRVPGRIIITSVIDNLVKGASGQAL 240
Query: 345 QNLNLIMGFPENLGLQYLPLF 365
QN+NL+ G PE L P+F
Sbjct: 241 QNMNLMFGLPEATALAQQPMF 261
>ref|ZP_00004400.1| COG0002: Acetylglutamate semialdehyde dehydrogenase [Rhodobacter
sphaeroides 2.4.1]
Length = 342
Score = 345 bits (884), Expect = 2e-93
Identities = 172/341 (50%), Positives = 231/341 (67%), Gaps = 5/341 (1%)
Query: 26 VGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKDA 85
+ +LGASGYTG+E++RL+A HP + ++ DRKAG A+S VFP L DLP L I++
Sbjct: 5 IAILGASGYTGAELVRLIATHPAMRIVALSGDRKAGMAMSEVFPFLRHLDLPRLQKIEEI 64
Query: 86 NFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQK 145
+FS VD FC LPH T+Q +I GLP+ LK+VDLSADFRLRD + YE WYG+PH AP+LQK
Sbjct: 65 DFSSVDLAFCALPHATSQAVIAGLPRDLKVVDLSADFRLRDPAAYETWYGKPHAAPELQK 124
Query: 146 EAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAG 205
EA+YGLTE R+EI+ ARLVA GC + Q + PLI+A +I +I++D K+GVSGAG
Sbjct: 125 EAVYGLTEFYRDEIRGARLVAGTGCNAATGQYAIRPLIEAGVIDLDDILIDLKAGVSGAG 184
Query: 206 RSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQS 264
RS KENLL E++EG +Y RHRH E +Q + AG V + FTPHL PM+RG+ +
Sbjct: 185 RSLKENLLHAELSEGTHAYSAGGRHRHLGEFDQEFSKIAGRPVQVRFTPHLTPMNRGILA 244
Query: 265 TIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIPG 324
+YV+ P + L Y E F+ +L G +P T ++GSNY V DR+PG
Sbjct: 245 NVYVKGDP----QAVHRALTERYLTETFLEVLPFGALPSTRDIRGSNYVHIGVIGDRVPG 300
Query: 325 RAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
A++++V+DNL KG+SGQA+QN NL++G E GL P+F
Sbjct: 301 CAMVVAVLDNLCKGSSGQAIQNANLMLGLDEAAGLGLAPVF 341
>gb|EAM67725.1| N-acetyl-gamma-glutamyl-phosphate reductase [Jannaschia sp. CCS1]
Length = 342
Score = 334 bits (857), Expect = 2e-90
Identities = 169/342 (49%), Positives = 227/342 (65%), Gaps = 5/342 (1%)
Query: 25 RVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKD 84
++ +LGASGYTG+E+ RL+A HP + +T DRKAGQ ++SV+P L LPDL+ I+D
Sbjct: 4 KIAILGASGYTGAELCRLIAGHPDMTIVALTGDRKAGQPMASVYPFLRGAGLPDLVRIED 63
Query: 85 ANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQ 144
+FS VD FC LPH T+Q +IK LP LK+VDLSADFRLRD ++Y +WYG+ H AP LQ
Sbjct: 64 VDFSGVDLAFCALPHATSQSVIKALPADLKVVDLSADFRLRDPADYAKWYGKDHDAPALQ 123
Query: 145 KEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGA 204
K+AIYGLTE R++I++ARLVA GC + Q L PLI+ ++ +II+D GVSGA
Sbjct: 124 KQAIYGLTEFYRDQIRDARLVAGTGCNAATGQFALRPLIENGVVDLDDIIIDLICGVSGA 183
Query: 205 GRSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQ 263
GR+ KENLL E++EG +Y + HRH E +Q + AG KV + FTPHL P +RG+
Sbjct: 184 GRALKENLLHAELSEGTHAYALGGTHRHLGEFDQEFSKVAGRKVEVQFTPHLAPFNRGIL 243
Query: 264 STIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIP 323
+T YV+ + L +Y E F+ +L G P TH V+G+NYC V DR
Sbjct: 244 ATCYVK----GDAQAIHATLTAAYRGEMFIEVLPFGAAPSTHDVRGTNYCHIGVVADRRS 299
Query: 324 GRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GRA I++ +DNL KG+SGQA+QN NL++G E GL L +F
Sbjct: 300 GRAQIVAALDNLCKGSSGQAVQNANLMLGLEETAGLLGLGVF 341
>gb|AAV95155.1| N-acetyl-gamma-glutamyl-phosphate reductase [Silicibacter pomeroyi
DSS-3] gi|56696752|ref|YP_167113.1|
N-acetyl-gamma-glutamyl-phosphate reductase
[Silicibacter pomeroyi DSS-3]
Length = 342
Score = 331 bits (848), Expect = 2e-89
Identities = 174/342 (50%), Positives = 223/342 (64%), Gaps = 5/342 (1%)
Query: 25 RVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKD 84
++ +LGASGYTG+E++RL+A HP + ++ +RKAGQ+++ VFPHL DLP L I +
Sbjct: 4 KIAILGASGYTGAELVRLIAEHPNMEIVALSGERKAGQSMAEVFPHLRHLDLPVLCKIDE 63
Query: 85 ANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQ 144
+F+ VD FC LPH T+QE+I+ LP LKIVDLSADFRLRD Y WYG H A + Q
Sbjct: 64 IDFAGVDLCFCALPHKTSQEVIRALPATLKIVDLSADFRLRDPEAYRTWYGNEHVALEQQ 123
Query: 145 KEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGA 204
EA+YGLTE R++I+ ARLVA GC + Q L PLI A +I II+D K VSGA
Sbjct: 124 AEAVYGLTEFYRDQIRTARLVAGTGCNAATGQYVLRPLIAAGVIDLDEIILDLKCAVSGA 183
Query: 205 GRSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQ 263
GRS KENLL E++EG ++Y V HRH E +Q + AG V + FTPHLIP +RG+
Sbjct: 184 GRSLKENLLHAELSEGANAYAVGGTHRHLGEFDQEFSALAGRPVQVQFTPHLIPANRGIL 243
Query: 264 STIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIP 323
+T YV + L +Y DE FV +L G P TH V+GSN+C V DRI
Sbjct: 244 ATTYVR----GDAQTVFQTLAAAYADEPFVHVLPFGETPSTHHVRGSNHCHIGVTGDRIA 299
Query: 324 GRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
GRAI+I+ +DNL KG+SGQALQN NL++G E GL PLF
Sbjct: 300 GRAIVIAALDNLTKGSSGQALQNANLMLGETETTGLTMAPLF 341
>ref|ZP_00339690.1| COG0002: Acetylglutamate semialdehyde dehydrogenase [Silicibacter
sp. TM1040]
Length = 342
Score = 330 bits (845), Expect = 5e-89
Identities = 172/341 (50%), Positives = 221/341 (64%), Gaps = 5/341 (1%)
Query: 26 VGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIKDA 85
V +LGASGYTG+E++RL++ HP + + A+RKAG ++ VFPHL LP L I +
Sbjct: 5 VAILGASGYTGAELIRLISQHPSITIKALAAERKAGMEMADVFPHLRHLSLPTLCKIDEI 64
Query: 86 NFSDVDAVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAPDLQK 145
+F+ +D FC LPH T+QE+I LP LKIVDLSADFRLRD YE+WYG PH A + Q+
Sbjct: 65 DFAQIDLCFCALPHKTSQEVIAKLPGDLKIVDLSADFRLRDPEAYEKWYGNPHAALEQQQ 124
Query: 146 EAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGVSGAG 205
EA+YGLTE R+EIK ARLVA GC + Q L PLI A +I II+D K VSGAG
Sbjct: 125 EAVYGLTEFYRDEIKGARLVAGTGCNAATGQFALRPLIAAGVIDLDEIILDMKCAVSGAG 184
Query: 206 RSAKENLLFTEVTEGMSSYGV-TRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRGMQS 264
R+ KENLL E++EG ++Y + HRH E +Q + AG V + FTPHL+P++RG+ +
Sbjct: 185 RALKENLLHAELSEGYNAYAIGGTHRHIGEFDQEFSAIAGRPVKVQFTPHLLPVNRGILA 244
Query: 265 TIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDRIPG 324
T YV+ + +Y DE FV LL G P TH V+GSN+ V DRI G
Sbjct: 245 TTYVK----GDAQAIYETFAKAYADEPFVELLPFGEAPSTHHVRGSNFVHIGVTADRIAG 300
Query: 325 RAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
RAI+I +DNL KG+SGQALQN NL++G E GL PLF
Sbjct: 301 RAIVIVALDNLTKGSSGQALQNANLMLGEDETAGLMMAPLF 341
>ref|ZP_00288771.1| COG0002: Acetylglutamate semialdehyde dehydrogenase [Magnetococcus
sp. MC-1]
Length = 349
Score = 326 bits (836), Expect = 6e-88
Identities = 173/346 (50%), Positives = 225/346 (65%), Gaps = 6/346 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
+RV +LGA+GYTG E++RLL HP ++ ++++R AGQ+I+ V+ HL Q + LI
Sbjct: 5 LRVAILGATGYTGGELIRLLHRHPGAELSFVSSERFAGQSIAKVYTHL--QAVGHLICQP 62
Query: 84 -DANFS--DVDAVFCCLPHGTTQEIIKGL-PKHLKIVDLSADFRLRDVSEYEEWYGQPHR 139
DA + D +FC LPH T+ E++ L + K+VDLSADFRL+ Y WYG H
Sbjct: 63 MDAKKACEAADFIFCALPHVTSMEVVPELLQRGAKVVDLSADFRLKSAETYAHWYGTQHL 122
Query: 140 APDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKS 199
AP+L +A YGL E+ RE IK A LVANPGCYPTS+QLPL PL++ +I +I D+KS
Sbjct: 123 APELLPQAAYGLPELFRESIKGANLVANPGCYPTSVQLPLFPLLQEGMIDPALVIADSKS 182
Query: 200 GVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMS 259
GVSGAGRS + LF E++EG +Y V HRH PE+EQ LA AAG ++ I FTPHL+P S
Sbjct: 183 GVSGAGRSPAQGTLFAELSEGFKAYKVEAHRHIPEMEQNLALAAGKEIKIRFTPHLLPQS 242
Query: 260 RGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFP 319
RG+ ST Y+ GV +R L Y DE FV +L +G +P T V+GSN C V
Sbjct: 243 RGILSTCYLRPTSGVNAQRVRDTLMTRYRDEPFVTVLPHGDMPATSDVRGSNACHMGVTE 302
Query: 320 DRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
D G II+SVIDNLVKGASG A+QN NL+ G+ E L LP+F
Sbjct: 303 DLRSGWLIIVSVIDNLVKGASGAAVQNFNLMTGWDETTALDSLPMF 348
>ref|ZP_00210446.1| COG0002: Acetylglutamate semialdehyde dehydrogenase [Ehrlichia
canis str. Jake]
Length = 346
Score = 323 bits (828), Expect = 5e-87
Identities = 166/348 (47%), Positives = 231/348 (65%), Gaps = 12/348 (3%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIAIK 83
V V V+GA+GY G E++RLL HP + + A + G+ +SS + H+ + I+I
Sbjct: 5 VSVAVVGATGYVGVELVRLLLAHPMVKIRYLCATQSTGKLLSSSYFHISQ----NYISIS 60
Query: 84 DANFSDVD-----AVFCCLPHGTTQEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPH 138
++F D+D VF CLPHG + EI+K + +K++DLSADFR++D Y++WYG H
Sbjct: 61 VSSFDDIDLDKLDVVFLCLPHGASSEIVKKIHNVVKVIDLSADFRIKDPDIYKQWYGV-H 119
Query: 139 RAPDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAK 198
PDL K+ +YGLTE+ E+I+ + +A PGCYPTS+ +PL PL++ L+++ NIIVDAK
Sbjct: 120 YCPDLIKDFVYGLTEIYYEDIQKSSFIACPGCYPTSVLIPLFPLLRLRLVKSNNIIVDAK 179
Query: 199 SGVSGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAA-GSKVTISFTPHLIP 257
SGVSGAGRS K++ LF EV + + SY V+ HRH PEIEQ L AA + + F P+LIP
Sbjct: 180 SGVSGAGRSVKQDNLFCEVYDAIKSYKVSNHRHIPEIEQELCFAACREDINLQFVPNLIP 239
Query: 258 MSRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNV 317
+ RGM S IY+E+ GV + D+R L L Y D FVV+ E I T SV G+NYC +
Sbjct: 240 VKRGMMSNIYLELEVGVSLTDVREALLLFYRDSFFVVVDEEKAI-TTRSVVGTNYCYLGI 298
Query: 318 FPDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
FP R+P +IISVIDNL+KGA+GQA+QN N++M + E L L +P F
Sbjct: 299 FPGRLPNTIVIISVIDNLLKGAAGQAVQNFNVMMSYEEKLALSNIPYF 346
>gb|AAQ61357.1| N-acetyl-gamma-glutamyl-phosphate reductase [Chromobacterium
violaceum ATCC 12472] gi|34499150|ref|NP_903365.1|
N-acetyl-gamma-glutamyl-phosphate reductase
[Chromobacterium violaceum ATCC 12472]
gi|39930966|sp|Q7NRT5|ARGC_CHRVO
N-acetyl-gamma-glutamyl-phosphate reductase (AGPR)
(N-acetyl-glutamate semialdehyde dehydrogenase) (NAGSA
dehydrogenase)
Length = 342
Score = 311 bits (797), Expect = 2e-83
Identities = 163/343 (47%), Positives = 222/343 (64%), Gaps = 6/343 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHL-GTQDLPDLIAI 82
V+VG++G +GYTG E+LRLLA HPQ V +T+ + AG + +FP L G DL
Sbjct: 2 VKVGIVGGTGYTGVELLRLLAKHPQACVTAVTSRKDAGIRVDEMFPSLRGRIDLA-FTTP 60
Query: 83 KDANFSDVDAVFCCLPHGTT-QEIIKGLPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
+A ++ D VF P+G Q+ L ++++DL+ADFR++DV E+E+WYG H +P
Sbjct: 61 DEARLAECDVVFFATPNGIAMQDAAALLDAGVRVIDLAADFRIKDVFEWEKWYGMQHASP 120
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
L +A+YGL EV RE I++ARLVANPGCYPT++QL +PLI+A +I T ++I D KSGV
Sbjct: 121 HLVADAVYGLPEVNREAIRSARLVANPGCYPTAVQLGFLPLIEAGVIDTASLIADCKSGV 180
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAAGSKVTISFTPHLIPMSRG 261
SGAGR + L E + +YGV HRH PEI QGLA A+G V ++F PHL PM RG
Sbjct: 181 SGAGRKGEIGALLAEAGDSFKAYGVAGHRHLPEIRQGLARASGKPVGLTFVPHLTPMIRG 240
Query: 262 MQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPDR 321
+ +T+Y + V DL+ + Y DE FV +L G P T SV+G+N C V +
Sbjct: 241 IHATLYARLTKDV---DLQRLFESRYADESFVDVLPAGSHPETRSVRGANLCRIAVHRPQ 297
Query: 322 IPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
+++SVIDNLVKGA+GQA+QNLNL+ G PEN GL +PL
Sbjct: 298 GGDTVVVLSVIDNLVKGAAGQAVQNLNLMFGLPENEGLDVVPL 340
>ref|YP_180646.1| N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia ruminantium
str. Welgevonden] gi|58418118|emb|CAI27322.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia
ruminantium str. Welgevonden]
gi|58417157|emb|CAI28270.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia
ruminantium str. Gardel] gi|57161589|emb|CAH58517.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia
ruminantium str. Welgevonden]
gi|58617545|ref|YP_196744.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia
ruminantium str. Gardel] gi|58579492|ref|YP_197704.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Ehrlichia
ruminantium str. Welgevonden]
Length = 347
Score = 309 bits (792), Expect = 8e-83
Identities = 162/343 (47%), Positives = 223/343 (64%), Gaps = 5/343 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQDLPDLIA-I 82
V V V+GA+GY G E++RLL HP + + A + G +SS + H+ +P I+
Sbjct: 5 VSVAVVGATGYVGVELVRLLLFHPMVKIKYLCATQSIGSLLSSHYDHVLKDSIPVSISCF 64
Query: 83 KDANFSDVDAVFCCLPHGTTQEIIKGLPKHLKIV-DLSADFRLRDVSEYEEWYGQPHRAP 141
+ S VD +F CLPHG + EI+K + +KI+ DLSADFR++D+ Y+EWYG H P
Sbjct: 65 SSIDLSKVDVIFLCLPHGQSNEIVKKIHNEVKIIIDLSADFRIKDIDTYKEWYGA-HCCP 123
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
DL ++ +YGLTE+ EEIK +R VA PGCY TS +PL PL++ L+++ NIIVDAKSGV
Sbjct: 124 DLIQDFVYGLTEIYWEEIKKSRFVACPGCYATSALVPLFPLLRLRLVKSQNIIVDAKSGV 183
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVTRHRHAPEIEQGLADAA-GSKVTISFTPHLIPMSR 260
SGAGRS + LF E+ + + SY +++HRH PEIEQ L AA + + F P+LIP+ R
Sbjct: 184 SGAGRSVDQKKLFCEIHDVIKSYNISKHRHIPEIEQELCFAACQENINVQFVPNLIPVKR 243
Query: 261 GMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
GM S+IY+E+ GV D+R L + Y+D +F+ + E I T SV G+NYC VFP
Sbjct: 244 GMLSSIYLELEEGVSPIDIREALLVFYKDSKFIFIDEEKAI-TTKSVIGTNYCYIGVFPG 302
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLP 363
RIP II+ IDNL+KGASGQA+QN N++M E L +P
Sbjct: 303 RIPNTVIIVCNIDNLLKGASGQAVQNFNIMMSCDETTALLNIP 345
>emb|CAG36054.1| probable N-acetyl-gamma-glutamyl-phosphate reductase [Desulfotalea
psychrophila LSv54] gi|51245177|ref|YP_065061.1|
N-acetyl-gamma-glutamyl-phosphate reductase
[Desulfotalea psychrophila LSv54]
Length = 346
Score = 306 bits (785), Expect = 5e-82
Identities = 161/346 (46%), Positives = 226/346 (64%), Gaps = 6/346 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLG--TQDLPDLIA 81
+++ ++GASGYTG E+ RLL NHPQ + +T+ + AG A+S VFP+L T + + +
Sbjct: 2 IKIAIIGASGYTGVELSRLLCNHPQVEITAVTSRQYAGVALSEVFPNLRGRTSLICENLT 61
Query: 82 IKDANFSDVDAVFCCLPHGTTQEIIKGL-PKHLKIVDLSADFRLRDVSEYEEWYGQPHRA 140
I++ D F +PH T I+ L K++DLSADFRL + YEEWY Q H A
Sbjct: 62 IEELCLR-ADLFFAAVPHKTAMNIVPQLLAAGKKVIDLSADFRLNSAAVYEEWY-QEHSA 119
Query: 141 PDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSG 200
+ +A+YGL E+ RE+I +L+ANPGCYPTSI L + PL++A +I+ +II D+KSG
Sbjct: 120 KEFLSQAVYGLPELYREQIAKTQLLANPGCYPTSIILGMAPLLRAGIIKPQSIIADSKSG 179
Query: 201 VSGAGRSAKENLLFTEVTEGMSSYGVTR-HRHAPEIEQGLADAAGSKVTISFTPHLIPMS 259
+GAGR AK LF EV +G +YGV R HRH PEIEQ L A + I+FTPHL+P+S
Sbjct: 180 TTGAGRGAKVGSLFCEVNDGFKAYGVGRKHRHTPEIEQELGKLANTDFNITFTPHLLPIS 239
Query: 260 RGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFP 319
RG+ STIY ++ + D+++ + Y+DE FV +L G P T V+GSNYC
Sbjct: 240 RGILSTIYADLNCEIEADEVQALYEEMYKDEPFVRVLPLGSAPATQYVRGSNYCDIGFAI 299
Query: 320 DRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPLF 365
D+ GR I++S IDN+VKGA+GQA+QN+N++ GF E GL+ +PLF
Sbjct: 300 DQTTGRIIVMSAIDNVVKGAAGQAVQNMNIMCGFAEQEGLEIVPLF 345
>ref|YP_076719.1| putative N-acetyl-gamma-glutamyl-phosphate reductase
[Symbiobacterium thermophilum IAM 14863]
gi|51857717|dbj|BAD41875.1| putative
N-acetyl-gamma-glutamyl-phosphate reductase
[Symbiobacterium thermophilum IAM 14863]
Length = 348
Score = 304 bits (779), Expect = 2e-81
Identities = 162/339 (47%), Positives = 216/339 (62%), Gaps = 4/339 (1%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLGTQ-DLPDLIAI 82
+RVG+ GA+GYTG E++RLLA HP+ V + + GQ +++V+PHL + DL A
Sbjct: 3 IRVGIAGATGYTGVELVRLLAQHPEAEVVVAGTESYQGQHLAAVYPHLRERVDLVGREAS 62
Query: 83 KDANFSDVDAVFCCLPHGTTQEIIKG-LPKHLKIVDLSADFRLRDVSEYEEWYGQPHRAP 141
+A + D VF LPHG T + L +++DL ADFRLRDV+ YE+WY + H AP
Sbjct: 63 PEA-LAGCDVVFTSLPHGLTMALAPAVLAAGGRLIDLGADFRLRDVTAYEQWYRKTHTAP 121
Query: 142 DLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKSGV 201
DL +EA+YGL E+ RE I+ ARLV NPGCYPT+ L PL++A +++T II DAKSGV
Sbjct: 122 DLMEEAVYGLPELYRERIRGARLVGNPGCYPTACALAAAPLLQAGVVETEGIIFDAKSGV 181
Query: 202 SGAGRSAKENLLFTEVTEGMSSYGVT-RHRHAPEIEQGLADAAGSKVTISFTPHLIPMSR 260
SGAGR + F+EV E +Y + HRH PEIEQ L+D AG V +SFTPHL+PM+R
Sbjct: 182 SGAGRGVNLGVHFSEVNENFKAYNIAGTHRHTPEIEQTLSDLAGRPVVVSFTPHLVPMTR 241
Query: 261 GMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVFPD 320
G+ +T Y + + L + Y E FV + G +P T V GSNYC + D
Sbjct: 242 GILATGYFTLKAERTTEQLVDLFREFYAGEPFVRVRPAGELPTTKQVWGSNYCDIGLQVD 301
Query: 321 RIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGL 359
R ++ISVIDNLVKGA+GQA+QN+NL+ G PE GL
Sbjct: 302 PRTRRVLVISVIDNLVKGAAGQAIQNMNLLFGLPETTGL 340
>ref|NP_953916.1| N-acetyl-gamma-glutamyl-phosphate reductase [Geobacter
sulfurreducens PCA] gi|39984910|gb|AAR36266.1|
N-acetyl-gamma-glutamyl-phosphate reductase [Geobacter
sulfurreducens PCA]
Length = 346
Score = 304 bits (779), Expect = 2e-81
Identities = 167/346 (48%), Positives = 224/346 (64%), Gaps = 8/346 (2%)
Query: 24 VRVGVLGASGYTGSEILRLLANHPQFGVALMTADRKAGQAISSVFPHLG---TQDLPDLI 80
++V V+GASGYTG E+LR+L HP+ V +T+++ AG+ +S +FP L Q L +L
Sbjct: 2 LKVAVVGASGYTGVELLRILHCHPEVAVTCITSEQSAGKPVSDLFPSLRGRYAQVLENLE 61
Query: 81 AIKDANFSDVDAVFCCLPHGTTQEIIKGLPKHLK-IVDLSADFRLRDVSEYEEWYGQPHR 139
I+ A +D+ +F LPH E++ K K ++DLSAD+R D + YE+WY +PH
Sbjct: 62 PIRVAEKADI--IFTALPHKAAMEVVPTFLKLGKRVIDLSADYRFNDAATYEQWY-EPHM 118
Query: 140 APDLQKEAIYGLTEVLREEIKNARLVANPGCYPTSIQLPLVPLIKANLIQTTNIIVDAKS 199
P L +A+YGL EV R +K A+LVANPGCYPTS+ L L PL+K LI+T II D+ S
Sbjct: 119 NPHLLPKAVYGLPEVRRAAVKGAKLVANPGCYPTSVILGLQPLLKHKLIETAGIISDSAS 178
Query: 200 GVSGAGRSAKENLLFTEVTEGMSSYGVTR-HRHAPEIEQGLADAAGSKVTISFTPHLIPM 258
GVSGAGR AK + L+ EV EG +YGV HRH PE+EQ L+ A ++TI+FTPHL PM
Sbjct: 179 GVSGAGRGAKVDNLYCEVNEGFKAYGVGGVHRHTPEMEQELSLLADERITITFTPHLAPM 238
Query: 259 SRGMQSTIYVEMAPGVRIDDLRHQLQLSYEDEEFVVLLENGVIPRTHSVKGSNYCLFNVF 318
RG+ STIY + +L Y E FV +L G +P T V+GSN+C V
Sbjct: 239 DRGILSTIYGRLLTPATTAELAGLYAEFYGGEPFVRVLPAGGVPSTAHVRGSNFCDIGVV 298
Query: 319 PDRIPGRAIIISVIDNLVKGASGQALQNLNLIMGFPENLGLQYLPL 364
D+ GR I++S IDNLVKGASGQA+QN+N++ G PE LGL+ L L
Sbjct: 299 VDQRTGRVIVVSAIDNLVKGASGQAVQNMNIMCGLPEGLGLEGLAL 344
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 606,054,374
Number of Sequences: 2540612
Number of extensions: 25306855
Number of successful extensions: 59743
Number of sequences better than 10.0: 514
Number of HSP's better than 10.0 without gapping: 316
Number of HSP's successfully gapped in prelim test: 198
Number of HSP's that attempted gapping in prelim test: 58397
Number of HSP's gapped (non-prelim): 556
length of query: 365
length of database: 863,360,394
effective HSP length: 129
effective length of query: 236
effective length of database: 535,621,446
effective search space: 126406661256
effective search space used: 126406661256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC145331.13


