

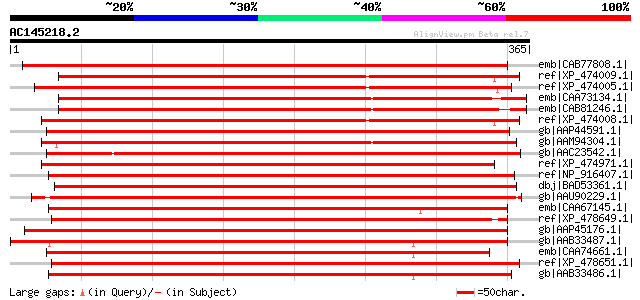
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145218.2 + phase: 1 /pseudo/partial
(365 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB77808.1| putative receptor kinase [Arabidopsis thaliana] ... 452 e-126
ref|XP_474009.1| OSJNBa0043L09.5 [Oryza sativa (japonica cultiva... 400 e-110
ref|XP_474005.1| OSJNBa0043L09.1 [Oryza sativa (japonica cultiva... 399 e-110
emb|CAA73134.1| serine/threonine kinase [Brassica oleracea] gi|7... 398 e-109
emb|CAB81246.1| serine/threonine kinase-like protein [Arabidopsi... 395 e-109
ref|XP_474008.1| OSJNBa0043L09.4 [Oryza sativa (japonica cultiva... 394 e-108
gb|AAP44591.1| putative receptor-like kinase [Oryza sativa (japo... 390 e-107
gb|AAM94304.1| receptor-like kinase [Sorghum bicolor] 389 e-107
gb|AAC23542.1| receptor protein kinase [Ipomoea trifida] 389 e-107
ref|XP_474971.1| OSJNBb0015G09.5 [Oryza sativa (japonica cultiva... 387 e-106
ref|NP_916407.1| putative receptor kinase [Oryza sativa (japonic... 387 e-106
dbj|BAD53361.1| putative receptor-like protein kinase ARK1 [Oryz... 385 e-106
gb|AAU90229.1| putative receptor-like protein kinase [Oryza sati... 385 e-105
emb|CAA67145.1| receptor-like kinase [Brassica oleracea var. ace... 382 e-105
ref|XP_478649.1| putative S-receptor kinase KIK1 precursor [Oryz... 382 e-105
gb|AAP45176.1| putative receptor protein kinase [Solanum bulboca... 382 e-105
gb|AAB33487.1| ARK3 product/receptor-like serine/threonine prote... 382 e-105
emb|CAA74661.1| SFR1 [Brassica oleracea] gi|7434410|pir||T14519 ... 382 e-105
ref|XP_478651.1| putative S-receptor kinase KIK1 precursor [Oryz... 381 e-104
gb|AAB33486.1| ARK2 product/receptor-like serine/threonine prote... 381 e-104
>emb|CAB77808.1| putative receptor kinase [Arabidopsis thaliana]
gi|15236268|ref|NP_192232.1| S-locus lectin protein
kinase family protein [Arabidopsis thaliana]
gi|4262151|gb|AAD14451.1| putative receptor kinase
[Arabidopsis thaliana] gi|25287707|pir||A85041 probable
receptor kinase [imported] - Arabidopsis thaliana
Length = 852
Score = 452 bits (1163), Expect = e-126
Identities = 219/342 (64%), Positives = 272/342 (79%), Gaps = 1/342 (0%)
Query: 10 LHDSEKHVRDLIGLGNIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGR 69
L DSE+H+++LI G ++DS+ I+VP + +I AT+NFS++NKLGQGG+GPVYKG
Sbjct: 491 LCDSERHIKELIESGRFKQDDSQGIDVPSFELETILYATSNFSNANKLGQGGFGPVYKGM 550
Query: 70 FPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSN 129
FPG QEIA+KRLS S QGL+EFKNE+VLIAKLQHRNLVRL GYC+ G+EK+LLYEYM +
Sbjct: 551 FPGDQEIAVKRLSRCSGQGLEEFKNEVVLIAKLQHRNLVRLLGYCVAGEEKLLLYEYMPH 610
Query: 130 KSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMI 189
KSLD FIFDR L WK+R +II+GIARG+LYLHQDSRLR+IHRDLKTSNILLD+EM
Sbjct: 611 KSLDFFIFDRKLCQRLDWKMRCNIILGIARGLLYLHQDSRLRIIHRDLKTSNILLDEEMN 670
Query: 190 PKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSG 249
PKISDFGLA+IFGG ET A+T RV+GTYGYMSPEYAL+G FS KSDVFSFGVV++E +SG
Sbjct: 671 PKISDFGLARIFGGSETSANTNRVVGTYGYMSPEYALEGLFSFKSDVFSFGVVVIETISG 730
Query: 250 KKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEP 309
K+NTGF ++ SLLG+AW LW + ++L+D AL E+C F+KC +GLLCVQ++P
Sbjct: 731 KRNTGFHEPEKSLSLLGHAWDLWKAERGIELLDQALQESCETEGFLKCLNVGLLCVQEDP 790
Query: 310 GNRPTMSNILTML-DGETATIPIPSQPTFFTTKHQSCSSSSS 350
+RPTMSN++ ML E AT+P P QP F + S S +SS
Sbjct: 791 NDRPTMSNVVFMLGSSEAATLPTPKQPAFVLRRCPSSSKASS 832
>ref|XP_474009.1| OSJNBa0043L09.5 [Oryza sativa (japonica cultivar-group)]
gi|38344785|emb|CAE02986.2| OSJNBa0043L09.5 [Oryza
sativa (japonica cultivar-group)]
Length = 814
Score = 400 bits (1028), Expect = e-110
Identities = 194/329 (58%), Positives = 249/329 (74%), Gaps = 6/329 (1%)
Query: 35 EVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKN 94
E P F + ATNNFSDSN LG+GG+G VYKG+ GG+EIA+KRLS+ STQGL+ F N
Sbjct: 483 EFPCINFEDVVTATNNFSDSNMLGEGGFGKVYKGKLGGGKEIAVKRLSTGSTQGLEHFTN 542
Query: 95 EIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDII 154
E+VLIAKLQH+NLVRL G CI GDEK+L+YEY+ NKSLD F+FD +L W RF II
Sbjct: 543 EVVLIAKLQHKNLVRLLGCCIHGDEKLLIYEYLPNKSLDHFLFDPASKFILDWPTRFKII 602
Query: 155 VGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVM 214
G+ARG+LYLHQDSRL +IHRDLKTSNILLD +M PKISDFG+A+IFGG + A+T RV+
Sbjct: 603 KGVARGLLYLHQDSRLTIIHRDLKTSNILLDADMSPKISDFGMARIFGGNQQEANTNRVV 662
Query: 215 GTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTE 274
GTYGYMSPEYA+DG FS+KSD++SFGV+LLEI+SG K + +LL YAWRLW +
Sbjct: 663 GTYGYMSPEYAMDGVFSVKSDIYSFGVILLEIVSGLK-ISLPQLMDFPNLLAYAWRLWKD 721
Query: 275 NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQ 334
+K +DL+DS+++E+C++NE + C IGLLCVQD P +RP MS+++ ML+ E A +P P Q
Sbjct: 722 DKTMDLVDSSIAESCSKNEVLLCIHIGLLCVQDNPNSRPLMSSVVFMLENEQAALPAPIQ 781
Query: 335 PTFFT-----TKHQSCSSSSSKLEISMQI 358
P +F TK ++SSS +S+ +
Sbjct: 782 PVYFAHRASETKQTGENTSSSNNNMSLTV 810
>ref|XP_474005.1| OSJNBa0043L09.1 [Oryza sativa (japonica cultivar-group)]
gi|38344781|emb|CAE02982.2| OSJNBa0043L09.1 [Oryza
sativa (japonica cultivar-group)]
Length = 827
Score = 399 bits (1026), Expect = e-110
Identities = 201/341 (58%), Positives = 251/341 (72%), Gaps = 6/341 (1%)
Query: 18 RDLIG-LGNIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEI 76
R ++G L E E++E+P+ +F I AATNNFSD N LGQGG+G VYKG G+E+
Sbjct: 486 RGILGYLSASNELGDENLELPFVSFGEIAAATNNFSDDNMLGQGGFGKVYKGMLDDGKEV 545
Query: 77 AIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFI 136
AIKRLS S QG +EF+NE+VLIAKLQHRNLVRL GYCI GDEK+L+YEY+ NKSLD FI
Sbjct: 546 AIKRLSKGSGQGAEEFRNEVVLIAKLQHRNLVRLLGYCIYGDEKLLIYEYLPNKSLDAFI 605
Query: 137 FDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFG 196
FD +L W RF II G+ARG+LYLHQDSRL VIHRDLK SNILLD +M PKISDFG
Sbjct: 606 FDHANKYVLDWPTRFKIIKGVARGLLYLHQDSRLTVIHRDLKPSNILLDVDMSPKISDFG 665
Query: 197 LAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFF 256
+A+IFGG + A+T RV+GTYGYMSPEYA+DG FS+KSD +SFGV+LLEI+S K
Sbjct: 666 MARIFGGNQHEANTNRVVGTYGYMSPEYAMDGAFSVKSDTYSFGVILLEIVSCLK-ISLP 724
Query: 257 RSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMS 316
R +LL YAW LW ++ +DLMDS++S++C+ E + C QIGLLCVQD P NRP MS
Sbjct: 725 RLTDFPNLLAYAWNLWKNDRAMDLMDSSISKSCSPTEVLLCIQIGLLCVQDNPNNRPLMS 784
Query: 317 NILTMLDGETATIPIPSQPTFFTTK----HQSCSSSSSKLE 353
++++ML+ ET T+ P QP +F + Q+ +S S LE
Sbjct: 785 SVVSMLENETTTLSAPIQPVYFAHRAFEGRQTGENSISLLE 825
>emb|CAA73134.1| serine/threonine kinase [Brassica oleracea] gi|7434416|pir||T14450
serine/threonine kinase (EC 2.7.1.-) BRLK - wild cabbage
Length = 850
Score = 398 bits (1022), Expect = e-109
Identities = 193/329 (58%), Positives = 251/329 (75%), Gaps = 7/329 (2%)
Query: 35 EVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKN 94
E+P + + I ATN+FS N+LG+GG+GPVYKG GQEIA+KRLS S QG+ EFKN
Sbjct: 513 ELPVFCLKVIVKATNDFSRENELGRGGFGPVYKGVLEDGQEIAVKRLSGKSGQGVDEFKN 572
Query: 95 EIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDII 154
EI+LIAKLQHRNLVRL G C +G+EK+L+YEYM NKSLD FIFD + L+ WKLRF II
Sbjct: 573 EIILIAKLQHRNLVRLLGCCFEGEEKMLVYEYMPNKSLDFFIFDEMKQELVDWKLRFAII 632
Query: 155 VGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVM 214
GIARG+LYLH+DSRLR+IHRDLK SN+LLD EM PKISDFG+A+IFGG + A+T RV+
Sbjct: 633 EGIARGLLYLHRDSRLRIIHRDLKVSNVLLDGEMNPKISDFGMARIFGGNQNEANTVRVV 692
Query: 215 GTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTE 274
GTYGYMSPEYA++G FS+KSDV+SFGV+LLEI+SGK+NT R+ + SL+GYAW L+T
Sbjct: 693 GTYGYMSPEYAMEGLFSVKSDVYSFGVLLLEIISGKRNTS-LRASEHGSLIGYAWFLYTH 751
Query: 275 NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQ 334
+ +L+D + TCN+ E ++C + +LCVQD RP M+ +L ML+ +TAT+P+P Q
Sbjct: 752 GRSEELVDPKIRATCNKREALRCIHVAMLCVQDSAAERPNMAAVLLMLESDTATLPVPRQ 811
Query: 335 PTFFTTKHQSCSSSSSKLEISMQIDSSYQ 363
PTF T S+ + ++++ +DSS Q
Sbjct: 812 PTFTT------STRRNSMDVNFALDSSQQ 834
>emb|CAB81246.1| serine/threonine kinase-like protein [Arabidopsis thaliana]
gi|3402758|emb|CAA20204.1| serine/threonine kinase-like
protein [Arabidopsis thaliana]
gi|15234429|ref|NP_193870.1| S-locus lectin protein
kinase family protein [Arabidopsis thaliana]
gi|7428021|pir||T05181 S-receptor kinase (EC 2.7.1.-)
T6K22.120 precursor - Arabidopsis thaliana
Length = 849
Score = 395 bits (1016), Expect = e-109
Identities = 190/329 (57%), Positives = 251/329 (75%), Gaps = 8/329 (2%)
Query: 35 EVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKN 94
E+P ++ +I ATN+F N+LG+GG+GPVYKG G+EIA+KRLS S QG+ EFKN
Sbjct: 513 ELPVFSLNAIAIATNDFCKENELGRGGFGPVYKGVLEDGREIAVKRLSGKSGQGVDEFKN 572
Query: 95 EIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDII 154
EI+LIAKLQHRNLVRL G C +G+EK+L+YEYM NKSLD F+FD T+ L+ WKLRF II
Sbjct: 573 EIILIAKLQHRNLVRLLGCCFEGEEKMLVYEYMPNKSLDFFLFDETKQALIDWKLRFSII 632
Query: 155 VGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVM 214
GIARG+LYLH+DSRLR+IHRDLK SN+LLD EM PKISDFG+A+IFGG + A+T RV+
Sbjct: 633 EGIARGLLYLHRDSRLRIIHRDLKVSNVLLDAEMNPKISDFGMARIFGGNQNEANTVRVV 692
Query: 215 GTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTE 274
GTYGYMSPEYA++G FS+KSDV+SFGV+LLEI+SGK+NT RS + SL+GYAW L+T
Sbjct: 693 GTYGYMSPEYAMEGLFSVKSDVYSFGVLLLEIVSGKRNTS-LRSSEHGSLIGYAWYLYTH 751
Query: 275 NKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQ 334
+ +L+D + TC++ E ++C + +LCVQD RP M+++L ML+ +TAT+ P Q
Sbjct: 752 GRSEELVDPKIRVTCSKREALRCIHVAMLCVQDSAAERPNMASVLLMLESDTATLAAPRQ 811
Query: 335 PTFFTTKHQSCSSSSSKLEISMQIDSSYQ 363
PTF +T+ S ++++ +DSS Q
Sbjct: 812 PTFTSTRRNS-------IDVNFALDSSQQ 833
>ref|XP_474008.1| OSJNBa0043L09.4 [Oryza sativa (japonica cultivar-group)]
gi|38344784|emb|CAE02985.2| OSJNBa0043L09.4 [Oryza
sativa (japonica cultivar-group)]
Length = 838
Score = 394 bits (1011), Expect = e-108
Identities = 193/341 (56%), Positives = 250/341 (72%), Gaps = 6/341 (1%)
Query: 23 LGNIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLS 82
L + E E++E+P+ +F I AATNNFSD N LGQGG+G VYKG +E+AIKRLS
Sbjct: 495 LSALNELGDENLELPFVSFGDIAAATNNFSDDNMLGQGGFGKVYKGMLGDNKEVAIKRLS 554
Query: 83 SVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT 142
S QG++EF+NE+VLIAKLQHRNLV+L G CI GDEK+L+YEY+ NKSL+ FIFD
Sbjct: 555 KGSGQGVEEFRNEVVLIAKLQHRNLVKLLGCCIHGDEKLLIYEYLPNKSLEAFIFDPASK 614
Query: 143 VLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFG 202
L W RF II G+ARG+LYLHQDSRL +IHRDLK+SNILLD +M PKISDFG+A+IFG
Sbjct: 615 YALDWPTRFKIIKGVARGLLYLHQDSRLTIIHRDLKSSNILLDVDMSPKISDFGMARIFG 674
Query: 203 GKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQIS 262
G + A+T RV+GTYGYMSPEYA+DG FS+KSD +S+GV+LLEI+SG K R
Sbjct: 675 GNQQEANTNRVVGTYGYMSPEYAMDGAFSVKSDTYSYGVILLEIVSGLK-ISLPRLMDFP 733
Query: 263 SLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
+LL YAW LW ++K +DL+DS+++E+C++ E + C IGLLCVQD P NRP MS+++ ML
Sbjct: 734 NLLAYAWSLWKDDKAMDLVDSSIAESCSKMEVLLCIHIGLLCVQDNPNNRPPMSSVVFML 793
Query: 323 DGETATIPIPSQPTFFT-----TKHQSCSSSSSKLEISMQI 358
+ E A +P P QP +F K ++SSS +S+ +
Sbjct: 794 ENEAAALPAPIQPVYFAHRASGAKQSGGNTSSSNNNMSLTV 834
>gb|AAP44591.1| putative receptor-like kinase [Oryza sativa (japonica
cultivar-group)] gi|34896978|ref|NP_909835.1| putative
receptor-like kinase [Oryza sativa (japonica
cultivar-group)]
Length = 868
Score = 390 bits (1001), Expect = e-107
Identities = 189/326 (57%), Positives = 249/326 (75%), Gaps = 1/326 (0%)
Query: 27 GENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST 86
G+ + + +++P + +I ATNNFS NKLGQGG+GPVY GR GQ+IA+KRLS ST
Sbjct: 528 GQGNHQDLDLPSFVIETILYATNNFSADNKLGQGGFGPVYMGRLDNGQDIAVKRLSRRST 587
Query: 87 QGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLG 146
QGL+EFKNE+ LIAKLQHRNLVRL G CI G E++L+YEYM N+SL+TF+F+ + +L
Sbjct: 588 QGLREFKNEVKLIAKLQHRNLVRLLGCCIDGSERMLIYEYMHNRSLNTFLFNEEKQSILN 647
Query: 147 WKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKET 206
W RF+II GIARG+LYLHQDS LR+IHRDLK SNILLD +M PKISDFG+A+IFG +T
Sbjct: 648 WSKRFNIINGIARGILYLHQDSALRIIHRDLKASNILLDRDMNPKISDFGVARIFGTDQT 707
Query: 207 GASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLG 266
A T++V+GTYGYMSPEYA+DG FS+KSDVFSFGV++LEI+SGKKN GF+ ++ +LL
Sbjct: 708 SAYTKKVVGTYGYMSPEYAMDGVFSMKSDVFSFGVLVLEIVSGKKNRGFYHNELDLNLLR 767
Query: 267 YAWRLWTENKLLDLMDSALSET-CNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
YAWRLW E + L+ +D +++ T N E ++C QIGLLCVQ++P +RPTMS + ML E
Sbjct: 768 YAWRLWKEGRSLEFLDQSIAGTSSNVTEVLRCIQIGLLCVQEQPRHRPTMSAVTMMLSSE 827
Query: 326 TATIPIPSQPTFFTTKHQSCSSSSSK 351
+ + P +P F T + S + +S+
Sbjct: 828 SPALLEPCEPAFCTGRSLSDDTEASR 853
>gb|AAM94304.1| receptor-like kinase [Sorghum bicolor]
Length = 839
Score = 389 bits (999), Expect = e-107
Identities = 190/337 (56%), Positives = 247/337 (72%), Gaps = 4/337 (1%)
Query: 23 LGNIGENDS---ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIK 79
LG++ E ++ E++++P+++F I +ATNNF++ N LGQGG+G VYKG +E+AIK
Sbjct: 493 LGHLDETNTLGDENLDLPFFSFDDIVSATNNFAEDNMLGQGGFGKVYKGILGENREVAIK 552
Query: 80 RLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDR 139
RLS S QG EF+NE+VLIAKLQHRNLVRL G CI GDEK+L+YEY+ NKSLD+FIFD
Sbjct: 553 RLSQGSGQGTDEFRNEVVLIAKLQHRNLVRLLGCCIHGDEKLLIYEYLPNKSLDSFIFDA 612
Query: 140 TRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAK 199
R +L W RF II GI+RG+LYLHQDSRL ++HRDLKTSNILLD +M PKISDFG+A+
Sbjct: 613 ARKNVLDWPTRFRIIKGISRGVLYLHQDSRLTIVHRDLKTSNILLDADMNPKISDFGMAR 672
Query: 200 IFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQ 259
IFGG + A+T RV+GTYGYMSPEYA+DG FS+ SD +S GV+LLEI+SG K T S
Sbjct: 673 IFGGNQQEANTNRVVGTYGYMSPEYAMDGAFSVMSDTYSLGVILLEIISGLKITS-THST 731
Query: 260 QISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNIL 319
SLL YAW LW + K +DL+DS + E+C+ NE ++C IGLLCVQD P +RP MS ++
Sbjct: 732 SFPSLLAYAWSLWNDGKAMDLVDSFVLESCSANEALRCIHIGLLCVQDNPNSRPLMSTVV 791
Query: 320 TMLDGETATIPIPSQPTFFTTKHQSCSSSSSKLEISM 356
ML+ ET + +P QP +F+ + + SM
Sbjct: 792 FMLENETTLLSVPKQPMYFSQWYLEAQGTGENTNSSM 828
>gb|AAC23542.1| receptor protein kinase [Ipomoea trifida]
Length = 853
Score = 389 bits (999), Expect = e-107
Identities = 193/335 (57%), Positives = 256/335 (75%), Gaps = 3/335 (0%)
Query: 27 GENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST 86
GE ++ E+P + F +I AT+NF+D NKLGQGG+G VYKG G +EIA+KRLS S
Sbjct: 510 GETMTDEFELPLFDFSTIVVATDNFADVNKLGQGGFGCVYKGMVEG-EEIAVKRLSKNSG 568
Query: 87 QGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLG 146
QG++EFKNE+ LIA+LQHRNLVRL G C+ +EKIL+YEYM NKSLD+ +F++ R+ LL
Sbjct: 569 QGVEEFKNELRLIARLQHRNLVRLLGCCVDMEEKILIYEYMENKSLDSTLFNKQRSSLLN 628
Query: 147 WKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKET 206
W+ RF+II GIARG+LYLHQDSR R+IHRDLK SNILLD EM PKISDFG+A+IFGG ET
Sbjct: 629 WQTRFNIICGIARGLLYLHQDSRFRIIHRDLKASNILLDKEMNPKISDFGMARIFGGDET 688
Query: 207 GA-STQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLL 265
A +T+RV+GTYGYMSPEYA+DG FS+KSDVFSFGV++LEI++GKKN GF+ +LL
Sbjct: 689 DANNTKRVVGTYGYMSPEYAMDGLFSVKSDVFSFGVLVLEIVTGKKNRGFYNQNNQQNLL 748
Query: 266 GYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGE 325
G+AWRLW E + +L+DSA+ E+ + E ++C Q+GLLCVQ++ +RP M+ ++ ML E
Sbjct: 749 GHAWRLWRERRGSELLDSAIGESYSLCEVMRCIQVGLLCVQEQAEDRPNMATVVLMLGSE 808
Query: 326 TATIPIPSQPTF-FTTKHQSCSSSSSKLEISMQID 359
+AT+P P P F ++ SS+S + S ++
Sbjct: 809 SATLPQPKHPGFCLGSRPADMDSSTSNCDESCTVN 843
>ref|XP_474971.1| OSJNBb0015G09.5 [Oryza sativa (japonica cultivar-group)]
gi|38346886|emb|CAE03911.2| OSJNBb0015G09.5 [Oryza
sativa (japonica cultivar-group)]
Length = 846
Score = 387 bits (994), Expect = e-106
Identities = 185/319 (57%), Positives = 238/319 (73%)
Query: 23 LGNIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLS 82
+ + E SE++E+ S+ ATNNFSD N LG+GG+G VYKG GG E+A+KRLS
Sbjct: 500 MNDSNEVGSENVELSSVDLDSVLTATNNFSDYNLLGKGGFGKVYKGVLEGGIEVAVKRLS 559
Query: 83 SVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRT 142
S QG++EF+NE+VLIAKLQHRNLVRL G CI DEK+L+YEY+ N+SLD F+FD R
Sbjct: 560 KGSGQGVEEFRNEVVLIAKLQHRNLVRLLGCCIHEDEKLLIYEYLPNRSLDAFLFDANRK 619
Query: 143 VLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFG 202
L W RF II G+ARG+LYLHQDSRL +IHRDLKTSNILLD EM PKISDFG+A+IFG
Sbjct: 620 NTLDWPTRFKIIKGVARGLLYLHQDSRLTIIHRDLKTSNILLDTEMSPKISDFGMARIFG 679
Query: 203 GKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQIS 262
G E A+T RV+GTYGYMSPEYALDG+FS+KSD +SFGV+LLE++SG K + S
Sbjct: 680 GNEQQANTTRVVGTYGYMSPEYALDGYFSVKSDTYSFGVILLEVVSGLKISSAHLKVDCS 739
Query: 263 SLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
+L+ YAW LW + D +DS++ E+C +E ++C +GLLC+QD+P RP MS+I+ ML
Sbjct: 740 NLIAYAWSLWKDGNARDFVDSSIVESCPLHEVLRCIHLGLLCIQDQPSARPLMSSIVFML 799
Query: 323 DGETATIPIPSQPTFFTTK 341
+ ETA +P P +P +FT +
Sbjct: 800 ENETAVLPAPKEPIYFTRR 818
>ref|NP_916407.1| putative receptor kinase [Oryza sativa (japonica cultivar-group)]
gi|20804899|dbj|BAB92579.1| putative receptor-like
kinase [Oryza sativa (japonica cultivar-group)]
Length = 856
Score = 387 bits (993), Expect = e-106
Identities = 181/328 (55%), Positives = 252/328 (76%)
Query: 28 ENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQ 87
E D + +++P ++I AAT++F+ SNK+G+GG+GPVY G+ GQE+A+KRLS S Q
Sbjct: 519 ECDEKDLDLPLLDLKAIVAATDDFAASNKIGEGGFGPVYMGKLEDGQEVAVKRLSRRSVQ 578
Query: 88 GLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGW 147
G+ EFKNE+ LIAKLQHRNLVRL G CI DE++L+YEYM N+SLDTFIFD + LL W
Sbjct: 579 GVVEFKNEVKLIAKLQHRNLVRLLGCCIDDDERMLVYEYMHNQSLDTFIFDEGKRKLLRW 638
Query: 148 KLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETG 207
RF+IIVG+ARG+LYLH+DSR R+IHRDLK SN+LLD M+PKISDFG+A++FGG +T
Sbjct: 639 SKRFEIIVGVARGLLYLHEDSRFRIIHRDLKASNVLLDRNMVPKISDFGIARMFGGDQTT 698
Query: 208 ASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGY 267
A T++V+GTYGYMSPEYA+DG FS+KSDV+SFGV++LEI++G++N GF+ ++ +LL Y
Sbjct: 699 AYTRKVIGTYGYMSPEYAMDGVFSMKSDVYSFGVLVLEIVTGRRNRGFYEAELDLNLLRY 758
Query: 268 AWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETA 327
+W LW E + +DL+D L + + +E ++C Q+ LLCV+ +P NRP MS+++ ML E A
Sbjct: 759 SWLLWKEGRSVDLLDQLLGGSFDYSEVLRCIQVALLCVEVQPRNRPLMSSVVMMLASENA 818
Query: 328 TIPIPSQPTFFTTKHQSCSSSSSKLEIS 355
T+P P++P +H S + SS L ++
Sbjct: 819 TLPEPNEPGVNIGRHASDTESSETLTVN 846
>dbj|BAD53361.1| putative receptor-like protein kinase ARK1 [Oryza sativa (japonica
cultivar-group)]
Length = 846
Score = 385 bits (990), Expect = e-106
Identities = 186/327 (56%), Positives = 248/327 (74%), Gaps = 2/327 (0%)
Query: 32 ESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQE 91
+ +E+P + +I AAT+ FS +NKLG+GG+GPVYKG+ GQEIA+K LS S QGL E
Sbjct: 508 DDLELPIFDLGTIAAATDGFSINNKLGEGGFGPVYKGKLEDGQEIAVKTLSKTSVQGLDE 567
Query: 92 FKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRF 151
FKNE++LIAKLQHRNLVRL G+ I G E+IL+YEYM+NKSLD F+F+++ +VLL W+ R+
Sbjct: 568 FKNEVMLIAKLQHRNLVRLLGFSISGQERILVYEYMANKSLDYFLFEKSNSVLLDWQARY 627
Query: 152 DIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQ 211
II GI RG+LYLHQDSR R+IHRDLK SN+LLD EM PKISDFG+A++FG +ET +T+
Sbjct: 628 RIIEGITRGLLYLHQDSRYRIIHRDLKASNVLLDKEMTPKISDFGMARMFGSEETEINTR 687
Query: 212 RVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRL 271
+V+GTYGYMSPEYA+DG FS+KSDVFSFGV+LLEI+SG++N G + +LLG+AW L
Sbjct: 688 KVVGTYGYMSPEYAMDGVFSVKSDVFSFGVLLLEIISGRRNRGVYSYSNHLNLLGHAWSL 747
Query: 272 WTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML-DGETATIP 330
W E K L+L D ++ + + +E +KC ++GLLCVQ+ P +RP MS +L ML + T+P
Sbjct: 748 WNEGKSLELADETMNGSFDSDEVLKCIRVGLLCVQENPDDRPLMSQVLLMLATTDATTLP 807
Query: 331 IPSQPTFFTTK-HQSCSSSSSKLEISM 356
P QP F + +SSSK + S+
Sbjct: 808 TPKQPGFAARRILMETDTSSSKPDCSI 834
>gb|AAU90229.1| putative receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 837
Score = 385 bits (989), Expect = e-105
Identities = 191/346 (55%), Positives = 253/346 (72%), Gaps = 5/346 (1%)
Query: 16 HVRDLIGLGNIGENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQE 75
H R+L GN + + +++P + +I +ATN FS NKLG+GG+GPVYKG GQE
Sbjct: 487 HSRELHSEGN---SHGDDLDLPLFDLETIASATNGFSADNKLGEGGFGPVYKGTLEDGQE 543
Query: 76 IAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTF 135
IA+K LS S QGL EF+NE++LIAKLQHRNLV+L GY + G EK+LLYE+M NKSLD F
Sbjct: 544 IAVKTLSKTSVQGLDEFRNEVMLIAKLQHRNLVQLIGYSVCGQEKMLLYEFMENKSLDCF 603
Query: 136 IFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDF 195
+FD++++ LL W+ R+ II GIARG+LYLHQDSR R+IHRDLKTSNILLD EM PKISDF
Sbjct: 604 LFDKSKSKLLDWQTRYHIIEGIARGLLYLHQDSRYRIIHRDLKTSNILLDKEMTPKISDF 663
Query: 196 GLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGF 255
G+A++FG +T +T RV+GTYGYM+PEYA+DG FS+KSDVFSFGV++LEI+SGK+N G
Sbjct: 664 GMARMFGSDDTEINTVRVVGTYGYMAPEYAMDGVFSVKSDVFSFGVIVLEIISGKRNRGV 723
Query: 256 FRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTM 315
+ +LL AW W+E LDL+D L+ + N+ E +KC ++GLLCVQ+ P +RP M
Sbjct: 724 YSYSSHLNLLARAWSSWSEGNSLDLVDKTLNGSFNQEEVLKCLKVGLLCVQENPDDRPLM 783
Query: 316 SNILTMLDGETAT-IPIPSQPTFFTTKHQSCSSSSSKLEISMQIDS 360
S +L ML AT +P P +P F + + +SSS+ + S +DS
Sbjct: 784 SQVLLMLASADATSLPDPRKPGFVARRAATEDTSSSRPDCSF-VDS 828
>emb|CAA67145.1| receptor-like kinase [Brassica oleracea var. acephala]
gi|7434413|pir||T14470 receptor-like kinase (EC 2.7.1.-)
SFR2 - wild cabbage
Length = 847
Score = 382 bits (982), Expect = e-105
Identities = 188/327 (57%), Positives = 243/327 (73%), Gaps = 4/327 (1%)
Query: 28 ENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQ 87
EN ++ +E+P F ++ AT+NFS++NKLGQGG+G VYKGR GQEIA+KRLS +S Q
Sbjct: 499 ENKTDDLELPLMDFEAVAIATDNFSNANKLGQGGFGIVYKGRLLDGQEIAVKRLSKMSVQ 558
Query: 88 GLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGW 147
G EFKNE+ LIA+LQH NLVRL G C+ EK+L+YEY+ N SLD+ +FD+TR+ L W
Sbjct: 559 GTDEFKNEVKLIARLQHINLVRLLGCCVDEGEKMLIYEYLENLSLDSHLFDKTRSCKLNW 618
Query: 148 KLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETG 207
+ RFDI GIARG+LYLHQDSR R+IHRDLK SN+LLD +M PKISDFG+A+IFG ET
Sbjct: 619 QKRFDITNGIARGLLYLHQDSRFRIIHRDLKASNVLLDKDMTPKISDFGMARIFGRDETE 678
Query: 208 ASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGY 267
A+T++V+GTYGYMSPEYA+DG FS KSDVFSFGV+LLEI+SGK+N GF+ S +LLG
Sbjct: 679 ANTRKVVGTYGYMSPEYAMDGIFSTKSDVFSFGVLLLEIISGKRNKGFYNSDHDLNLLGC 738
Query: 268 AWRLWTENKLLDLMDSALSE----TCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLD 323
WR W + K LD++D + + T E ++C +IGLLCVQ+ +RPTMS+++ ML
Sbjct: 739 VWRNWKKGKGLDIVDPIILDSSPSTYRPLEILRCIKIGLLCVQERANDRPTMSSVVMMLG 798
Query: 324 GETATIPIPSQPTFFTTKHQSCSSSSS 350
ETA IP P QP + + + SSS
Sbjct: 799 SETAAIPQPEQPGYCVGRSPLDTDSSS 825
>ref|XP_478649.1| putative S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)] gi|28971965|dbj|BAC65366.1| putative
S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)] gi|50510068|dbj|BAD30706.1| putative
S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)]
Length = 865
Score = 382 bits (981), Expect = e-105
Identities = 180/321 (56%), Positives = 244/321 (75%), Gaps = 4/321 (1%)
Query: 30 DSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGL 89
+ ++ E+P + F ++ AT+NFS SNKLG+GG+G VYKGR PGG+EIA+KRLS S QGL
Sbjct: 523 EGKNCELPLFAFETLATATDNFSISNKLGEGGFGHVYKGRLPGGEEIAVKRLSRSSGQGL 582
Query: 90 QEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKL 149
+EFKNE++LIAKLQHRNLVRL G CI+G+EKIL+YEYM NKSLD F+FD R LL W+
Sbjct: 583 EEFKNEVILIAKLQHRNLVRLLGCCIQGEEKILVYEYMPNKSLDAFLFDPERRGLLDWRT 642
Query: 150 RFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGAS 209
RF II G+ARG+LYLH+DSRLRV+HRDLK SNILLD +M PKISDFG+A+IFGG + +
Sbjct: 643 RFQIIEGVARGLLYLHRDSRLRVVHRDLKASNILLDRDMNPKISDFGMARIFGGDQNQVN 702
Query: 210 TQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAW 269
T RV+GT GYMSPEYA++G FS++SDV+SFG+++LEI++G+KN+ F + +++GYAW
Sbjct: 703 TNRVVGTLGYMSPEYAMEGLFSVRSDVYSFGILILEIITGQKNSSFHHMEGSLNIVGYAW 762
Query: 270 RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI 329
+LW ++ +L+D A+ TC E ++C + LLCVQD +RP + ++ L +++ +
Sbjct: 763 QLWNGDRGQELIDPAIRGTCPAKEALRCVHMALLCVQDHAHDRPDIPYVVLTLGSDSSVL 822
Query: 330 PIPSQPTFFTTKHQSCSSSSS 350
P P PTF C+SSSS
Sbjct: 823 PTPRPPTFTL----QCTSSSS 839
>gb|AAP45176.1| putative receptor protein kinase [Solanum bulbocastanum]
Length = 711
Score = 382 bits (981), Expect = e-105
Identities = 191/341 (56%), Positives = 251/341 (73%), Gaps = 1/341 (0%)
Query: 11 HDSEKHVRDLIGLGNIGENDSESIE-VPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGR 69
H E + D N+ E D ++ + + F I AATNNFS NKLG+GG+GPVYKG+
Sbjct: 352 HIREMNAADSFNNTNLKEEDVREVQDLKIFGFGLIMAATNNFSSDNKLGEGGFGPVYKGQ 411
Query: 70 FPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSN 129
FP G+E+AIKRLS S QGL EFKNE++LIAK+QHRNLVR+ G CI GDEK+L+YEYM N
Sbjct: 412 FPDGREVAIKRLSRTSGQGLAEFKNELILIAKVQHRNLVRVLGCCIHGDEKMLIYEYMPN 471
Query: 130 KSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMI 189
KSLD F+FD R LL W+ RF+II GIA+G+LYLH+ SR+RVIHRDLK SN+LLD+ M
Sbjct: 472 KSLDFFLFDPERKKLLDWQKRFEIIEGIAQGLLYLHKYSRMRVIHRDLKASNVLLDENMN 531
Query: 190 PKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSG 249
PKI+DFGLA+IF ET A T+RV+GTYGYM+PE+A++G FSIKSDVFSFGV++LEILSG
Sbjct: 532 PKIADFGLARIFKQNETEAVTRRVVGTYGYMAPEFAMEGAFSIKSDVFSFGVLMLEILSG 591
Query: 250 KKNTGFFRSQQISSLLGYAWRLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEP 309
++N + + +L+GYAW LW E L+L D L + + +F++ +GLLCVQ+
Sbjct: 592 RRNASLQQFNRPLNLIGYAWELWKEGCGLELKDPDLEDLYDTEQFLRVIHVGLLCVQEGA 651
Query: 310 GNRPTMSNILTMLDGETATIPIPSQPTFFTTKHQSCSSSSS 350
+RPTMS++++ML + ++PI QP FFT + + S SSS
Sbjct: 652 TDRPTMSDVISMLCNGSMSLPIAKQPAFFTGRDEIESYSSS 692
>gb|AAB33487.1| ARK3 product/receptor-like serine/threonine protein kinase ARK3
[Arabidopsis thaliana, Columbia, Peptide, 851 aa]
Length = 851
Score = 382 bits (981), Expect = e-105
Identities = 195/362 (53%), Positives = 256/362 (69%), Gaps = 12/362 (3%)
Query: 1 KVSIQIQESLHDSEKHVRDLIGLGNI--------GENDSESIEVPYYTFRSIQAATNNFS 52
K SI I+ + D + RDL+ + EN+++ +E+P F + ATNNFS
Sbjct: 468 KRSILIETPIVDHQLRSRDLLMNEVVISSRRHISRENNTDDLELPLMEFEEVAMATNNFS 527
Query: 53 DSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGLQEFKNEIVLIAKLQHRNLVRLRG 112
++NKLGQGG+G VYKG+ GQE+A+KRLS S QG EFKNE+ LIA+LQH NLVRL
Sbjct: 528 NANKLGQGGFGIVYKGKLLDGQEMAVKRLSKTSVQGTDEFKNEVKLIARLQHINLVRLLA 587
Query: 113 YCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKLRFDIIVGIARGMLYLHQDSRLRV 172
C+ EK+L+YEY+ N SLD+ +FD++R L W++RFDII GIARG+LYLHQDSR R+
Sbjct: 588 CCVDAGEKMLIYEYLENLSLDSHLFDKSRNSKLNWQMRFDIINGIARGLLYLHQDSRFRI 647
Query: 173 IHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGASTQRVMGTYGYMSPEYALDGFFSI 232
IHRDLK SNILLD M PKISDFG+A+IFG ET A+T++V+GTYGYMSPEYA+DG FS+
Sbjct: 648 IHRDLKASNILLDKYMTPKISDFGMARIFGRDETEANTRKVVGTYGYMSPEYAMDGIFSM 707
Query: 233 KSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAWRLWTENKLLDLMD----SALSET 288
KSDVFSFGV+LLEI+S K+N GF+ S + +LLG WR W E K L+++D +LS T
Sbjct: 708 KSDVFSFGVLLLEIISSKRNKGFYNSDRDLNLLGCVWRNWKEGKGLEIIDPIITDSLSST 767
Query: 289 CNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATIPIPSQPTFFTTKHQSCSSS 348
++E ++C QIGLLCVQ+ +RPTMS ++ ML E+ TIP P P + + + S
Sbjct: 768 FRQHEILRCIQIGLLCVQERAEDRPTMSLVILMLGSESTTIPQPKAPGYCLERSLLDTDS 827
Query: 349 SS 350
SS
Sbjct: 828 SS 829
>emb|CAA74661.1| SFR1 [Brassica oleracea] gi|7434410|pir||T14519 probable S-receptor
kinase (EC 2.7.1.-) SFR1 - wild cabbage
Length = 849
Score = 382 bits (981), Expect = e-105
Identities = 188/315 (59%), Positives = 238/315 (74%), Gaps = 4/315 (1%)
Query: 27 GENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVST 86
G+ +E +E+P F +I AT+NFS +NKLGQGG+G VYKGR G+EIA+KRLS +S
Sbjct: 503 GDMKTEDLELPLMDFEAIATATHNFSSTNKLGQGGFGIVYKGRLLDGKEIAVKRLSKMSL 562
Query: 87 QGLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLG 146
QG EFKNE+ LIA+LQH NLVRL G C+ EK+L+YEY+ N SLD+ +FD++R L
Sbjct: 563 QGTDEFKNEVRLIARLQHINLVRLLGCCVDKGEKMLIYEYLENLSLDSHLFDKSRRSNLN 622
Query: 147 WKLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKET 206
W+LRFDI GIARG+LYLHQDSR R+IHRDLK SNILLD MIPKISDFG+A+IF ET
Sbjct: 623 WQLRFDIANGIARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFRRDET 682
Query: 207 GASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLG 266
A+T++V+GTYGYMSPEYA++G FS+KSDVFSFGV+LLEI+SGK++TGF+ S SLLG
Sbjct: 683 EANTRKVVGTYGYMSPEYAMNGIFSVKSDVFSFGVLLLEIISGKRSTGFYNSSGDLSLLG 742
Query: 267 YAWRLWTENKLLDLMD----SALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTML 322
WR W E K LD++D +LS T +E ++C IGLLCVQ+ +RP MS+++ ML
Sbjct: 743 CVWRNWKERKGLDIIDPIIIDSLSSTFKTHEILRCIHIGLLCVQERAEDRPAMSSVMVML 802
Query: 323 DGETATIPIPSQPTF 337
ET T+P P QP F
Sbjct: 803 GSETTTLPEPKQPAF 817
>ref|XP_478651.1| putative S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)] gi|28971966|dbj|BAC65367.1| putative
S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)] gi|50510070|dbj|BAD30708.1| putative
S-receptor kinase KIK1 precursor [Oryza sativa (japonica
cultivar-group)]
Length = 853
Score = 381 bits (978), Expect = e-104
Identities = 181/329 (55%), Positives = 246/329 (74%)
Query: 30 DSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQGL 89
D +S E+ Y+F I+AAT NFSDSNKLG GG+GPVY G+ PGG+E+A+KRL S QGL
Sbjct: 514 DGKSHELKVYSFDRIKAATCNFSDSNKLGAGGFGPVYMGKLPGGEEVAVKRLCRKSGQGL 573
Query: 90 QEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGWKL 149
+EFKNE++LIAKLQHRNLVRL G CI+G+EKIL+YEYM NKSLD F+F+ + LL W+
Sbjct: 574 EEFKNEVILIAKLQHRNLVRLLGCCIQGEEKILVYEYMPNKSLDAFLFNPEKQGLLDWRK 633
Query: 150 RFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETGAS 209
RFDII GIARG+LYLH+DSRLRV+HRDLK SNILLD +M PKISDFG+A++FGG + +
Sbjct: 634 RFDIIEGIARGLLYLHRDSRLRVVHRDLKASNILLDKDMNPKISDFGMARMFGGDQNQFN 693
Query: 210 TQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGYAW 269
T RV+GT+GYMSPEYA++G FS+KSD++SFGV++LEI++GK+ F Q ++ G+AW
Sbjct: 694 TNRVVGTFGYMSPEYAMEGIFSVKSDIYSFGVLMLEIITGKRALSFHGQQDSLNIAGFAW 753
Query: 270 RLWTENKLLDLMDSALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLDGETATI 329
R W E+K +L+D + +C+ + ++C I LLCVQD RP + ++ ML +++++
Sbjct: 754 RQWNEDKGEELIDPLIRASCSLRQVLRCIHIALLCVQDHAQERPDIPAVILMLSSDSSSL 813
Query: 330 PIPSQPTFFTTKHQSCSSSSSKLEISMQI 358
P+P PT + +S SS+ + S I
Sbjct: 814 PMPRPPTLMLHGRSAETSKSSEKDQSHSI 842
>gb|AAB33486.1| ARK2 product/receptor-like serine/threonine protein kinase ARK2
[Arabidopsis thaliana, Columbia, Peptide, 850 aa]
Length = 850
Score = 381 bits (978), Expect = e-104
Identities = 189/330 (57%), Positives = 244/330 (73%), Gaps = 4/330 (1%)
Query: 28 ENDSESIEVPYYTFRSIQAATNNFSDSNKLGQGGYGPVYKGRFPGGQEIAIKRLSSVSTQ 87
EN ++ +E+P ++++ ATNNFS NKLGQGG+G VYKG G+EIA+KRLS +S+Q
Sbjct: 503 ENKTDYLELPLMEWKALAMATNNFSTDNKLGQGGFGIVYKGMLLDGKEIAVKRLSKMSSQ 562
Query: 88 GLQEFKNEIVLIAKLQHRNLVRLRGYCIKGDEKILLYEYMSNKSLDTFIFDRTRTVLLGW 147
G EF NE+ LIAKLQH NLVRL G C+ EK+L+YEY+ N SLD+ +FD+TR+ L W
Sbjct: 563 GTDEFMNEVRLIAKLQHINLVRLLGCCVDKGEKMLIYEYLENLSLDSHLFDQTRSSNLNW 622
Query: 148 KLRFDIIVGIARGMLYLHQDSRLRVIHRDLKTSNILLDDEMIPKISDFGLAKIFGGKETG 207
+ RFDII GIARG+LYLHQDSR R+IHRDLK SN+LLD M PKISDFG+A+IFG +ET
Sbjct: 623 QKRFDIINGIARGLLYLHQDSRCRIIHRDLKASNVLLDKNMTPKISDFGMARIFGREETE 682
Query: 208 ASTQRVMGTYGYMSPEYALDGFFSIKSDVFSFGVVLLEILSGKKNTGFFRSQQISSLLGY 267
A+T+RV+GTYGYMSPEYA+DG FS+KSDVFSFGV+LLEI+SGK+N GF+ S + +LLG+
Sbjct: 683 ANTRRVVGTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISGKRNKGFYNSNRDLNLLGF 742
Query: 268 AWRLWTENKLLDLMD----SALSETCNENEFVKCAQIGLLCVQDEPGNRPTMSNILTMLD 323
WR W E K L+++D ALS +E ++C QIGLLCVQ+ +RP MS+++ ML
Sbjct: 743 VWRHWKEGKELEIVDPINIDALSSEFPTHEILRCIQIGLLCVQERAEDRPVMSSVMVMLG 802
Query: 324 GETATIPIPSQPTFFTTKHQSCSSSSSKLE 353
ET IP P +P F + SSS +
Sbjct: 803 SETTAIPQPKRPGFCVGRSSLEVDSSSSTQ 832
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.137 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 609,373,666
Number of Sequences: 2540612
Number of extensions: 25969623
Number of successful extensions: 107452
Number of sequences better than 10.0: 20012
Number of HSP's better than 10.0 without gapping: 10809
Number of HSP's successfully gapped in prelim test: 9204
Number of HSP's that attempted gapping in prelim test: 65529
Number of HSP's gapped (non-prelim): 22721
length of query: 365
length of database: 863,360,394
effective HSP length: 129
effective length of query: 236
effective length of database: 535,621,446
effective search space: 126406661256
effective search space used: 126406661256
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC145218.2


