

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.7 - phase: 0
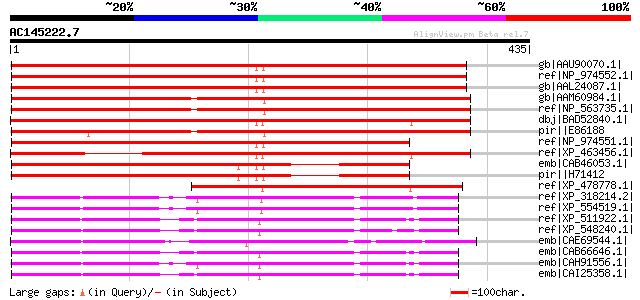
(435 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAU90070.1| At4g14950 [Arabidopsis thaliana] gi|18414346|ref|... 583 e-165
ref|NP_974552.1| expressed protein [Arabidopsis thaliana] 583 e-165
gb|AAL24087.1| unknown protein [Arabidopsis thaliana] 580 e-164
gb|AAM60984.1| unknown [Arabidopsis thaliana] 540 e-152
ref|NP_563735.1| expressed protein [Arabidopsis thaliana] 538 e-151
dbj|BAD52840.1| putative vacuole membrane protein 1 [Oryza sativ... 530 e-149
pir||E86188 hypothetical protein [imported] - Arabidopsis thalia... 528 e-148
ref|NP_974551.1| expressed protein [Arabidopsis thaliana] 508 e-142
ref|XP_463456.1| P0010B10.6 [Oryza sativa (japonica cultivar-gro... 422 e-116
emb|CAB46053.1| hypothetical protein [Arabidopsis thaliana] gi|7... 396 e-109
pir||H71412 hypothetical protein - Arabidopsis thaliana 396 e-109
ref|XP_478778.1| putative vacuole Membrane Protein 1 [Oryza sati... 279 1e-73
ref|XP_318214.2| ENSANGP00000024974 [Anopheles gambiae str. PEST... 225 2e-57
ref|XP_554519.1| ENSANGP00000028469 [Anopheles gambiae str. PEST... 225 2e-57
ref|XP_511922.1| PREDICTED: similar to hypothetical protein DKFZ... 223 7e-57
ref|XP_548240.1| PREDICTED: similar to hypothetical protein DKFZ... 223 1e-56
emb|CAE69544.1| Hypothetical protein CBG15756 [Caenorhabditis br... 223 1e-56
emb|CAB66646.1| hypothetical protein [Homo sapiens] gi|49065468|... 222 2e-56
emb|CAH91556.1| hypothetical protein [Pongo pygmaeus] 222 2e-56
emb|CAI25358.1| novel protein [Mus musculus] gi|56237870|emb|CAI... 220 8e-56
>gb|AAU90070.1| At4g14950 [Arabidopsis thaliana] gi|18414346|ref|NP_567450.1|
expressed protein [Arabidopsis thaliana]
Length = 416
Score = 583 bits (1503), Expect = e-165
Identities = 281/389 (72%), Positives = 325/389 (83%), Gaps = 7/389 (1%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 25 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 84
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE LEY R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 85 HGKHVEEVLEYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 144
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 145 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 204
Query: 182 LPPYFISRAARLSGSRMDAMEELD---SEDKGV----LNQIKCWFFSHTQHLNFFTILVL 234
LPPYFISRAA +SGS +D MEELD +ED G LN++K W +H+QHLNFFT+LVL
Sbjct: 205 LPPYFISRAASISGSTVDGMEELDGSSTEDSGFMATHLNRVKRWLLTHSQHLNFFTVLVL 264
Query: 235 ASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENE 294
ASVPNPLFDLAGIMCGQFGIPFW FFLATLIGKAIIKTHIQT+FII VCNNQLLDW+ENE
Sbjct: 265 ASVPNPLFDLAGIMCGQFGIPFWEFFLATLIGKAIIKTHIQTIFIICVCNNQLLDWMENE 324
Query: 295 FIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLM 354
IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV + K KKWDFSF SIWN +VWLM
Sbjct: 325 LIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPSHIKVKKWDFSFASIWNGIVWLM 384
Query: 355 LMNFFIKIVNSTAQTHLKKQQESEVAALT 383
L+NFF+KIV +TAQ HLKK+QE E+A LT
Sbjct: 385 LLNFFVKIVTATAQRHLKKKQEKEMATLT 413
>ref|NP_974552.1| expressed protein [Arabidopsis thaliana]
Length = 406
Score = 583 bits (1503), Expect = e-165
Identities = 281/389 (72%), Positives = 325/389 (83%), Gaps = 7/389 (1%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 15 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 74
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE LEY R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 75 HGKHVEEVLEYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 134
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 135 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 194
Query: 182 LPPYFISRAARLSGSRMDAMEELD---SEDKGV----LNQIKCWFFSHTQHLNFFTILVL 234
LPPYFISRAA +SGS +D MEELD +ED G LN++K W +H+QHLNFFT+LVL
Sbjct: 195 LPPYFISRAASISGSTVDGMEELDGSSTEDSGFMATHLNRVKRWLLTHSQHLNFFTVLVL 254
Query: 235 ASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENE 294
ASVPNPLFDLAGIMCGQFGIPFW FFLATLIGKAIIKTHIQT+FII VCNNQLLDW+ENE
Sbjct: 255 ASVPNPLFDLAGIMCGQFGIPFWEFFLATLIGKAIIKTHIQTIFIICVCNNQLLDWMENE 314
Query: 295 FIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLM 354
IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV + K KKWDFSF SIWN +VWLM
Sbjct: 315 LIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPSHIKVKKWDFSFASIWNGIVWLM 374
Query: 355 LMNFFIKIVNSTAQTHLKKQQESEVAALT 383
L+NFF+KIV +TAQ HLKK+QE E+A LT
Sbjct: 375 LLNFFVKIVTATAQRHLKKKQEKEMATLT 403
>gb|AAL24087.1| unknown protein [Arabidopsis thaliana]
Length = 416
Score = 580 bits (1494), Expect = e-164
Identities = 280/389 (71%), Positives = 324/389 (82%), Gaps = 7/389 (1%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 25 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 84
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE L Y R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 85 HGKHVEEVLGYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 144
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 145 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 204
Query: 182 LPPYFISRAARLSGSRMDAMEELD---SEDKGV----LNQIKCWFFSHTQHLNFFTILVL 234
LPPYFISRAA +SGS +D MEELD +ED G LN++K W +H+QHLNFFT+LVL
Sbjct: 205 LPPYFISRAASISGSTVDGMEELDGSSTEDSGFMATHLNRVKRWLLTHSQHLNFFTVLVL 264
Query: 235 ASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENE 294
ASVPNPLFDLAGIMCGQFGIPFW FFLATLIGKAIIKTHIQT+FII VCNNQLLDW+ENE
Sbjct: 265 ASVPNPLFDLAGIMCGQFGIPFWEFFLATLIGKAIIKTHIQTIFIICVCNNQLLDWMENE 324
Query: 295 FIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLM 354
IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV + K KKWDFSF SIWN +VWLM
Sbjct: 325 LIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPSHIKVKKWDFSFASIWNGMVWLM 384
Query: 355 LMNFFIKIVNSTAQTHLKKQQESEVAALT 383
L+NFF+KIV +TAQ HLKK+QE E+A LT
Sbjct: 385 LLNFFVKIVTATAQRHLKKKQEKEMATLT 413
>gb|AAM60984.1| unknown [Arabidopsis thaliana]
Length = 416
Score = 540 bits (1391), Expect = e-152
Identities = 270/389 (69%), Positives = 306/389 (78%), Gaps = 8/389 (2%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
+LE LTLT+QPFKTL+ +AV Y+++ YLLA GWL+LF +L+TL
Sbjct: 24 DLEKLTLTSQPFKTLRLFVVAVFLYVRRWSSYLLANVGWLILFCSIFVAFAALLVTLDGP 83
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE EY RFGLWW+ LGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK
Sbjct: 84 HVKHVEELSEYTRFGLWWIFLGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 143
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SA YDTIQL R PSWLDK C +FG P+F S VPLSSILPQVQ+EA+LWGLGTA+GE
Sbjct: 144 SAVYDTIQLKRSPSWLDKPCHEFGSPVFSSG----VPLSSILPQVQIEAILWGLGTALGE 199
Query: 182 LPPYFISRAARLSGSRMDAMEELDSEDKGVL----NQIKCWFFSHTQHLNFFTILVLASV 237
LPPYFISRAA LSG +M +E +D G + NQIK W SH+Q+LNFFTIL+LASV
Sbjct: 200 LPPYFISRAASLSGGKMKELETCSGDDNGFIAIRVNQIKSWLLSHSQYLNFFTILILASV 259
Query: 238 PNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENEFIW 297
PNPLFDLAGIMCGQF PFW FFLATLIGKAIIKTHIQTVFII VCNNQLLDW+ENE I+
Sbjct: 260 PNPLFDLAGIMCGQFEKPFWEFFLATLIGKAIIKTHIQTVFIICVCNNQLLDWVENELIY 319
Query: 298 VLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLMLMN 357
+LS +PGFAS LP+LTA L MK+KYL A PVS + KKWD SF S+WN VVWLML+N
Sbjct: 320 ILSFVPGFASALPELTAKLRLMKEKYLIASPPVSSDINVKKWDLSFASVWNGVVWLMLLN 379
Query: 358 FFIKIVNSTAQTHLKKQQESEVAALTKKS 386
FF +IV STAQ +LKKQQE E+ ALT KS
Sbjct: 380 FFGQIVTSTAQRYLKKQQEEELDALTNKS 408
>ref|NP_563735.1| expressed protein [Arabidopsis thaliana]
Length = 416
Score = 538 bits (1386), Expect = e-151
Identities = 269/389 (69%), Positives = 305/389 (78%), Gaps = 8/389 (2%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
+LE LTLT+QPFKTL+ +AV Y+++ YLLA GWL+LF +L+TL
Sbjct: 24 DLEKLTLTSQPFKTLRLFVVAVFLYVRRWSSYLLANVGWLILFCSIFVAFAALLVTLDGP 83
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE EY RFGLWW+ LGVASSIGLGSGLHTFVLYLGPHIALFTIK MQCGRVDLK
Sbjct: 84 HVKHVEELSEYTRFGLWWIFLGVASSIGLGSGLHTFVLYLGPHIALFTIKVMQCGRVDLK 143
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SA YDTIQL R PSWLDK C +FG P+F S VPLSSILPQVQ+EA+LWGLGTA+GE
Sbjct: 144 SAIYDTIQLKRSPSWLDKPCHEFGSPVFSSG----VPLSSILPQVQIEAILWGLGTALGE 199
Query: 182 LPPYFISRAARLSGSRMDAMEELDSEDKGVL----NQIKCWFFSHTQHLNFFTILVLASV 237
LPPYFISRAA LSG +M +E +D G + NQIK W SH+Q+LNFFTIL+LASV
Sbjct: 200 LPPYFISRAASLSGGKMKELETCSGDDNGFIAKRVNQIKSWLLSHSQYLNFFTILILASV 259
Query: 238 PNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENEFIW 297
PNPLFDLAGIMCGQF PFW FFLATLIGKAIIKTHIQTVFII VCNNQLLDW+ENE I+
Sbjct: 260 PNPLFDLAGIMCGQFEKPFWEFFLATLIGKAIIKTHIQTVFIICVCNNQLLDWVENELIY 319
Query: 298 VLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVWLMLMN 357
+LS +PGFAS LP+LTA L MK+KYL A PVS + KKWD SF S+WN VVWLML+N
Sbjct: 320 ILSFVPGFASALPELTAKLRLMKEKYLIASPPVSSDINVKKWDLSFASVWNGVVWLMLLN 379
Query: 358 FFIKIVNSTAQTHLKKQQESEVAALTKKS 386
FF +IV STAQ +LKKQQE E+ ALT KS
Sbjct: 380 FFGQIVTSTAQRYLKKQQEEELDALTNKS 408
>dbj|BAD52840.1| putative vacuole membrane protein 1 [Oryza sativa (japonica
cultivar-group)]
Length = 438
Score = 530 bits (1364), Expect = e-149
Identities = 257/396 (64%), Positives = 316/396 (78%), Gaps = 10/396 (2%)
Query: 1 MELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGC 60
+ELENLTLT PF+TL++ LA++QY+K+ Y+L+KG ++ V V G++L
Sbjct: 35 LELENLTLTKHPFRTLRFFMLAMLQYLKRLATYILSKGALFVVLIVLVLAPGILLAVTDG 94
Query: 61 LHEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDL 120
LH+KH++EFL Y RF LWWV+LGVASSIGLGSGLHTFVLYLGPHIALFTIKA+QCGR+DL
Sbjct: 95 LHKKHVQEFLNYARFVLWWVSLGVASSIGLGSGLHTFVLYLGPHIALFTIKAVQCGRIDL 154
Query: 121 KSAPYDTIQLNRGPSWLDKNCSQFGPPLFQ-SEYGTRVPLSSILPQVQMEAVLWGLGTAI 179
K+APYDTIQL +GPSWLDK CS FGPP++Q S + R+P+ +LPQVQ+EAVLWG+GTA+
Sbjct: 155 KTAPYDTIQLKQGPSWLDKKCSDFGPPVYQASAHSVRIPVFELLPQVQLEAVLWGIGTAL 214
Query: 180 GELPPYFISRAARLSGSRMDAMEELD---SEDKG----VLNQIKCWFFSHTQHLNFFTIL 232
GELPPYFISRAARLSGS +A++ELD S++ G LN+ K W SH+QHLNF TIL
Sbjct: 215 GELPPYFISRAARLSGSEPEAVKELDAAASDEHGPIASTLNRTKRWLLSHSQHLNFITIL 274
Query: 233 VLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIE 292
+LASVPNPLFDLAGIMCGQFGIPFW FF ATLIGKAIIKTHIQT+FIIS+CNNQLL +E
Sbjct: 275 ILASVPNPLFDLAGIMCGQFGIPFWEFFFATLIGKAIIKTHIQTLFIISLCNNQLLYLME 334
Query: 293 NEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTK--GKKWDFSFTSIWNTV 350
E IW+ HIPGF++ LP + A LH+ KDKYL P + ++K +W+FSFT +WNTV
Sbjct: 335 KELIWIFGHIPGFSASLPSVIAKLHSAKDKYLSTPTSATSSSKMEDTQWNFSFTLVWNTV 394
Query: 351 VWLMLMNFFIKIVNSTAQTHLKKQQESEVAALTKKS 386
VWL+L+NFFIKIV STAQ +LKKQQ+ E+ +T S
Sbjct: 395 VWLVLVNFFIKIVTSTAQEYLKKQQDIEMELITDSS 430
>pir||E86188 hypothetical protein [imported] - Arabidopsis thaliana
gi|2388561|gb|AAB71442.1| Similar to Arabidopsis
hypothetical protein PID:e326839 (gb|Z97337).
[Arabidopsis thaliana]
Length = 430
Score = 528 bits (1361), Expect = e-148
Identities = 269/403 (66%), Positives = 305/403 (74%), Gaps = 22/403 (5%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
+LE LTLT+QPFKTL+ +AV Y+++ YLLA GWL+LF +L+TL
Sbjct: 24 DLEKLTLTSQPFKTLRLFVVAVFLYVRRWSSYLLANVGWLILFCSIFVAFAALLVTLDGP 83
Query: 62 HEK--------------HLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIAL 107
H K H+EE EY RFGLWW+ LGVASSIGLGSGLHTFVLYLGPHIAL
Sbjct: 84 HVKMKKTVKFSNFLLVQHVEELSEYTRFGLWWIFLGVASSIGLGSGLHTFVLYLGPHIAL 143
Query: 108 FTIKAMQCGRVDLKSAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQ 167
FTIK MQCGRVDLKSA YDTIQL R PSWLDK C +FG P+F S VPLSSILPQVQ
Sbjct: 144 FTIKVMQCGRVDLKSAIYDTIQLKRSPSWLDKPCHEFGSPVFSSG----VPLSSILPQVQ 199
Query: 168 MEAVLWGLGTAIGELPPYFISRAARLSGSRMDAMEELDSEDKGVL----NQIKCWFFSHT 223
+EA+LWGLGTA+GELPPYFISRAA LSG +M +E +D G + NQIK W SH+
Sbjct: 200 IEAILWGLGTALGELPPYFISRAASLSGGKMKELETCSGDDNGFIAKRVNQIKSWLLSHS 259
Query: 224 QHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVC 283
Q+LNFFTIL+LASVPNPLFDLAGIMCGQF PFW FFLATLIGKAIIKTHIQTVFII VC
Sbjct: 260 QYLNFFTILILASVPNPLFDLAGIMCGQFEKPFWEFFLATLIGKAIIKTHIQTVFIICVC 319
Query: 284 NNQLLDWIENEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSF 343
NNQLLDW+ENE I++LS +PGFAS LP+LTA L MK+KYL A PVS + KKWD SF
Sbjct: 320 NNQLLDWVENELIYILSFVPGFASALPELTAKLRLMKEKYLIASPPVSSDINVKKWDLSF 379
Query: 344 TSIWNTVVWLMLMNFFIKIVNSTAQTHLKKQQESEVAALTKKS 386
S+WN VVWLML+NFF +IV STAQ +LKKQQE E+ ALT KS
Sbjct: 380 ASVWNGVVWLMLLNFFGQIVTSTAQRYLKKQQEEELDALTNKS 422
>ref|NP_974551.1| expressed protein [Arabidopsis thaliana]
Length = 355
Score = 508 bits (1307), Expect = e-142
Identities = 246/341 (72%), Positives = 284/341 (83%), Gaps = 7/341 (2%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 15 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 74
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE LEY R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 75 HGKHVEEVLEYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 134
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 135 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 194
Query: 182 LPPYFISRAARLSGSRMDAMEELD---SEDKGV----LNQIKCWFFSHTQHLNFFTILVL 234
LPPYFISRAA +SGS +D MEELD +ED G LN++K W +H+QHLNFFT+LVL
Sbjct: 195 LPPYFISRAASISGSTVDGMEELDGSSTEDSGFMATHLNRVKRWLLTHSQHLNFFTVLVL 254
Query: 235 ASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIENE 294
ASVPNPLFDLAGIMCGQFGIPFW FFLATLIGKAIIKTHIQT+FII VCNNQLLDW+ENE
Sbjct: 255 ASVPNPLFDLAGIMCGQFGIPFWEFFLATLIGKAIIKTHIQTIFIICVCNNQLLDWMENE 314
Query: 295 FIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTK 335
IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV + K
Sbjct: 315 LIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPSHIK 355
>ref|XP_463456.1| P0010B10.6 [Oryza sativa (japonica cultivar-group)]
Length = 406
Score = 422 bits (1085), Expect = e-116
Identities = 216/396 (54%), Positives = 272/396 (68%), Gaps = 57/396 (14%)
Query: 1 MELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGC 60
+ELENLTLT PF+TL++ LA++QY+K+ Y+L+KG ++ V V G++L
Sbjct: 50 LELENLTLTKHPFRTLRFFMLAMLQYLKRLATYILSKGALFVVLIVLVLAPGILLAVTDG 109
Query: 61 LHEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDL 120
LH+ KA+QCGR+DL
Sbjct: 110 LHK-----------------------------------------------KAVQCGRIDL 122
Query: 121 KSAPYDTIQLNRGPSWLDKNCSQFGPPLFQ-SEYGTRVPLSSILPQVQMEAVLWGLGTAI 179
K+APYDTIQL +GPSWLDK CS FGPP++Q S + R+P+ +LPQVQ+EAVLWG+GTA+
Sbjct: 123 KTAPYDTIQLKQGPSWLDKKCSDFGPPVYQASAHSVRIPVFELLPQVQLEAVLWGIGTAL 182
Query: 180 GELPPYFISRAARLSGSRMDAMEELD---SEDKG----VLNQIKCWFFSHTQHLNFFTIL 232
GELPPYFISRAARLSGS +A++ELD S++ G LN+ K W SH+QHLNF TIL
Sbjct: 183 GELPPYFISRAARLSGSEPEAVKELDAAASDEHGPIASTLNRTKRWLLSHSQHLNFITIL 242
Query: 233 VLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIE 292
+LASVPNPLFDLAGIMCGQFGIPFW FF ATLIGKAIIKTHIQT+FIIS+CNNQLL +E
Sbjct: 243 ILASVPNPLFDLAGIMCGQFGIPFWEFFFATLIGKAIIKTHIQTLFIISLCNNQLLYLME 302
Query: 293 NEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTK--GKKWDFSFTSIWNTV 350
E IW+ HIPGF++ LP + A LH+ KDKYL P + ++K +W+FSFT +WNTV
Sbjct: 303 KELIWIFGHIPGFSASLPSVIAKLHSAKDKYLSTPTSATSSSKMEDTQWNFSFTLVWNTV 362
Query: 351 VWLMLMNFFIKIVNSTAQTHLKKQQESEVAALTKKS 386
VWL+L+NFFIKIV STAQ +LKKQQ+ E+ +T S
Sbjct: 363 VWLVLVNFFIKIVTSTAQEYLKKQQDIEMELITDSS 398
>emb|CAB46053.1| hypothetical protein [Arabidopsis thaliana]
gi|7268241|emb|CAB78537.1| hypothetical protein
[Arabidopsis thaliana] gi|25407532|pir||C85164
hypothetical protein dl3515w [imported] - Arabidopsis
thaliana
Length = 797
Score = 396 bits (1018), Expect = e-109
Identities = 208/363 (57%), Positives = 246/363 (67%), Gaps = 68/363 (18%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 8 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 67
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE LEY R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 68 HGKHVEEVLEYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 127
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 128 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 187
Query: 182 LPPYFISRA----------------------ARLSGSRMDAMEELD---SEDKGV----L 212
LPPYFISRA A +SGS +D MEELD +ED G L
Sbjct: 188 LPPYFISRAGLYTDSLYIFLFDLCSRPCMCIASISGSTVDGMEELDGSSTEDSGFMATHL 247
Query: 213 NQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKT 272
N++K W +H+QHLNFFT+LVLAS
Sbjct: 248 NRVKRWLLTHSQHLNFFTVLVLAS------------------------------------ 271
Query: 273 HIQTVFIISVCNNQLLDWIENEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSP 332
T+FII VCNNQLLDW+ENE IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV
Sbjct: 272 ---TIFIICVCNNQLLDWMENELIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPS 328
Query: 333 NTK 335
+ K
Sbjct: 329 HIK 331
>pir||H71412 hypothetical protein - Arabidopsis thaliana
Length = 797
Score = 396 bits (1018), Expect = e-109
Identities = 208/363 (57%), Positives = 246/363 (67%), Gaps = 68/363 (18%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+ENLTLTTQP TLK A IQYIK+++ YLLA GGW +L + + G +L+T+
Sbjct: 8 EVENLTLTTQPLNTLKLFVEATIQYIKRSISYLLAHGGWFILITTLLVVSGGLLVTVDGP 67
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H KH+EE LEY R+GLWW+ALGVASSIGLGSGLHTFVLYLGPHIALFT+KA CGRVDLK
Sbjct: 68 HGKHVEEVLEYVRYGLWWIALGVASSIGLGSGLHTFVLYLGPHIALFTLKATLCGRVDLK 127
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
SAPYDTIQL R PSWLDK+CS+FGPPL S G+RVPL+SILPQVQ+EA+LWG+GTA+GE
Sbjct: 128 SAPYDTIQLKRVPSWLDKSCSEFGPPLMISAAGSRVPLTSILPQVQLEAILWGIGTALGE 187
Query: 182 LPPYFISRA----------------------ARLSGSRMDAMEELD---SEDKGV----L 212
LPPYFISRA A +SGS +D MEELD +ED G L
Sbjct: 188 LPPYFISRAGLYTDSLYIFLFDLCSRPCMCIASISGSTVDGMEELDGSSTEDSGFMATHL 247
Query: 213 NQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKT 272
N++K W +H+QHLNFFT+LVLAS
Sbjct: 248 NRVKRWLLTHSQHLNFFTVLVLAS------------------------------------ 271
Query: 273 HIQTVFIISVCNNQLLDWIENEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSP 332
T+FII VCNNQLLDW+ENE IW+LSH+PG AS+LP LTA LHAMK+KY+ AP PV
Sbjct: 272 ---TIFIICVCNNQLLDWMENELIWILSHVPGLASMLPGLTAKLHAMKEKYIDAPSPVPS 328
Query: 333 NTK 335
+ K
Sbjct: 329 HIK 331
>ref|XP_478778.1| putative vacuole Membrane Protein 1 [Oryza sativa (japonica
cultivar-group)] gi|34394214|dbj|BAC84666.1| putative
vacuole Membrane Protein 1 [Oryza sativa (japonica
cultivar-group)]
Length = 255
Score = 279 bits (714), Expect = 1e-73
Identities = 133/235 (56%), Positives = 175/235 (73%), Gaps = 8/235 (3%)
Query: 153 YGTRVPLSSILPQVQMEAVLWGLGTAIGELPPYFISRAARLSGSRMDAMEELDSEDKG-- 210
Y +P S IL +V +EAVLWG+GTA+GELPPYF+SRAA +SG ++D +EELD+ G
Sbjct: 2 YQETIPFSKILHEVHLEAVLWGIGTALGELPPYFLSRAASMSGRKLDELEELDASVSGEG 61
Query: 211 ----VLNQIKCWFFSHTQHLNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIG 266
L++ K W SH+Q+LNF TIL+LASVPNPLFDLAGI+CGQF IPFW FFLATLIG
Sbjct: 62 FLSSTLHRAKRWLMSHSQYLNFPTILLLASVPNPLFDLAGILCGQFNIPFWKFFLATLIG 121
Query: 267 KAIIKTHIQTVFIISVCNNQLLDWIENEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKA 326
KA+IK +IQT +I++CNNQLLD +E +WV ++P +SVLP L A L K K+L +
Sbjct: 122 KAVIKVYIQTTLVITLCNNQLLDLVEKRIMWVFGNVPMVSSVLPSLVAKLKTAKSKFLSS 181
Query: 327 PHPVSPNT--KGKKWDFSFTSIWNTVVWLMLMNFFIKIVNSTAQTHLKKQQESEV 379
S ++ K KW+ SF+ IWNTVVWL++MNF ++I+ STAQ++LK+QQE E+
Sbjct: 182 SVAASASSVVKETKWNLSFSLIWNTVVWLLIMNFIVQIITSTAQSYLKRQQELEI 236
>ref|XP_318214.2| ENSANGP00000024974 [Anopheles gambiae str. PEST]
gi|55236792|gb|EAA43728.2| ENSANGP00000024974 [Anopheles
gambiae str. PEST]
Length = 443
Score = 225 bits (573), Expect = 2e-57
Identities = 145/392 (36%), Positives = 215/392 (53%), Gaps = 44/392 (11%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+++L L P KTLKY++L VI ++ + LL + L + + + V + T G
Sbjct: 62 EIKSLVLWASPMKTLKYASLEVIYLLQTNLSRLLHRRAQLTALLIVICSTAVAIYTPG-- 119
Query: 62 HEKHLEEFLEYFR-FGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDL 120
+ L ++L++ F ++W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C ++
Sbjct: 120 QHQILVQYLKHKGIFVIYWLGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECSSLNF 179
Query: 121 KSAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTR---VP-LSSILPQVQMEAVLWGLG 176
PY P+ + L E GT VP L SI+ +V+ EA LWG G
Sbjct: 180 PEPPY--------PNEI----------LCPDENGTAELIVPSLWSIMTKVRYEAFLWGAG 221
Query: 177 TAIGELPPYFISRAARLSGSRMDAMEELDSEDK-----------GVLNQIKCWFFSHTQH 225
TA+GELPPYF+++AARLSG+ + +E L +K + ++ K +
Sbjct: 222 TALGELPPYFMAKAARLSGNNTEELENLQELEKLQKRKEKGEKLNLFDKGKLMMEDIVER 281
Query: 226 LNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNN 285
+ FF IL+ AS+PNPLFDLAGI CG F +PFW FF ATLIGKA+IK HIQ +F+I N
Sbjct: 282 VGFFGILLCASIPNPLFDLAGITCGHFLVPFWKFFGATLIGKAVIKMHIQKIFVIISFNE 341
Query: 286 QLLDWIENEFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFT 344
L+D +FI +L+ IP L K K + + H S N+
Sbjct: 342 SLVD----KFIDILALIPMVGVRLQKPFKGFFETQKQRLHRNSHKQSINSSDGN---LLA 394
Query: 345 SIWNTVVWLMLMNFFIKIVNSTAQTHLKKQQE 376
S++ V+ M+ F IVN+ AQ++ K+ ++
Sbjct: 395 SLFEYFVFAMICYFLTSIVNTFAQSYCKRMRK 426
>ref|XP_554519.1| ENSANGP00000028469 [Anopheles gambiae str. PEST]
gi|55236791|gb|EAL39418.1| ENSANGP00000028469 [Anopheles
gambiae str. PEST]
Length = 399
Score = 225 bits (573), Expect = 2e-57
Identities = 145/392 (36%), Positives = 215/392 (53%), Gaps = 44/392 (11%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E+++L L P KTLKY++L VI ++ + LL + L + + + V + T G
Sbjct: 35 EIKSLVLWASPMKTLKYASLEVIYLLQTNLSRLLHRRAQLTALLIVICSTAVAIYTPG-- 92
Query: 62 HEKHLEEFLEYFR-FGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDL 120
+ L ++L++ F ++W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C ++
Sbjct: 93 QHQILVQYLKHKGIFVIYWLGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECSSLNF 152
Query: 121 KSAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTR---VP-LSSILPQVQMEAVLWGLG 176
PY P+ + L E GT VP L SI+ +V+ EA LWG G
Sbjct: 153 PEPPY--------PNEI----------LCPDENGTAELIVPSLWSIMTKVRYEAFLWGAG 194
Query: 177 TAIGELPPYFISRAARLSGSRMDAMEELDSEDK-----------GVLNQIKCWFFSHTQH 225
TA+GELPPYF+++AARLSG+ + +E L +K + ++ K +
Sbjct: 195 TALGELPPYFMAKAARLSGNNTEELENLQELEKLQKRKEKGEKLNLFDKGKLMMEDIVER 254
Query: 226 LNFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNN 285
+ FF IL+ AS+PNPLFDLAGI CG F +PFW FF ATLIGKA+IK HIQ +F+I N
Sbjct: 255 VGFFGILLCASIPNPLFDLAGITCGHFLVPFWKFFGATLIGKAVIKMHIQKIFVIISFNE 314
Query: 286 QLLDWIENEFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFT 344
L+D +FI +L+ IP L K K + + H S N+
Sbjct: 315 SLVD----KFIDILALIPMVGVRLQKPFKGFFETQKQRLHRNSHKQSINSSDGN---LLA 367
Query: 345 SIWNTVVWLMLMNFFIKIVNSTAQTHLKKQQE 376
S++ V+ M+ F IVN+ AQ++ K+ ++
Sbjct: 368 SLFEYFVFAMICYFLTSIVNTFAQSYCKRMRK 399
>ref|XP_511922.1| PREDICTED: similar to hypothetical protein DKFZp566I133 [Pan
troglodytes] gi|17221829|gb|AAL36461.1| TDC1 [Homo
sapiens] gi|14602501|gb|AAH09758.1| Transmembrane
protein 49 [Homo sapiens] gi|20070349|ref|NP_112200.2|
transmembrane protein 49 [Homo sapiens]
Length = 406
Score = 223 bits (569), Expect = 7e-57
Identities = 137/384 (35%), Positives = 211/384 (54%), Gaps = 35/384 (9%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E +N+ L QP TL+Y +L ++ +K+ L + ++ F + + L G +
Sbjct: 40 ERQNIVLWRQPLITLQYFSLEILVILKEWTSKLWHRQSIVVSFLLLLAVLIATYYVEG-V 98
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H+++++ + F +W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C V+
Sbjct: 99 HQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFP 158
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
PY Q P + GT + L SI+ +V++EA +WG+GTAIGE
Sbjct: 159 EPPYP---------------DQIICPDEEGTEGT-ISLWSIISKVRIEACMWGIGTAIGE 202
Query: 182 LPPYFISRAARLSGSRMDAMEELDSED--------KGVLNQIKCWFFSHTQHLNFFTILV 233
LPPYF++RAARLSG+ D E + E+ + ++ K Q + FF IL
Sbjct: 203 LPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQDFASRAKLAVQKLVQKVGFFGILA 262
Query: 234 LASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIEN 293
AS+PNPLFDLAGI CG F +PFW FF ATLIGKAIIK HIQ +F+I + +++
Sbjct: 263 CASIPNPLFDLAGITCGHFLVPFWTFFGATLIGKAIIKMHIQKIFVIITFSKHIVE---- 318
Query: 294 EFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVW 352
+ + + +PG L K L A + K +P +G+ W + ++ +V
Sbjct: 319 QMVAFIGAVPGIGPSLQKPFQEYLEAQRQKLHHKSEMGTP--QGENW---LSWMFEKLVV 373
Query: 353 LMLMNFFIKIVNSTAQTHLKKQQE 376
+M+ F + I+NS AQ++ K+ Q+
Sbjct: 374 VMVCYFILSIINSMAQSYAKRIQQ 397
>ref|XP_548240.1| PREDICTED: similar to hypothetical protein DKFZp566I133 [Canis
familiaris]
Length = 660
Score = 223 bits (567), Expect = 1e-56
Identities = 139/385 (36%), Positives = 210/385 (54%), Gaps = 37/385 (9%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E +N+ L QP TL+Y +L ++ +K L + ++ F + + L T G
Sbjct: 294 ERQNIVLWRQPLITLQYFSLEILVILKDWTSKLWHRQSIVVSFLLLLAVLTATYYTEGA- 352
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H+++++ + F +W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C V+
Sbjct: 353 HQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFP 412
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
PY Q P GT + L SI+ +V++EA +WG+GTAIGE
Sbjct: 413 EPPYP---------------DQIICPDEDGIEGT-ISLWSIISKVRIEACMWGIGTAIGE 456
Query: 182 LPPYFISRAARLSGSRMDAMEELDSED--------KGVLNQIKCWFFSHTQHLNFFTILV 233
LPPYF++RAARLSG+ D E + E+ + ++ K + Q + FF IL
Sbjct: 457 LPPYFMARAARLSGAEPDDEEYQEFEEMLEHAETAQDFASRAKLAVQNLVQKVGFFGILA 516
Query: 234 LASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIEN 293
AS+PNPLFDLAGI CG F +PFW FF ATLIGKAIIK HIQ +F+I + +++
Sbjct: 517 CASIPNPLFDLAGITCGHFLVPFWTFFGATLIGKAIIKMHIQKIFVIITFSKHIVE---- 572
Query: 294 EFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNT-KGKKWDFSFTSIWNTVV 351
+ + + +PG L K L A + K H T +G+ W + ++ +V
Sbjct: 573 QMVAFIGAVPGIGPSLQKPFQEYLEAQRQ---KLHHRREMGTQQGENW---LSWMFEKLV 626
Query: 352 WLMLMNFFIKIVNSTAQTHLKKQQE 376
+M+ F + I+NS AQ++ K+ Q+
Sbjct: 627 VVMVCYFILSIINSMAQSYAKRMQQ 651
>emb|CAE69544.1| Hypothetical protein CBG15756 [Caenorhabditis briggsae]
Length = 470
Score = 223 bits (567), Expect = 1e-56
Identities = 143/405 (35%), Positives = 220/405 (54%), Gaps = 41/405 (10%)
Query: 1 MELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGC 60
+E E + + +P T+ Y+ + + + L +L+ L LF +++G T G
Sbjct: 46 LERETIVIWKKPITTIYYALMEIANLSIELFLKILSHKFLLGLFFISLGLAIYGYRTPGA 105
Query: 61 LHEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDL 120
H++H++ ++ + WWV LGV SSIGLGSGLHTF++YLGPHIA T+ A +C +D
Sbjct: 106 -HQEHVQTIEKHILWWSWWVLLGVLSSIGLGSGLHTFLIYLGPHIAAVTMAAYECQSLDF 164
Query: 121 KSAPY-DTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAI 179
PY ++IQ PS ++ V I+ +V++E++LWG GTA+
Sbjct: 165 PEPPYPESIQC---PS---------------TKSSIAVTFWQIVAKVRVESLLWGAGTAL 206
Query: 180 GELPPYFISRAARLSGSR------------MDAMEELDSEDKGVLNQ-IKCWFFSHTQHL 226
GELPPYF++RAAR+SG M+A +E D E K + + IK W + L
Sbjct: 207 GELPPYFMARAARISGQEPDDEEYREFLELMNADKEQDGEQKLSMGERIKSWVEHNIHRL 266
Query: 227 NFFTILVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQ 286
F IL+ AS+PNPLFDLAGI CG F +PFW+FF ATLIGKA++K H+Q F+I +
Sbjct: 267 GFPGILLFASIPNPLFDLAGITCGHFLVPFWSFFGATLIGKALVKMHVQMGFVILAFS-- 324
Query: 287 LLDWIENEFIWVLSHIPGFASVLPKLTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSI 346
D F+ +L IP +V P + + + +K KA H K + + ++
Sbjct: 325 --DHHAETFVKLLESIP---AVGPHIRKPISDLLEKQRKALHTTPGEHKEQSTNLLAFAL 379
Query: 347 WNTVVWLMLMNFFIKIVNSTAQTHLKKQQESEVAALTKKSDSDTQ 391
+ +V +M++ FF+ IVNS A+ + K+ E + +D + Q
Sbjct: 380 -SAMVTVMILFFFLSIVNSLAKDYHKRLWERKRRLNKTLTDEENQ 423
>emb|CAB66646.1| hypothetical protein [Homo sapiens] gi|49065468|emb|CAG38552.1|
VMP1 [Homo sapiens]
Length = 406
Score = 222 bits (566), Expect = 2e-56
Identities = 137/384 (35%), Positives = 211/384 (54%), Gaps = 35/384 (9%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E +N+ L QP TL+Y +L ++ +K+ L + ++ F + + L G +
Sbjct: 40 ERQNIVLWRQPLITLQYFSLEILVILKEWTSKLWHRQSIVVSFLLLLAVLIATYYVEG-V 98
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H+++++ + F +W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C V+
Sbjct: 99 HQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFP 158
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
PY Q P + GT + L SI+ +V++EA +WG+GTAIGE
Sbjct: 159 EPPYP---------------DQIICPDEEGTEGT-IFLWSIISKVRIEACMWGIGTAIGE 202
Query: 182 LPPYFISRAARLSGSRMDAMEELDSED--------KGVLNQIKCWFFSHTQHLNFFTILV 233
LPPYF++RAARLSG+ D E + E+ + ++ K Q + FF IL
Sbjct: 203 LPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQDFASRAKLAVQKLVQKVGFFGILA 262
Query: 234 LASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIEN 293
AS+PNPLFDLAGI CG F +PFW FF ATLIGKAIIK HIQ +F+I + +++
Sbjct: 263 CASIPNPLFDLAGITCGHFLVPFWTFFGATLIGKAIIKMHIQKIFVIITFSKHIVE---- 318
Query: 294 EFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVW 352
+ + + +PG L K L A + K +P +G+ W + ++ +V
Sbjct: 319 QMVAFIGAVPGIGPSLQKPFQEYLEAQRQKLHHKSEMGTP--QGENW---LSWMFEKLVV 373
Query: 353 LMLMNFFIKIVNSTAQTHLKKQQE 376
+M+ F + I+NS AQ++ K+ Q+
Sbjct: 374 VMVCYFILSIINSMAQSYAKRIQQ 397
>emb|CAH91556.1| hypothetical protein [Pongo pygmaeus]
Length = 406
Score = 222 bits (565), Expect = 2e-56
Identities = 137/386 (35%), Positives = 211/386 (54%), Gaps = 39/386 (10%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E +N+ L QP TL+Y +L ++ +K+ L + ++ F + + L G
Sbjct: 40 ERQNIVLWRQPLITLQYFSLEILVILKEWTSKLWHRQSIVVSFLLLLAVLIATYYVEGA- 98
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H+++++ + F +W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C V+
Sbjct: 99 HQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFP 158
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTR--VPLSSILPQVQMEAVLWGLGTAI 179
PY P + + E GT + L SI+ +V++EA +WG+GTAI
Sbjct: 159 EPPY--------PDQI----------ICPDEGGTEGTISLWSIISKVRIEACMWGIGTAI 200
Query: 180 GELPPYFISRAARLSGSRMDAMEELDSED--------KGVLNQIKCWFFSHTQHLNFFTI 231
GELPPYF++RAARLSG+ D E + E+ + ++ K Q + FF I
Sbjct: 201 GELPPYFMARAARLSGAEPDDEEYQEFEEMLEHAESAQDFASRAKLAVQKLVQKVGFFGI 260
Query: 232 LVLASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWI 291
L AS+PNPLFDLAGI CG F +PFW FF ATLIGKAIIK HIQ +F+I + +++
Sbjct: 261 LACASIPNPLFDLAGITCGHFLVPFWTFFGATLIGKAIIKMHIQKIFVIITFSKHIVE-- 318
Query: 292 ENEFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTV 350
+ + + +PG L K L A + K +P +G+ W + ++ +
Sbjct: 319 --QMVAFIGAVPGIGPSLQKPFQEYLEAQRQKLHHKSEMGTP--QGENW---LSWMFEKL 371
Query: 351 VWLMLMNFFIKIVNSTAQTHLKKQQE 376
V +M+ F + I+NS AQ++ K+ Q+
Sbjct: 372 VVVMVCYFILSIINSMAQSYAKRIQQ 397
>emb|CAI25358.1| novel protein [Mus musculus] gi|56237870|emb|CAI25797.1| novel
protein [Mus musculus] gi|26346304|dbj|BAC36803.1|
unnamed protein product [Mus musculus]
gi|27753991|ref|NP_083754.2| vacuole Membrane Protein 1
[Mus musculus] gi|26335511|dbj|BAC31456.1| unnamed
protein product [Mus musculus]
Length = 406
Score = 220 bits (560), Expect = 8e-56
Identities = 136/384 (35%), Positives = 206/384 (53%), Gaps = 35/384 (9%)
Query: 2 ELENLTLTTQPFKTLKYSTLAVIQYIKKTMLYLLAKGGWLMLFSVAVGTLGVVLMTLGCL 61
E +N+ L QP TL+Y +L + +K+ L + ++ F + + L G
Sbjct: 40 ERQNIVLWRQPLITLQYFSLETLVVLKEWTSKLWHRQSIVVSFLLLLAALVATYYVEGA- 98
Query: 62 HEKHLEEFLEYFRFGLWWVALGVASSIGLGSGLHTFVLYLGPHIALFTIKAMQCGRVDLK 121
H+++++ + F +W+ LG+ SS+GLG+GLHTF+LYLGPHIA T+ A +C V+
Sbjct: 99 HQQYVQRIEKQFLLYAYWIGLGILSSVGLGTGLHTFLLYLGPHIASVTLAAYECNSVNFP 158
Query: 122 SAPYDTIQLNRGPSWLDKNCSQFGPPLFQSEYGTRVPLSSILPQVQMEAVLWGLGTAIGE 181
PY Q P + G + L SI+ +V++EA +WG+GTAIGE
Sbjct: 159 EPPYP---------------DQIICPEEEGAEGA-ISLWSIISKVRIEACMWGIGTAIGE 202
Query: 182 LPPYFISRAARLSGSRMDAMEELDSED--------KGVLNQIKCWFFSHTQHLNFFTILV 233
LPPYF++RAARLSG+ D E + E+ + ++ K Q + FF IL
Sbjct: 203 LPPYFMARAARLSGAEPDDEEYQEFEEMLEHAEAAQDFASRAKLAVQKLVQKVGFFGILA 262
Query: 234 LASVPNPLFDLAGIMCGQFGIPFWNFFLATLIGKAIIKTHIQTVFIISVCNNQLLDWIEN 293
AS+PNPLFDLAGI CG F +PFW FF ATLIGKAIIK HIQ +F+I + +++
Sbjct: 263 CASIPNPLFDLAGITCGHFLVPFWTFFGATLIGKAIIKMHIQKIFVIVTFSKHIVE---- 318
Query: 294 EFIWVLSHIPGFASVLPK-LTANLHAMKDKYLKAPHPVSPNTKGKKWDFSFTSIWNTVVW 352
+ + + +PG L K L A + K +P +G+ W + ++ +V
Sbjct: 319 QMVTFIGAVPGIGPSLQKPFQEYLEAQRQKLHHRSEAGTP--QGENW---LSWMFEKLVV 373
Query: 353 LMLMNFFIKIVNSTAQTHLKKQQE 376
M+ F + I+NS AQ + K+ Q+
Sbjct: 374 AMVCYFVLSIINSMAQNYAKRIQQ 397
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.331 0.143 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 728,084,130
Number of Sequences: 2540612
Number of extensions: 30223113
Number of successful extensions: 82278
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 82110
Number of HSP's gapped (non-prelim): 70
length of query: 435
length of database: 863,360,394
effective HSP length: 131
effective length of query: 304
effective length of database: 530,540,222
effective search space: 161284227488
effective search space used: 161284227488
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC145222.7


