

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145109.3 - phase: 0
(190 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
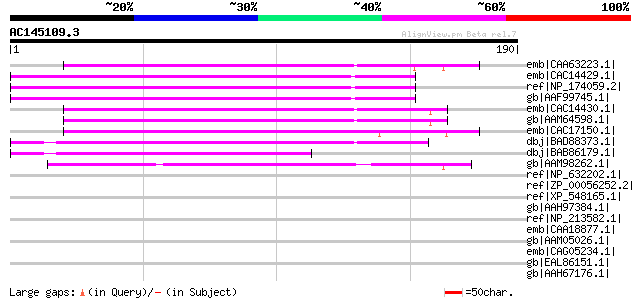
Score E
Sequences producing significant alignments: (bits) Value
emb|CAA63223.1| TOM20 [Solanum tuberosum] gi|7459240|pir||T07679... 111 1e-23
emb|CAC14429.1| TOM20-2 protein [Arabidopsis thaliana] gi|136318... 107 1e-22
ref|NP_174059.2| mitochondrial import receptor subunit TOM20-2 (... 107 2e-22
gb|AAF99745.1| F17L21.18 [Arabidopsis thaliana] 107 2e-22
emb|CAC14430.1| TOM20-3 protein [Arabidopsis thaliana] gi|193108... 106 3e-22
gb|AAM64598.1| putative TOM20 [Arabidopsis thaliana] 106 3e-22
emb|CAC17150.1| TOM20-1 protein [Arabidopsis thaliana] gi|927963... 100 2e-20
dbj|BAD88373.1| putative TOM20 [Oryza sativa (japonica cultivar-... 100 2e-20
dbj|BAB86179.1| putative mitochondrial import receptor subunit T... 86 7e-16
gb|AAM98262.1| At5g40930/MMG1_2 [Arabidopsis thaliana] gi|101774... 80 2e-14
ref|NP_632202.1| hypothetical protein MM0178 [Methanosarcina maz... 40 0.034
ref|ZP_00056252.2| COG4235: Cytochrome c biogenesis factor [Magn... 37 0.37
ref|XP_548165.1| PREDICTED: similar to Oxysterol binding protein... 36 0.49
gb|AAH97384.1| Interferon-induced protein with tetratricopeptide... 36 0.49
ref|NP_213582.1| hypothetical protein aq_854 [Aquifex aeolicus V... 36 0.64
emb|CAA18877.1| SPBC23E6.09 [Schizosaccharomyces pombe] gi|19113... 35 0.83
gb|AAM05026.1| TPR-domain containing protein [Methanosarcina ace... 35 0.83
emb|CAG05234.1| unnamed protein product [Tetraodon nigroviridis] 35 1.1
gb|EAL86151.1| oxidoreductase, short chain dehydrogenase/reducta... 35 1.4
gb|AAH67176.1| Small glutamine-rich tetratricopeptide repeat (TP... 35 1.4
>emb|CAA63223.1| TOM20 [Solanum tuberosum] gi|7459240|pir||T07679 protein import
receptor TOM20, mitochondrial - potato
gi|13631844|sp|P92792|TOM20_SOLTU Mitochondrial import
receptor subunit TOM20 (Translocase of outer membrane 20
kDa subunit)
Length = 204
Score = 111 bits (277), Expect = 1e-23
Identities = 69/165 (41%), Positives = 94/165 (56%), Gaps = 10/165 (6%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y +NP DA+NLTRWG ALL LSQ +S I + SKLEEA ++NP D W
Sbjct: 20 AETTYAQNPLDADNLTRWGGALLELSQFQPVAESKQMISDATSKLEEALTVNPEKHDALW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
LG A T LTPD +AK++F+ A F++AF DPS+ Y+ SLE+ K E H +
Sbjct: 80 CLGNAHTSHVFLTPDMDEAKVYFEKATQCFQQAFDADPSNDLYRKSLEV-TAKAPELHME 138
Query: 141 IVNHGLGQQS----KGSSSATKVIK-----HYKFLVFLFTLLVIG 176
I HG QQ+ +S++TK K K+ +F + +L +G
Sbjct: 139 IHRHGPMQQTMAAEPSTSTSTKSSKKTKSSDLKYDIFGWVILAVG 183
>emb|CAC14429.1| TOM20-2 protein [Arabidopsis thaliana]
gi|13631839|sp|P82873|TO202_ARATH Mitochondrial import
receptor subunit TOM20-2 (Translocase of outer membrane
20 kDa subunit 2)
Length = 210
Score = 107 bits (268), Expect = 1e-22
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERFIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>ref|NP_174059.2| mitochondrial import receptor subunit TOM20-2 (TOM20-2)
[Arabidopsis thaliana]
Length = 210
Score = 107 bits (266), Expect = 2e-22
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERLIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>gb|AAF99745.1| F17L21.18 [Arabidopsis thaliana]
Length = 209
Score = 107 bits (266), Expect = 2e-22
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERLIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>emb|CAC14430.1| TOM20-3 protein [Arabidopsis thaliana] gi|19310823|gb|AAL85142.1|
putative TOM20 protein [Arabidopsis thaliana]
gi|14532806|gb|AAK64184.1| putative TOM20 protein
[Arabidopsis thaliana] gi|9279631|dbj|BAB01089.1|
TOM20-like protein [Arabidopsis thaliana]
gi|13631842|sp|P82874|TO203_ARATH Mitochondrial import
receptor subunit TOM20-3 (Translocase of outer membrane
20 kDa subunit 3) gi|15232079|ref|NP_189344.1|
mitochondrial import receptor subunit TOM20-3 /
translocase of outer membrane 20 kDa subunit 3 (TOM20-3)
[Arabidopsis thaliana]
Length = 202
Score = 106 bits (265), Expect = 3e-22
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>gb|AAM64598.1| putative TOM20 [Arabidopsis thaliana]
Length = 202
Score = 106 bits (265), Expect = 3e-22
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>emb|CAC17150.1| TOM20-1 protein [Arabidopsis thaliana] gi|9279630|dbj|BAB01088.1|
TOM20-like protein [Arabidopsis thaliana]
gi|13631831|sp|P82872|TO201_ARATH Mitochondrial import
receptor subunit TOM20-1 (Translocase of outer membrane
20 kDa subunit 1) gi|15232078|ref|NP_189343.1|
mitochondrial import receptor subunit TOM20-1 /
translocase of outer membrane 20 kDa subunit 1 (TOM20-1)
[Arabidopsis thaliana]
Length = 188
Score = 100 bits (250), Expect = 2e-20
Identities = 65/164 (39%), Positives = 87/164 (52%), Gaps = 8/164 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE+ Y NP DA+NL RWG ALL LSQ + DSL IQ +ISKLE+A ++P D W
Sbjct: 14 AEETYKLNPEDADNLMRWGEALLELSQFQNVIDSLKMIQDAISKLEDAILIDPMKHDAVW 73
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQ--- 137
LG A T A LTPD A+L+F A ++F A Q P + Y SLE+ D
Sbjct: 74 CLGNAYTSYARLTPDDTQARLNFGLAYLFFGIAVAQQPDNQVYHKSLEMADKAPQLHTGF 133
Query: 138 HPKIVNHGLGQQSKGSSSATKVIKH-----YKFLVFLFTLLVIG 176
H + LG + + KV+K+ K++V + +L IG
Sbjct: 134 HKNRLLSLLGGVETLAIPSPKVVKNKKSSDEKYIVMGWVILAIG 177
>dbj|BAD88373.1| putative TOM20 [Oryza sativa (japonica cultivar-group)]
Length = 202
Score = 100 bits (249), Expect = 2e-20
Identities = 62/157 (39%), Positives = 86/157 (54%), Gaps = 5/157 (3%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M P R F F L C A+ Y +NPHDA+NLTRWG ALL LSQ + P+SL ++
Sbjct: 6 MSDPERMFFFDLA----CQNAKVTYEQNPHDADNLTRWGGALLELSQMRNGPESLKCLED 61
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ SKLEEA ++P D W LG A T T D+ A F+ A F++A +P++
Sbjct: 62 AESKLEEALKIDPMKADALWCLGNAQTSHGFFTSDTVKANEFFEKATQCFQKAVDVEPAN 121
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKGSSSAT 157
Y+ SL+L +K E H +I Q S+ +SS +
Sbjct: 122 DLYRKSLDL-SSKAPELHMEIHRQMASQASQAASSTS 157
>dbj|BAB86179.1| putative mitochondrial import receptor subunit TOM20 (translocase
of outer membrane 20 KDA subunit) [Oryza sativa
(japonica cultivar-group)]
Length = 237
Score = 85.5 bits (210), Expect = 7e-16
Identities = 49/113 (43%), Positives = 63/113 (55%), Gaps = 4/113 (3%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M P R F F L C A+ Y +NPHDA+NLTRWG ALL LSQ + P+SL ++
Sbjct: 52 MSDPERMFFFDLA----CQNAKVTYEQNPHDADNLTRWGGALLELSQMRNGPESLKCLED 107
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRA 113
+ SKLEEA ++P D W LG A T T D+ A F+ A F++A
Sbjct: 108 AESKLEEALKIDPMKADALWCLGNAQTSHGFFTSDTVKANEFFEKATQCFQKA 160
>gb|AAM98262.1| At5g40930/MMG1_2 [Arabidopsis thaliana] gi|10177431|dbj|BAB10523.1|
protein import receptor TOM20, mitochondrial-like
[Arabidopsis thaliana] gi|26450710|dbj|BAC42464.1|
protein import receptor TOM20, mitochondrial-like
[Arabidopsis thaliana] gi|18087635|gb|AAL58947.1|
AT5g40930/MMG1_2 [Arabidopsis thaliana]
gi|15237520|ref|NP_198909.1| mitochondrial import
receptor subunit TOM20-4 / translocase of outer membrane
20 kDa subunit 4 [Arabidopsis thaliana]
gi|13631824|sp|P82805|TO204_ARATH Mitochondrial import
receptor subunit TOM20-4 (Translocase of outer membrane
20 kDa subunit 4)
Length = 187
Score = 80.5 bits (197), Expect = 2e-14
Identities = 56/161 (34%), Positives = 80/161 (48%), Gaps = 9/161 (5%)
Query: 15 KHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPN 74
+H +AE Y KNP DAENLTRW ALL LSQ + P + I +I KL EA ++P
Sbjct: 14 EHARKVAEATYVKNPLDAENLTRWAGALLELSQFQTEPKQM--ILEAILKLGEALVIDPK 71
Query: 75 NPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKD 134
D WL+G A L+ D +A +F+ A +F+ A + P Y+ SL L
Sbjct: 72 KHDALWLIGNAHLSFGFLSSDQTEASDNFEKASQFFQLAVEEQPESELYRKSLTLA---- 127
Query: 135 HEQHPKIVNHGLGQQSKGSSSATKVIK--HYKFLVFLFTLL 173
+ P++ G S S+ K K +K+ VF + +L
Sbjct: 128 -SKAPELHTGGTAGPSSNSAKTMKQKKTSEFKYDVFGWVIL 167
>ref|NP_632202.1| hypothetical protein MM0178 [Methanosarcina mazei Go1]
gi|20904523|gb|AAM29874.1| conserved protein
[Methanosarcina mazei Goe1]
Length = 1711
Score = 40.0 bits (92), Expect = 0.034
Identities = 24/75 (32%), Positives = 41/75 (54%), Gaps = 14/75 (18%)
Query: 17 ECILAEQRYNK----NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLN 72
EC AE+ + + NP D ++L G++LL+L + + ++ LE+ SL
Sbjct: 453 ECEKAEEAFAEVLKINPEDIDSLYNRGISLLKLGRK----------ETALEYLEKVVSLR 502
Query: 73 PNNPDVHWLLGMALT 87
P+ PD+ + LG+ALT
Sbjct: 503 PDYPDLSYSLGVALT 517
>ref|ZP_00056252.2| COG4235: Cytochrome c biogenesis factor [Magnetospirillum
magnetotacticum MS-1]
Length = 275
Score = 36.6 bits (83), Expect = 0.37
Identities = 25/88 (28%), Positives = 42/88 (47%), Gaps = 14/88 (15%)
Query: 21 AEQRYNKN----PHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNP 76
A++ Y K P D + + LL ++ S PD +IS+LEE +L+P+ P
Sbjct: 173 AKEAYRKAVTLLPADTQTRLELAIMLLEEAEGDSLPDE------AISRLEEVLALSPDQP 226
Query: 77 DVHWLLGMALTMQALLTPDSHDAKLHFD 104
D + G++ M+ DS AK ++
Sbjct: 227 DALYFTGLSAAMKG----DSARAKARWN 250
>ref|XP_548165.1| PREDICTED: similar to Oxysterol binding protein-related protein 7
(OSBP-related protein 7) (ORP-7) [Canis familiaris]
Length = 1257
Score = 36.2 bits (82), Expect = 0.49
Identities = 37/137 (27%), Positives = 59/137 (43%), Gaps = 22/137 (16%)
Query: 19 ILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAF-SLNPNNPD 77
+ AE K+ H L+R GV S+A P LH++ VS ++F SLNP +
Sbjct: 656 LTAEWDRLKDLHQGSELSRTGV-----SEASGGPRRLHSLSVSSDTTADSFSSLNPEEQE 710
Query: 78 VHWLLGMALTMQ-----ALLTPDSHDAKLHFDSADVYFKRAFRQD---------PSDPTY 123
++ G LT Q L DSH FD+ +V + ++ S+ T
Sbjct: 711 ALYMKGRELTPQLSQSSILSLADSHTE--FFDACEVLLSASSSENEGSEEEESCTSEITT 768
Query: 124 QISLELPDTKDHEQHPK 140
+S E+ D + + + K
Sbjct: 769 SLSEEVLDLRGADHYQK 785
>gb|AAH97384.1| Interferon-induced protein with tetratricopeptide repeats 2 [Rattus
norvegicus] gi|67078506|ref|NP_001019924.1|
interferon-induced protein with tetratricopeptide
repeats 2 [Rattus norvegicus]
Length = 464
Score = 36.2 bits (82), Expect = 0.49
Identities = 28/98 (28%), Positives = 44/98 (44%), Gaps = 19/98 (19%)
Query: 22 EQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWL 81
E+ K+P + E+ + W +A RL + D I LE+A SL+P+N V L
Sbjct: 162 EKALEKDPKNPESTSGWAIANYRLDDWPASNDY-------IDSLEQAISLSPDNTYVKVL 214
Query: 82 LGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPS 119
L M L ++H + A + A ++DPS
Sbjct: 215 LAMKL------------EEVHENRAKELVEEALKKDPS 240
>ref|NP_213582.1| hypothetical protein aq_854 [Aquifex aeolicus VF5]
gi|2983399|gb|AAC06984.1| hypothetical protein [Aquifex
aeolicus VF5] gi|7459517|pir||B70374 conserved
hypothetical protein aq_854 - Aquifex aeolicus
Length = 545
Score = 35.8 bits (81), Expect = 0.64
Identities = 21/71 (29%), Positives = 34/71 (47%), Gaps = 8/71 (11%)
Query: 53 DSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKR 112
D+L I+ + L +A L+P NPD + LG +L + K + A+ K+
Sbjct: 400 DNLGDIKNAEKALRKAIELDPENPDYYNYLGYSLLLWY--------GKERVEEAEELIKK 451
Query: 113 AFRQDPSDPTY 123
A +DP +P Y
Sbjct: 452 ALEKDPENPAY 462
>emb|CAA18877.1| SPBC23E6.09 [Schizosaccharomyces pombe]
gi|19113401|ref|NP_596609.1| hypothetical protein
SPBC23E6.09 [Schizosaccharomyces pombe 972h-]
gi|31340484|sp|O60184|YG49_SCHPO Protein C23E6.09 in
chromosome II gi|7491517|pir||T39943 hypothetical
protein SPBC23E6.09 - fission yeast
(Schizosaccharomyces pombe)
Length = 1102
Score = 35.4 bits (80), Expect = 0.83
Identities = 37/134 (27%), Positives = 55/134 (40%), Gaps = 26/134 (19%)
Query: 18 CILAEQRYNKNPH--------DAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAF 69
C +A+Q+YNK D N T W + Q + + D+L A
Sbjct: 593 CYVAQQKYNKAYEAYQQAVYRDGRNPTFWCSIGVLYYQINQYQDALDAYS-------RAI 645
Query: 70 SLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLEL 129
LNP +V + LG D+ DA ++RA DP++P + L+L
Sbjct: 646 RLNPYISEVWYDLGTLYESCHNQISDALDA----------YQRAAELDPTNPHIKARLQL 695
Query: 130 PDTKDHEQHPKIVN 143
++EQH KIVN
Sbjct: 696 LRGPNNEQH-KIVN 708
>gb|AAM05026.1| TPR-domain containing protein [Methanosarcina acetivorans str. C2A]
gi|20090471|ref|NP_616546.1| TPR-domain containing
protein [Methanosarcina acetivorans C2A]
Length = 1885
Score = 35.4 bits (80), Expect = 0.83
Identities = 20/59 (33%), Positives = 33/59 (55%), Gaps = 10/59 (16%)
Query: 28 NPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMAL 86
NP D + L G++LLRL + + ++ LE+ SL+P+ PD+ + LG+AL
Sbjct: 579 NPEDLDALYNRGISLLRLGRN----------ETALEYLEKVVSLSPDYPDLAYSLGVAL 627
>emb|CAG05234.1| unnamed protein product [Tetraodon nigroviridis]
Length = 337
Score = 35.0 bits (79), Expect = 1.1
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 12/82 (14%)
Query: 66 EEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQI 125
E A ++P + +G+ALT A + A YFK+A DP + TY+
Sbjct: 138 ERAIGIDPTYSKAYGRMGLALT-----------AMSKYPEAISYFKKALVLDPENDTYKS 186
Query: 126 SLELPDTKDHEQHPKIVNHGLG 147
+L++ + K H++ + GLG
Sbjct: 187 NLKIAEQK-HKEATSPIAAGLG 207
>gb|EAL86151.1| oxidoreductase, short chain dehydrogenase/reductase family family
[Aspergillus fumigatus Af293]
Length = 588
Score = 34.7 bits (78), Expect = 1.4
Identities = 27/91 (29%), Positives = 38/91 (41%)
Query: 43 LRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLH 102
LR S S L T+ + ISK A SL+ N + L QALL + ++
Sbjct: 484 LRKSSKQSDQILLKTVSIKISKDNAAHSLHSNLVEEAKKLRTPFDRQALLGMNCGQSEER 543
Query: 103 FDSADVYFKRAFRQDPSDPTYQISLELPDTK 133
S ++ DPS+P I + L TK
Sbjct: 544 ISSPQLFRHLFMEMDPSEPVVLIEVALTTTK 574
>gb|AAH67176.1| Small glutamine-rich tetratricopeptide repeat (TPR)-containing
[Danio rerio] gi|47086521|ref|NP_997929.1| small
glutamine-rich tetratricopeptide repeat (TPR)-containing
[Danio rerio]
Length = 320
Score = 34.7 bits (78), Expect = 1.4
Identities = 23/88 (26%), Positives = 41/88 (46%), Gaps = 14/88 (15%)
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLH-FDSADVYFKRAFRQDPS 119
++ E A ++ N + +G+AL A L+ + A Y+K+A DP
Sbjct: 144 AVQDCERAIGIDANYSKAYGRMGLAL------------ASLNKYSEAVSYYKKALELDPD 191
Query: 120 DPTYQISLELPDTKDHEQHPKIVNHGLG 147
+ TY+++L++ + K E P GLG
Sbjct: 192 NDTYKVNLQVAEQKVKETQPSTAG-GLG 218
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 331,313,161
Number of Sequences: 2540612
Number of extensions: 12523847
Number of successful extensions: 28433
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 28396
Number of HSP's gapped (non-prelim): 63
length of query: 190
length of database: 863,360,394
effective HSP length: 121
effective length of query: 69
effective length of database: 555,946,342
effective search space: 38360297598
effective search space used: 38360297598
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC145109.3


