

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.11 + phase: 0 /pseudo
(394 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
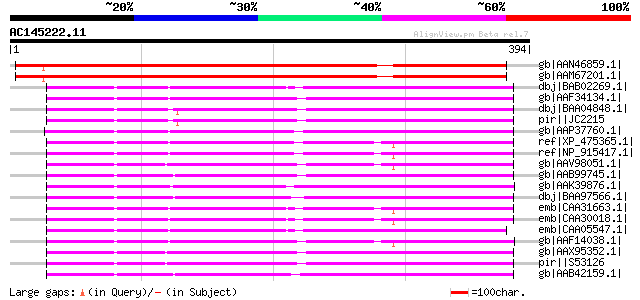
gb|AAN46859.1| At2g32120/F22D22.13 [Arabidopsis thaliana] gi|217... 564 e-159
gb|AAM67201.1| 70kD heat shock protein [Arabidopsis thaliana] 562 e-159
dbj|BAB02269.1| 70 kDa heat shock protein [Arabidopsis thaliana]... 192 1e-47
gb|AAF34134.1| high molecular weight heat shock protein [Malus x... 191 3e-47
dbj|BAA04848.1| HSP70 [Lilium longiflorum] 191 4e-47
pir||JC2215 dnaK-type molecular chaperone LIM18 - trumpet lily 191 4e-47
gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|... 190 6e-47
ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar... 190 7e-47
ref|NP_915417.1| putative HSP70 [Oryza sativa (japonica cultivar... 189 2e-46
gb|AAV98051.1| heat shock protein 70 [Medicago sativa] 187 4e-46
gb|AAB99745.1| HSP70 [Triticum aestivum] 187 4e-46
gb|AAK39876.1| heat shock protein 70KD [Guillardia theta] gi|252... 187 5e-46
dbj|BAA97566.1| hsp70 [Blastocystis hominis] 187 6e-46
emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida] 186 8e-46
emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida] gi|123... 186 8e-46
emb|CAA05547.1| heat shock protein 70 [Arabidopsis thaliana] 186 1e-45
gb|AAF14038.1| heat-shock protein (At-hsc70-3) [Arabidopsis thal... 185 2e-45
gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragm... 185 2e-45
pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment) 185 2e-45
gb|AAB42159.1| Hsc70 gi|2119719|pir||JC4786 dnaK-type molecular ... 184 3e-45
>gb|AAN46859.1| At2g32120/F22D22.13 [Arabidopsis thaliana]
gi|21703121|gb|AAM74502.1| At2g32120/F22D22.13
[Arabidopsis thaliana] gi|4263707|gb|AAD15393.1| 70kD
heat shock protein [Arabidopsis thaliana]
gi|15225187|ref|NP_180771.1| heat shock protein 70
family protein / HSP70 family protein [Arabidopsis
thaliana] gi|30685183|ref|NP_850183.1| heat shock
protein 70 family protein / HSP70 family protein
[Arabidopsis thaliana] gi|25295956|pir||B84729 70kD heat
shock protein [imported] - Arabidopsis thaliana
Length = 563
Score = 564 bits (1454), Expect = e-159
Identities = 283/376 (75%), Positives = 324/376 (85%), Gaps = 14/376 (3%)
Query: 5 EPAYTVASDSETTGEEKSSP---LPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLM 61
E AYTVASDSE TGEEKSS LPEIA+GIDIGTSQCS+AVWNGS+V +L+N RNQKL+
Sbjct: 3 EAAYTVASDSENTGEEKSSSSPSLPEIALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLI 62
Query: 62 KSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTL 121
KSFVTFKDE P+GGV++Q +HE EML G IFNMKRL+GRVDTDPVVHASKNLPFLVQTL
Sbjct: 63 KSFVTFKDEVPAGGVSNQLAHEQEMLTGAAIFNMKRLVGRVDTDPVVHASKNLPFLVQTL 122
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
DIGVRPFIAALVNN WRSTTPEEVLA+FLVELRLM E LKRP+RNVVLTVPVSFSRFQL
Sbjct: 123 DIGVRPFIAALVNNAWRSTTPEEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQL 182
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
TR ERACAMAGLHVLRLMPEPTA+ALLY QQQQ + + MGSGSE++A+IFNMGAGYCDV
Sbjct: 183 TRFERACAMAGLHVLRLMPEPTAIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDV 242
Query: 242 AVTATAGGVSQIKALAGSTIGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQ 301
AVTATAGGVSQIKALAGS IGGED+LQN +RH+ P +E ++ LLRVA Q
Sbjct: 243 AVTATAGGVSQIKALAGSPIGGEDILQNTIRHIAPPNE-----------EASGLLRVAAQ 291
Query: 302 DAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVG 361
DAIH+L+ Q +VQ++VDLGN KI KV+DR EFEEVN++VFE+CE L++QCL DA+V G
Sbjct: 292 DAIHRLTDQENVQIEVDLGNGNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGG 351
Query: 362 DINDVILVGGCSYIPQ 377
DI+D+I+VGGCSYIP+
Sbjct: 352 DIDDLIMVGGCSYIPK 367
>gb|AAM67201.1| 70kD heat shock protein [Arabidopsis thaliana]
Length = 563
Score = 562 bits (1449), Expect = e-159
Identities = 282/376 (75%), Positives = 324/376 (86%), Gaps = 14/376 (3%)
Query: 5 EPAYTVASDSETTGEEKSSP---LPEIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLM 61
E AYTVASDSE TGEEKSS LPEIA+GIDIGTSQCS+AVWNGS+V +L+N RNQKL+
Sbjct: 3 EAAYTVASDSENTGEEKSSSSPSLPEIALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLI 62
Query: 62 KSFVTFKDEAPSGGVTSQFSHEHEMLFGDTIFNMKRLIGRVDTDPVVHASKNLPFLVQTL 121
KSFVTFKDE P+GGV++Q +HE EML G IFNMKRL+GRVDTDPVVHASKNLPFLVQTL
Sbjct: 63 KSFVTFKDEVPAGGVSNQLAHEQEMLTGAAIFNMKRLVGRVDTDPVVHASKNLPFLVQTL 122
Query: 122 DIGVRPFIAALVNNVWRSTTPEEVLAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQL 181
DIGVRPFIAALVNN WRSTTPEEVLA+FLVELRLM E LKRP+RNVVLTVPVSFSRFQL
Sbjct: 123 DIGVRPFIAALVNNAWRSTTPEEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQL 182
Query: 182 TRLERACAMAGLHVLRLMPEPTAVALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDV 241
TR ERACAMAGLHVLRLMPEPTA+ALLY QQQQ + + MGSGSE++A+IFNMGAGYCDV
Sbjct: 183 TRFERACAMAGLHVLRLMPEPTAIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDV 242
Query: 242 AVTATAGGVSQIKALAGSTIGGEDLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQ 301
AVTATAGGVSQIKALAGS IGGED+LQ+ +RH+ P +E ++ LLRVA Q
Sbjct: 243 AVTATAGGVSQIKALAGSPIGGEDILQSTIRHIAPPNE-----------EASGLLRVAAQ 291
Query: 302 DAIHQLSTQSSVQVDVDLGNSLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVG 361
DAIH+L+ Q +VQ++VDLGN KI KV+DR EFEEVN++VFE+CE L++QCL DA+V G
Sbjct: 292 DAIHRLTDQENVQIEVDLGNGNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGG 351
Query: 362 DINDVILVGGCSYIPQ 377
DI+D+I+VGGCSYIP+
Sbjct: 352 DIDDLIMVGGCSYIPK 367
>dbj|BAB02269.1| 70 kDa heat shock protein [Arabidopsis thaliana]
gi|16649031|gb|AAL24367.1| 70 kDa heat shock protein
[Arabidopsis thaliana] gi|15809846|gb|AAL06851.1|
AT3g12580/T2E22_110 [Arabidopsis thaliana]
gi|15809832|gb|AAL06844.1| AT3g12580/T2E22_110
[Arabidopsis thaliana] gi|12321973|gb|AAG51030.1| heat
shock protein 70; 34105-36307 [Arabidopsis thaliana]
gi|15230534|ref|NP_187864.1| heat shock protein 70,
putative / HSP70, putative [Arabidopsis thaliana]
Length = 650
Score = 192 bits (488), Expect = 1e-47
Identities = 122/357 (34%), Positives = 199/357 (55%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKN-LPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V A K+ PF V + G +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVS-GPGEKPMIVVNHKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L P++N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 MVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLL 356
>gb|AAF34134.1| high molecular weight heat shock protein [Malus x domestica]
Length = 650
Score = 191 bits (486), Expect = 3e-47
Identities = 121/357 (33%), Positives = 195/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V K+ PF V G +P I + + EE+ +
Sbjct: 67 VNTVFDAKRLIGRRYSDPSVQGDMKHWPFKVIP-GPGDKPMIVVIYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M LV++R + E +L I+N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 126 MVLVKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKHKKDISSSPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGVDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 356
>dbj|BAA04848.1| HSP70 [Lilium longiflorum]
Length = 649
Score = 191 bits (484), Expect = 4e-47
Identities = 126/359 (35%), Positives = 196/359 (54%), Gaps = 16/359 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVR--PFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DP V A L PF V + G R P I + EEV
Sbjct: 67 TNTVFDAKRLIGRHFSDPSVQADMKLWPFKVIS---GTRDKPMIVVQYKGEEKQFAVEEV 123
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAV 205
+M LV++R + +L R I N V+TVP F+ Q + A A+AGL+VLR++ EPTA
Sbjct: 124 SSMVLVKMREIAVAYLGRSINNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAA 183
Query: 206 ALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGE 264
A+ YG ++ S SEK LIF++G G DV++ G+ ++KA AG T +GGE
Sbjct: 184 AIAYGLDKKATSP------SEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 237
Query: 265 DLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSL 323
D M+ H + + K+ +++ LR A + A LS+ + +++D L + +
Sbjct: 238 DFDNRMVNHFAQEFKRKHKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLCDGI 297
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 298 DFYSTITRARFEELNIDLFRKCMDPVEKCLTDAKMDKSSVHDVVLVGGSTRIPKVQQLL 356
>pir||JC2215 dnaK-type molecular chaperone LIM18 - trumpet lily
Length = 651
Score = 191 bits (484), Expect = 4e-47
Identities = 126/359 (35%), Positives = 196/359 (54%), Gaps = 16/359 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 11 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 68
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVR--PFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DP V A L PF V + G R P I + EEV
Sbjct: 69 TNTVFDAKRLIGRHFSDPSVQADMKLWPFKVIS---GTRDKPMIVVQYKGEEKQFAVEEV 125
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAV 205
+M LV++R + +L R I N V+TVP F+ Q + A A+AGL+VLR++ EPTA
Sbjct: 126 SSMVLVKMREIAVAYLGRSINNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAA 185
Query: 206 ALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGE 264
A+ YG ++ S SEK LIF++G G DV++ G+ ++KA AG T +GGE
Sbjct: 186 AIAYGLDKKATSP------SEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGE 239
Query: 265 DLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSL 323
D M+ H + + K+ +++ LR A + A LS+ + +++D L + +
Sbjct: 240 DFDNRMVNHFAQEFKRKHKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLCDGI 299
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 DFYSTITRARFEELNIDLFRKCMDPVEKCLTDAKMDKSSVHDVVLVGGSTRIPKVQQLL 358
>gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|NP_173055.1| heat
shock protein 70, putative / HSP70, putative
[Arabidopsis thaliana] gi|6587810|gb|AAF18501.1|
Identical to gb|AJ002551 heat shock protein 70 from
Arabidopsis thaliana and contains a PF|00012 HSP 70
domain. EST gb|F13893 comes from this gene
gi|25082767|gb|AAN71999.1| heat shock protein hsp70,
putative [Arabidopsis thaliana] gi|25295976|pir||B86295
hypothetical protein T24D18.14 [imported] - Arabidopsis
thaliana
Length = 646
Score = 190 bits (483), Expect = 6e-47
Identities = 119/359 (33%), Positives = 197/359 (54%), Gaps = 12/359 (3%)
Query: 27 EIAIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEM 86
E AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ +
Sbjct: 6 EKAIGIDLGTTYSCVGVWMNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAK--NQVAL 63
Query: 87 LFGDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEV 145
+T+F+ KRLIGR +DP V + + PF V + G +P I N + +PEE+
Sbjct: 64 NPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVS-GPGEKPMIVVSYKNEEKQFSPEEI 122
Query: 146 LAMFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAV 205
+M LV+++ + E L R ++N V+TVP F+ Q + A A++GL+VLR++ EPTA
Sbjct: 123 SSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAA 182
Query: 206 ALLYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGE 264
A+ YG ++ EK LIF++G G DV++ GV ++KA AG T +GGE
Sbjct: 183 AIAYGLDKKGT------KAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGE 236
Query: 265 DLLQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSL 323
D ++ H + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 237 DFDNRLVNHFVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGI 296
Query: 324 KICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + + L DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 297 DFYATISRARFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLL 355
>ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar-group)]
gi|47900318|gb|AAT39165.1| putative hsp70 [Oryza sativa
(japonica cultivar-group)]
Length = 646
Score = 190 bits (482), Expect = 7e-47
Identities = 122/361 (33%), Positives = 197/361 (53%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 66 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVVP-GPGDKPMIVVQYKGEEKQFAAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 MVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 294
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 295 GIDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 354
Query: 382 L 382
L
Sbjct: 355 L 355
>ref|NP_915417.1| putative HSP70 [Oryza sativa (japonica cultivar-group)]
gi|21104622|dbj|BAB93214.1| putative HSP70 [Oryza sativa
(japonica cultivar-group)] gi|15623835|dbj|BAB67894.1|
putative HSP70 [Oryza sativa (japonica cultivar-group)]
Length = 648
Score = 189 bits (479), Expect = 2e-46
Identities = 122/361 (33%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + + EE+ +
Sbjct: 66 INTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIP-GPGDKPMIVVQYKGEEKQFSAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 125 MVLIKMREIAEAYLGSNIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKAT------SSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 239 DNRMVNHFVLE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 294
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 295 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 354
Query: 382 L 382
L
Sbjct: 355 L 355
>gb|AAV98051.1| heat shock protein 70 [Medicago sativa]
Length = 649
Score = 187 bits (476), Expect = 4e-46
Identities = 123/361 (34%), Positives = 197/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V T G +P I + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRISDASVQSDMKLWPFKV-TAGPGEKPMIGVNYKGEEKLFASEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+VLR++ EPTA A+
Sbjct: 126 MVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFE 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 296 GVDFYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQL 355
Query: 382 L 382
L
Sbjct: 356 L 356
>gb|AAB99745.1| HSP70 [Triticum aestivum]
Length = 648
Score = 187 bits (476), Expect = 4e-46
Identities = 119/357 (33%), Positives = 193/357 (53%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 8 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 65
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V +P I + EE+ +
Sbjct: 66 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPAD-KPMIVVNYKGEEKQFAAEEISS 124
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E L ++N V+TVP F+ Q + A A+AGL+VLR++ EPTA A+
Sbjct: 125 MVLIKMREIAEAFLGNSVKNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPTAAAI 184
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 185 AYGLDKKATST------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 238
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 239 DNRMVNHFVQEFKRKHKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGVDF 298
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 299 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLL 355
>gb|AAK39876.1| heat shock protein 70KD [Guillardia theta] gi|25295948|pir||D90093
heat shock protein 70KD [imported] - Guillardia theta
nucleomorph gi|13812189|ref|NP_113319.1| heat shock
protein 70KD [Guillardia theta]
Length = 650
Score = 187 bits (475), Expect = 5e-46
Identities = 118/358 (32%), Positives = 200/358 (54%), Gaps = 11/358 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V +W VE++ N + + S+V F + G +++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGIWQHDRVEIIANDQGNRTTPSYVAFTETERLIGDSAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR DP V K+ PF V D G +P I + PEE+ A
Sbjct: 67 HNTVFDAKRLIGRRFQDPAVQDDIKHFPFKVICKD-GDKPAIEVKFKGETKVFAPEEISA 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E+ L + ++N V+TVP F+ Q + A A+ GL+VLR++ EPTA A+
Sbjct: 126 MVLMKMKEIAESFLGKDVKNAVITVPAYFNDSQRQATKDAGAITGLNVLRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++T GS SE+ LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-----LDKKTAGSKSERNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 240
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
++ + + + + FK+ +S+ LR A + A LS+ + V++D L + +
Sbjct: 241 DSRLVNYFVSEFKRKFKKDVTTNARSLRRLRTACERAKRTLSSTTQTTVEIDSLVDGIDF 300
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRILL 383
+ RA+FEE+ ++F + + L D+K+ +I+DV+LVGG + IP+ ++L+
Sbjct: 301 YSSITRAKFEELCMDLFRGTLDPVEKVLRDSKIAKSEIDDVVLVGGSTRIPKVQQLLI 358
>dbj|BAA97566.1| hsp70 [Blastocystis hominis]
Length = 656
Score = 187 bits (474), Expect = 6e-46
Identities = 114/357 (31%), Positives = 199/357 (54%), Gaps = 15/357 (4%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
++GID+GT+ V VW +VE++ N + Q+ S+V F D G ++ ++ M
Sbjct: 9 SVGIDLGTTYSCVGVWQNDKVEIIANDQGQRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHAS-KNLPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR DP V + K+ PF ++ G +P I ++ +PEE+ +
Sbjct: 67 TNTVFDAKRLIGRKFDDPSVQSDMKHWPFTLKAGPDG-KPLIEVEYKGEKKTFSPEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L + ++N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLSKMKQIAEAYLGKEVKNAVITVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG Q+ G EK LIF++G G DV++ + G+ ++KA AG T +GGED
Sbjct: 186 AYGLDQK---------GGEKNILIFDLGGGTFDVSLLSIEDGIFEVKATAGDTHLGGEDF 236
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ + + + F++ +++ LR A + A LS+ + +++D L +
Sbjct: 237 DNRMVEYFCTEFQRKFRKDLTTSQRALRRLRTACERAKRTLSSSTQATIEIDSLFEGIDF 296
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N + F K + + + L DAK+ ++++V+LVGG S IP+ ++L
Sbjct: 297 NSQITRARFEELNMDYFRKTMAPVEKVLRDAKLSKSEVDEVVLVGGSSRIPKVQKLL 353
>emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida]
Length = 646
Score = 186 bits (473), Expect = 8e-46
Identities = 124/361 (34%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 4 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 61
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 62 INTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIP-GPGDKPMIVVTYKGEEKQFAAEEISS 120
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 121 MVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 180
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 181 AYG-LDKKAS-----SAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 234
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 235 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 290
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 291 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 350
Query: 382 L 382
L
Sbjct: 351 L 351
>emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida]
gi|123650|sp|P09189|HSP7C_PETHY Heat shock cognate 70
kDa protein
Length = 651
Score = 186 bits (473), Expect = 8e-46
Identities = 124/361 (34%), Positives = 198/361 (54%), Gaps = 20/361 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V G +P I + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIP-GPGDKPMIVVTYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L +++ + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYE 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++
Sbjct: 296 GIDFYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQL 355
Query: 382 L 382
L
Sbjct: 356 L 356
>emb|CAA05547.1| heat shock protein 70 [Arabidopsis thaliana]
Length = 650
Score = 186 bits (472), Expect = 1e-45
Identities = 118/352 (33%), Positives = 195/352 (54%), Gaps = 12/352 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKN-LPFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V A K+ PF V + G +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVS-GPGEKPMIVVNHKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
+ L+++R + E L P++N V+ VP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 IVLIKMREIAEAFLGSPVKNAVVIVPAYFNDSQRQGTKDAGVISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG +KAS S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYG-LDKKAS-----SVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQ 377
+ RA FEE+N ++F KC + +CL DAK++ ++DV++VGG + IP+
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVVVGGSTRIPK 351
>gb|AAF14038.1| heat-shock protein (At-hsc70-3) [Arabidopsis thaliana]
gi|24111393|gb|AAN46823.1| At3g09440/F11F8.1
[Arabidopsis thaliana] gi|3043415|emb|CAA76606.1|
At-hsc70-3 [Arabidopsis thaliana]
gi|20465683|gb|AAM20310.1| putative heat-shock protein
[Arabidopsis thaliana] gi|15292925|gb|AAK92833.1|
putative heat-shock protein At-hsc70-3 [Arabidopsis
thaliana] gi|20856798|gb|AAM26685.1| At3g09440/F11F8.1
[Arabidopsis thaliana] gi|6682224|gb|AAF23276.1| heat
shock cognate 70kD protein [Arabidopsis thaliana]
gi|18206367|sp|O65719|HSP73_ARATH Heat shock cognate 70
kDa protein 3 (Hsc70.3) gi|15232682|ref|NP_187555.1|
heat shock cognate 70 kDa protein 3 (HSC70-3) (HSP70-3)
[Arabidopsis thaliana]
Length = 649
Score = 185 bits (470), Expect = 2e-45
Identities = 121/362 (33%), Positives = 200/362 (54%), Gaps = 20/362 (5%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR TD V + L PF +++ +P I + + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKS-GPAEKPMIVVNYKGEDKEFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEI----KSMALLRVATQDAIHQLSTQSSVQVDVD-LGN 321
M+ H + + FKR ++I +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQE----FKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFD 295
Query: 322 SLKICKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRI 381
+ + RA FEE+N ++F KC + +CL DAK++ I+DV+LVGG + IP+ ++
Sbjct: 296 GIDFYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQL 355
Query: 382 LL 383
L+
Sbjct: 356 LV 357
>gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragment) [Oryza sativa
(japonica cultivar-group)]
Length = 649
Score = 185 bits (469), Expect = 2e-45
Identities = 118/357 (33%), Positives = 193/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V G +P I + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFSDASVQSDIKLWPFKV-IAGPGDKPMIVVQYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPRVQQLL 356
>pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment)
Length = 649
Score = 185 bits (469), Expect = 2e-45
Identities = 118/357 (33%), Positives = 193/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +D V + L PF V G +P I + EE+ +
Sbjct: 67 INTVFDAKRLIGRRFSDASVQSDIKLWPFKV-IAGPGDKPMIVVQYKGEEKQFAAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L+++R + E +L I+N V+TVP F+ Q + A +AGL+V+R++ EPTA A+
Sbjct: 126 MVLIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKKATSV------GEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++DV+LVGG + IP+ ++L
Sbjct: 300 YSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPRVQQLL 356
>gb|AAB42159.1| Hsc70 gi|2119719|pir||JC4786 dnaK-type molecular chaperone hsc70-3
- tomato
Length = 651
Score = 184 bits (468), Expect = 3e-45
Identities = 115/357 (32%), Positives = 194/357 (54%), Gaps = 12/357 (3%)
Query: 29 AIGIDIGTSQCSVAVWNGSEVELLKNKRNQKLMKSFVTFKDEAPSGGVTSQFSHEHEMLF 88
AIGID+GT+ V VW VE++ N + + S+V F D G ++ ++ M
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAK--NQVAMNP 66
Query: 89 GDTIFNMKRLIGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNVWRSTTPEEVLA 147
+T+F+ KRLIGR +DP V + L PF V +P I + + EE+ +
Sbjct: 67 TNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPAD-KPMIVVNYKGEEKQFSAEEISS 125
Query: 148 MFLVELRLMTETHLKRPIRNVVLTVPVSFSRFQLTRLERACAMAGLHVLRLMPEPTAVAL 207
M L++++ + E L I+N V+TVP F+ Q + A ++GL+V+R++ EPTA A+
Sbjct: 126 MVLIKMKEIAEAFLGTTIKNAVVTVPAYFNDSQRQATKDAGTISGLNVMRIINEPTAAAI 185
Query: 208 LYGQQQQKASQETMGSGSEKIALIFNMGAGYCDVAVTATAGGVSQIKALAGST-IGGEDL 266
YG ++ + S EK LIF++G G DV++ G+ ++KA AG T +GGED
Sbjct: 186 AYGLDKK------ISSTGEKTVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDF 239
Query: 267 LQNMMRHLLPDSENIFKRHGVEEIKSMALLRVATQDAIHQLSTQSSVQVDVD-LGNSLKI 325
M+ H + + + K+ +++ LR A + A LS+ + +++D L +
Sbjct: 240 DNRMVNHFVQEFKRKHKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLYEGIDF 299
Query: 326 CKVVDRAEFEEVNKEVFEKCESLIIQCLHDAKVEVGDINDVILVGGCSYIPQGWRIL 382
+ RA FEE+N ++F KC + +CL DAK++ ++D++LVGG + IP+ ++L
Sbjct: 300 YTTITRARFEELNMDLFRKCMEPVEKCLRDAKIDKSGVHDIVLVGGSTRIPKVQQLL 356
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 627,491,852
Number of Sequences: 2540612
Number of extensions: 25071345
Number of successful extensions: 70865
Number of sequences better than 10.0: 2433
Number of HSP's better than 10.0 without gapping: 2203
Number of HSP's successfully gapped in prelim test: 230
Number of HSP's that attempted gapping in prelim test: 61235
Number of HSP's gapped (non-prelim): 2818
length of query: 394
length of database: 863,360,394
effective HSP length: 130
effective length of query: 264
effective length of database: 533,080,834
effective search space: 140733340176
effective search space used: 140733340176
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC145222.11


