

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.9 + phase: 0
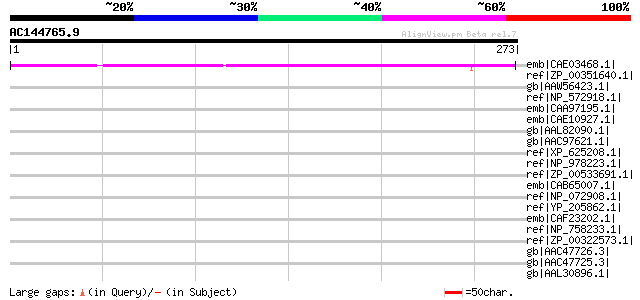
(273 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAE03468.1| OSJNBa0083N12.5 [Oryza sativa (japonica cultivar... 165 1e-39
ref|ZP_00351640.1| COG0457: FOG: TPR repeat [Anabaena variabilis... 39 0.12
gb|AAW56423.1| polyprotein [Bean common mosaic virus] 37 0.60
ref|NP_572918.1| CG10990-PA [Drosophila melanogaster] gi|2283221... 36 1.0
emb|CAA97195.1| unnamed protein product [Saccharomyces cerevisia... 35 1.7
emb|CAE10927.1| hypothetical protein [Wolinella succinogenes] gi... 35 1.7
gb|AAL82090.1| hypothetical protein [Pyrococcus furiosus DSM 363... 35 1.7
gb|AAC97621.1| ORF MSV061 putative LINE reverse transcriptase, s... 35 2.3
ref|XP_625208.1| PREDICTED: similar to ENSANGP00000014748 [Apis ... 34 3.9
ref|NP_978223.1| tagatose-6-phosphate kinase [Bacillus cereus AT... 34 5.0
ref|ZP_00533691.1| hypothetical protein Cphamn1DRAFT_0696 [Chlor... 33 6.6
emb|CAB65007.1| transmembrane protein [Erysiphe pisi] 33 6.6
ref|NP_072908.1| hypothetical protein MG242 [Mycoplasma genitali... 33 6.6
ref|YP_205862.1| hypothetical protein VF2479 [Vibrio fischeri ES... 33 6.6
emb|CAF23202.1| probable DNA-directed DNA polymerase III subunit... 33 8.6
ref|NP_758233.1| hypothetical protein MYPE8450 [Mycoplasma penet... 33 8.6
ref|ZP_00322573.1| COG0542: ATPases with chaperone activity, ATP... 33 8.6
gb|AAC47726.3| myosin-C [Toxoplasma gondii] 33 8.6
gb|AAC47725.3| myosin-B [Toxoplasma gondii] 33 8.6
gb|AAL30896.1| myosin B [Toxoplasma gondii] 33 8.6
>emb|CAE03468.1| OSJNBa0083N12.5 [Oryza sativa (japonica cultivar-group)]
gi|50928447|ref|XP_473751.1| OSJNBa0083N12.5 [Oryza
sativa (japonica cultivar-group)]
Length = 2646
Score = 165 bits (418), Expect = 1e-39
Identities = 102/276 (36%), Positives = 156/276 (55%), Gaps = 8/276 (2%)
Query: 1 MAAQVYARGSFFSDCLNVCAKGGLLDTGSHYIQCWKQNERADPGWANSHDLYAIEQKFME 60
+AA Y R ++ CL++C+KG L G +Q +++ + + AI F+E
Sbjct: 1719 LAADAYFRAKCYAKCLSMCSKGKLFQKGLLLLQQLEEHLLENSSLVK---VAAIRNTFLE 1775
Query: 61 NCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAK 120
+CAL+YF+ D + MM FV++F S+D R FL S NL DELL +E + GNF+EAA IAK
Sbjct: 1776 DCALHYFECGDIKHMMPFVKSFSSMDHIRVFLNSKNLVDELLSVEMDMGNFVEAA-GIAK 1834
Query: 121 TMGDILREADLLGKAGEFLDAYELVFFYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTF 180
G+IL EADLL KAG +A +L+ +F SLW+ S WP K+F +K LL KA
Sbjct: 1835 HTGNILLEADLLEKAGFLENATQLILLQLFVNSLWASHSTGWPPKRFAEKEQLLAKAKEM 1894
Query: 181 AKEVSSSFYELASTKVE-LSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRLN 239
++ VS SFY L ++ + LS++H ++ I L ++ E++ +LD H +
Sbjct: 1895 SRNVSESFYCLVCSEADALSDEHKSLASITYNLIEGNKCGNLLVELIASRLILDVHLQAE 1954
Query: 240 SSKYVWQD---SMFDVSVEGMIMKNQLSVETLFCCW 272
+S Y ++ S + M++ NQ+S+ETL W
Sbjct: 1955 ASGYCFESEPGSHDGRHCKDMLVLNQISLETLVYDW 1990
>ref|ZP_00351640.1| COG0457: FOG: TPR repeat [Anabaena variabilis ATCC 29413]
Length = 689
Score = 39.3 bits (90), Expect = 0.12
Identities = 33/116 (28%), Positives = 54/116 (46%), Gaps = 11/116 (9%)
Query: 24 LLDTGSHYIQCWKQNERADPGWANSHDLYA------IEQKFMENCALNYFDKNDYRSMMK 77
L + G+ Y+ C KQ + A + S +++ E K + N L Y Y+ +K
Sbjct: 496 LANLGNIYL-CLKQYQAAIEFYKKSWEIFRKIGDRDAESKSLGNLGLVYLSLGQYQRAIK 554
Query: 78 FVRAFHSI----DLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREA 129
F + I ++K FL +L L LE + + F + ++ IAK +GDI EA
Sbjct: 555 FFQQSLVISNDRNVKCSFLSNLGLAHYYLEQYQRAIEFYQQSLEIAKEIGDIRGEA 610
>gb|AAW56423.1| polyprotein [Bean common mosaic virus]
Length = 3201
Score = 37.0 bits (84), Expect = 0.60
Identities = 36/153 (23%), Positives = 62/153 (39%), Gaps = 28/153 (18%)
Query: 97 LPDELLELEEESGNFMEAAVNIAKTMGDILREADLLGKAGE------------FLDAYEL 144
L D+L + +G+ ++ +TM + +DLL E + D Y+L
Sbjct: 967 LVDQLETINNSAGHLLDILKECPRTMHSYVPASDLLTLYLERQVSNCQLLSNGYTDIYDL 1026
Query: 145 VF-----FYVF-------AKSLWSGGSKAWPLKQFT--QKAGLLGKALTFAKEVSSSFYE 190
++ Y+ A SLW S W LKQF+ + GL+ + +E S+SF
Sbjct: 1027 LYVKMEKIYISRLEQEWRALSLWKKSSLTWHLKQFSIATEKGLIKRVTEGRREFSASFVS 1086
Query: 191 --LASTKVELSNKHDNIFEIVNQLKSSRIHSSI 221
+ K L ++ D I +L S + +
Sbjct: 1087 ECFTTAKSHLGSRKDMILRACEKLGHSIVRKCV 1119
>ref|NP_572918.1| CG10990-PA [Drosophila melanogaster] gi|22832218|gb|AAF48312.2|
CG10990-PA [Drosophila melanogaster]
Length = 509
Score = 36.2 bits (82), Expect = 1.0
Identities = 41/139 (29%), Positives = 69/139 (49%), Gaps = 17/139 (12%)
Query: 86 DLKRGFLQSL-NLPDELLELEEES---GNFMEAAVN--------IAKTMGDILREADLLG 133
D+++GF L NLPD +L+ E GNFM AV +AK+ G+ LR L
Sbjct: 268 DIEKGFNMLLANLPDLVLDTPEAPIMLGNFMARAVADDCIPPKFVAKS-GEELRHLGLGE 326
Query: 134 KAGEFLD-AYELVFFYVFAK--SLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYE 190
A + L A L++ +V+A ++W G P+K T + LL K +++V+ +
Sbjct: 327 HAEQALRRADSLIYKHVWAHLDNVWGMGGPLRPVKTITMQMELLLKEYLSSRDVAEAQRC 386
Query: 191 LASTKVELSNKHDNIFEIV 209
L + +V H+ ++E +
Sbjct: 387 LRALEVP-HYHHELVYEAI 404
>emb|CAA97195.1| unnamed protein product [Saccharomyces cerevisiae]
gi|6321608|ref|NP_011685.1| Pseudouridine synthase
responsible for modification of cytoplasmic and
mitochondrial tRNAs at position 31; mutation of Asp168
to Ala abolishes enzyme activity; not essential for
viability; Pus6p [Saccharomyces cerevisiae]
gi|1723726|sp|P53294|YG3X_YEAST Hypothetical 46.8 kDa
protein in CLC1-PDS2 intergenic region
Length = 404
Score = 35.4 bits (80), Expect = 1.7
Identities = 26/74 (35%), Positives = 34/74 (45%), Gaps = 11/74 (14%)
Query: 43 PGWANSHDLYAIE-----QKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNL 97
P W N DL A E QKF++ KN+ R+M F H++DLK L L L
Sbjct: 322 PRWENQQDLDAEELKVRFQKFVDET------KNNCRTMETFCPECHTVDLKDPVLSDLEL 375
Query: 98 PDELLELEEESGNF 111
+ EE +G F
Sbjct: 376 WLHAWKYEEINGKF 389
>emb|CAE10927.1| hypothetical protein [Wolinella succinogenes]
gi|34558212|ref|NP_908027.1| hypothetical protein WS1918
[Wolinella succinogenes DSM 1740]
Length = 480
Score = 35.4 bits (80), Expect = 1.7
Identities = 40/194 (20%), Positives = 80/194 (40%), Gaps = 49/194 (25%)
Query: 67 FDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLPD------------ELLELEEESG----N 110
+DK R +++ I+ +GF++S L + +LL +E++ +
Sbjct: 165 YDKVFLRQQGNYLKLEEKIEATQGFIRSATLKERPLADRLKEKEKKLLAIEDKKSQAYFD 224
Query: 111 FMEAAVNIAKTMGDILR----EADLLGKAG------------EFLDAYELVFFYVFAKSL 154
F + + K D+L + DLL + EF+D Y+ + + K L
Sbjct: 225 FEKEVKELRKRYVDLLHFISLQRDLLAELSALSKMFVERYFQEFVDLYQPLSMELERKLL 284
Query: 155 WSGGSKAWPL--------------KQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSN 200
W +KA+ L +QF +G+ G T++ + +Y + K +L N
Sbjct: 285 WLLNTKAYELDSNLWDRAKQSKLIRQFFMDSGITG---TYSSKTFLKYYLRSLDKDKLRN 341
Query: 201 KHDNIFEIVNQLKS 214
++ +F ++ L+S
Sbjct: 342 ENKELFSLLAYLES 355
>gb|AAL82090.1| hypothetical protein [Pyrococcus furiosus DSM 3638]
gi|18978338|ref|NP_579695.1| hypothetical protein PF1966
[Pyrococcus furiosus DSM 3638]
Length = 334
Score = 35.4 bits (80), Expect = 1.7
Identities = 40/160 (25%), Positives = 66/160 (41%), Gaps = 28/160 (17%)
Query: 93 QSLNLPDELLELEEESGNFMEAAVNIAK--TMGDILREAD----LLGKAGEFLDAYELVF 146
++L + ELLE+E+E GN E A+ +A + D L EAD L+ +A + +
Sbjct: 108 EALKIYQELLEIEKELGNEREIALTLANIAVVKDELGEADEAIKLMSEAKDIFEKINDTK 167
Query: 147 FYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKHDNIF 206
Y A L F + G KA+ +EV L N D+
Sbjct: 168 NYHIA---------LIDLAHFYYELGEYEKAMQIIEEV-------------LRNPLDDEI 205
Query: 207 EIVNQLKSSRIHSSIRGEILCLWELLDSHFRLNSSKYVWQ 246
E+ +L S IHS++ L ++ R +Y+++
Sbjct: 206 EVHGRLVESEIHSALENYKKAALSLRNALLRSGDDEYLFE 245
>gb|AAC97621.1| ORF MSV061 putative LINE reverse transcriptase, similar to
Caenorhabditis elegans GB:U00063 [Melanoplus sanguinipes
entomopoxvirus] gi|11361248|pir||T28222 hypothetical
protein ORF61 - Melanoplus sanguinipes entomopoxvirus
gi|9631308|ref|NP_048132.1| ORF MSV061 putative LINE
reverse transcriptase, similar to Caenorhabditis elegans
GB:U00063 [Melanoplus sanguinipes entomopoxvirus]
Length = 531
Score = 35.0 bits (79), Expect = 2.3
Identities = 19/73 (26%), Positives = 40/73 (54%), Gaps = 5/73 (6%)
Query: 188 FYELASTKVELSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRLNSSKYVWQD 247
FYEL+S + + +H+ F + + + I +IL +++L+D ++N SKY W++
Sbjct: 459 FYELSSEEENIITRHEMHF--LKNILDFNLEDDIIRQILDVYKLVD---KINESKYKWKE 513
Query: 248 SMFDVSVEGMIMK 260
+ + + + MK
Sbjct: 514 RLSSMDYDRLPMK 526
>ref|XP_625208.1| PREDICTED: similar to ENSANGP00000014748 [Apis mellifera]
Length = 292
Score = 34.3 bits (77), Expect = 3.9
Identities = 23/75 (30%), Positives = 33/75 (43%), Gaps = 3/75 (4%)
Query: 150 FAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKHD---NIF 206
F SL+ G SK + Q+A + K S+FYE A + N+HD N
Sbjct: 25 FFGSLFGGSSKFEEAVECYQRAANMFKMAKNWSSAGSAFYEAAELHAKAGNRHDAATNYV 84
Query: 207 EIVNQLKSSRIHSSI 221
+ N K S I+ +I
Sbjct: 85 DAANCFKKSNINEAI 99
>ref|NP_978223.1| tagatose-6-phosphate kinase [Bacillus cereus ATCC 10987]
gi|42736897|gb|AAS40831.1| tagatose-6-phosphate kinase
[Bacillus cereus ATCC 10987]
Length = 307
Score = 33.9 bits (76), Expect = 5.0
Identities = 30/134 (22%), Positives = 59/134 (43%), Gaps = 7/134 (5%)
Query: 89 RGFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREADLLG--KAGEFLDAYELVF 146
RG L++L++ DE + ++EE+ N + + + ++L + + F+ YE +
Sbjct: 68 RGKLENLSIVDEFVSIKEETRNCIALITKKSASATEVLEAGPTISEDEIALFIKKYEKIV 127
Query: 147 ----FYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKH 202
F V + SL G SK++ KQ QKA GK F+ + +
Sbjct: 128 KDFEFIVASGSLPKGISKSF-YKQLAQKAAENGKKFILDTSGEPLFHGIQGKPFLIKPNR 186
Query: 203 DNIFEIVNQLKSSR 216
I +++ + + S+
Sbjct: 187 QEICQLLQKKRVSQ 200
>ref|ZP_00533691.1| hypothetical protein Cphamn1DRAFT_0696 [Chlorobium phaeobacteroides
BS1] gi|67912179|gb|EAM61945.1| hypothetical protein
Cphamn1DRAFT_0696 [Chlorobium phaeobacteroides BS1]
Length = 351
Score = 33.5 bits (75), Expect = 6.6
Identities = 29/92 (31%), Positives = 50/92 (53%), Gaps = 19/92 (20%)
Query: 173 LLGKALTFAKE--VSSSFYELASTKVELSNKHDNI-----------FEIVNQLKSSRIHS 219
L G A TF KE VS SF AST++E++NK+ +I F+I Q+KS++
Sbjct: 4 LSGIAQTFEKERSVSRSFKTTASTEIEVTNKYGDIHLIPWDQDSVRFDIRLQVKSTK--- 60
Query: 220 SIRGEILCLWELLDSHFRLNSSKYVWQDSMFD 251
+ ++ ++ +D F+ N + YV ++F+
Sbjct: 61 --QSKLDKTFDFIDFDFKAN-NYYVIAQTVFE 89
>emb|CAB65007.1| transmembrane protein [Erysiphe pisi]
Length = 1475
Score = 33.5 bits (75), Expect = 6.6
Identities = 22/71 (30%), Positives = 33/71 (45%), Gaps = 2/71 (2%)
Query: 39 ERADPGWANSHDLYAIEQKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGFLQSLNLP 98
E A+P W + LY I F +N +FD ND+R + A S+D + + N
Sbjct: 492 ENANPKW--NETLYLIVTTFTDNLTFQFFDYNDFRKDKEIGTATLSLDTIEEYPELENEQ 549
Query: 99 DELLELEEESG 109
E+L + SG
Sbjct: 550 LEVLMNGKSSG 560
>ref|NP_072908.1| hypothetical protein MG242 [Mycoplasma genitalium G-37]
gi|3844846|gb|AAC71463.1| conserved hypothetical protein
[Mycoplasma genitalium G-37] gi|1361643|pir||G64226
hypothetical protein MG242 - Mycoplasma genitalium
gi|1351523|sp|P47484|Y242_MYCGE Hypothetical protein
MG242
Length = 630
Score = 33.5 bits (75), Expect = 6.6
Identities = 23/81 (28%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 36 KQNERADPGWANSHDLYA----IEQKFMENCALNYFDKNDYRSMMKFVRAFHSIDLKRGF 91
K R DP DLY IEQ +++ ++K + + + + + I+L+
Sbjct: 291 KNTLREDPDQNPETDLYELIQFIEQNYLKKDKKTSWNKKKVQDLEQLLEEINKINLETKN 350
Query: 92 LQSLNLPDELLELEEESGNFM 112
+SL PDE+ ELE ++ NF+
Sbjct: 351 -ESLAYPDEITELEIDNDNFV 370
>ref|YP_205862.1| hypothetical protein VF2479 [Vibrio fischeri ES114]
gi|59481187|gb|AAW86974.1| hypothetical exported protein
[Vibrio fischeri ES114]
Length = 652
Score = 33.5 bits (75), Expect = 6.6
Identities = 28/120 (23%), Positives = 58/120 (48%), Gaps = 16/120 (13%)
Query: 159 SKAWPLKQFTQK--------AGLLGKALTFAKEVSSSFYELASTKVELSNKHDNIFEIV- 209
+K W + QFT + A ++GK L + +S + +++L N + + +IV
Sbjct: 213 NKLWVIPQFTLRRFNLESDNADIIGKPLQW---LSEQHFTYQFKRIDLLNINLELPQIVT 269
Query: 210 NQLKSSRIHSSIRGEILCLWELLDSHFRLNSSKYVWQDSMFDVSVEGMIM-KNQLSVETL 268
N L S + + + LWE +++ LN+ VW ++M + + + + K + ++E L
Sbjct: 270 NDLSLSAENILLPYD---LWEQTNANISLNADSIVWNETMIEKPIMDVTLNKKEANIENL 326
>emb|CAF23202.1| probable DNA-directed DNA polymerase III subunits gamma/tau dnaX
[Parachlamydia sp. UWE25] gi|46446112|ref|YP_007477.1|
probable DNA-directed DNA polymerase III subunits
gamma/tau dnaX [Parachlamydia sp. UWE25]
Length = 522
Score = 33.1 bits (74), Expect = 8.6
Identities = 42/161 (26%), Positives = 70/161 (43%), Gaps = 20/161 (12%)
Query: 90 GFLQSLNLPDELLELEEESGNFMEAAVNIAKTMGDILREA----DLLGKAGEFLDAYELV 145
G + +L D+++ E N A + +G + REA DL GK G F+ A+E+
Sbjct: 215 GLRDAESLFDQIVAFSEGKIN----AATVQSILGLMPREAYFELDLAGKEGNFVKAFEIA 270
Query: 146 FFYVFAKSLWSGGSKAWPLKQF-----TQKAGLLGKALTFAKEVSSSFYELAS---TKVE 197
+ +FA+ + F Q AG+ LT + + FYE A+ T+ +
Sbjct: 271 -YRIFAEGKDLNHFIEGLIDHFRHILLIQLAGITSPLLTISPK-EKQFYERAAHLYTQEQ 328
Query: 198 LSNKHDNIFEIVNQLKSSRIHSSIRGEILCLWELLDSHFRL 238
+ D + E Q++ + HS L ++ SHFRL
Sbjct: 329 CLDLIDYLIESAQQIRFT--HSGKIALEAILLHVIRSHFRL 367
>ref|NP_758233.1| hypothetical protein MYPE8450 [Mycoplasma penetrans HF-2]
gi|26454308|dbj|BAC44637.1| conserved hypothetical
protein [Mycoplasma penetrans HF-2]
Length = 1084
Score = 33.1 bits (74), Expect = 8.6
Identities = 20/72 (27%), Positives = 38/72 (52%), Gaps = 1/72 (1%)
Query: 143 ELVFFYVFAKSLWSGGSKAWPLKQFTQKAGLLGKALTFAKEVSSSFYELASTKVELSNKH 202
+L +Y+ A S + + + KQ+ K + A +F+ E S + Y +S+ ++L+N
Sbjct: 266 QLFAYYIDASSYSNTNNTNYKYKQYLSKVSISSNAFSFSGEKSLNDYVSSSSTLDLNNFK 325
Query: 203 DNIFEIV-NQLK 213
+ F +V N LK
Sbjct: 326 NIAFPVVINNLK 337
>ref|ZP_00322573.1| COG0542: ATPases with chaperone activity, ATP-binding subunit
[Pediococcus pentosaceus ATCC 25745]
Length = 739
Score = 33.1 bits (74), Expect = 8.6
Identities = 19/67 (28%), Positives = 37/67 (54%), Gaps = 2/67 (2%)
Query: 84 SIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIA--KTMGDILREADLLGKAGEFLDA 141
++ L + ++Q LPD+ ++L +E+G+ +NIA +T+ + EA+ KA +
Sbjct: 320 AVKLSKRYIQDRFLPDKAIDLLDEAGSKKNLTINIADPETIQKKIDEAEAQKKAALQSED 379
Query: 142 YELVFFY 148
YE +Y
Sbjct: 380 YEKAAYY 386
>gb|AAC47726.3| myosin-C [Toxoplasma gondii]
Length = 1101
Score = 33.1 bits (74), Expect = 8.6
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query: 22 GGLLDTGSHYIQCWKQNERADP-GWANSHDLYAI-EQKFMENCALNYFDKNDYRSMMKFV 79
G + T +H+I+C K NE P GW S L + +E L R+ +F
Sbjct: 594 GIVAQTEAHFIRCLKPNEEKKPLGWNGSKVLNQLFSLSILEALQLRQVGYAYRRNFSEFC 653
Query: 80 RAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTM 122
F +DL G + S E+ +L E E++ I KTM
Sbjct: 654 SHFRWLDL--GLVNSDRDRKEVAQLLLEQSGIPESSWVIGKTM 694
>gb|AAC47725.3| myosin-B [Toxoplasma gondii]
Length = 1004
Score = 33.1 bits (74), Expect = 8.6
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query: 22 GGLLDTGSHYIQCWKQNERADP-GWANSHDLYAI-EQKFMENCALNYFDKNDYRSMMKFV 79
G + T +H+I+C K NE P GW S L + +E L R+ +F
Sbjct: 594 GIVAQTEAHFIRCLKPNEEKKPLGWNGSKVLNQLFSLSILEALQLRQVGYAYRRNFSEFC 653
Query: 80 RAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTM 122
F +DL G + S E+ +L E E++ I KTM
Sbjct: 654 SHFRWLDL--GLVNSDRDRKEVAQLLLEQSGIPESSWVIGKTM 694
>gb|AAL30896.1| myosin B [Toxoplasma gondii]
Length = 1074
Score = 33.1 bits (74), Expect = 8.6
Identities = 30/103 (29%), Positives = 44/103 (42%), Gaps = 4/103 (3%)
Query: 22 GGLLDTGSHYIQCWKQNERADP-GWANSHDLYAI-EQKFMENCALNYFDKNDYRSMMKFV 79
G + T +H+I+C K NE P GW S L + +E L R+ +F
Sbjct: 664 GIVAQTEAHFIRCLKPNEEKKPLGWNGSKVLNQLFSLSILEALQLRQVGYAYRRNFSEFC 723
Query: 80 RAFHSIDLKRGFLQSLNLPDELLELEEESGNFMEAAVNIAKTM 122
F +DL G + S E+ +L E E++ I KTM
Sbjct: 724 SHFRWLDL--GLVNSDRDRKEVAQLLLEQSGIPESSWVIGKTM 764
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.135 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 429,531,683
Number of Sequences: 2540612
Number of extensions: 16856280
Number of successful extensions: 40956
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 40951
Number of HSP's gapped (non-prelim): 25
length of query: 273
length of database: 863,360,394
effective HSP length: 126
effective length of query: 147
effective length of database: 543,243,282
effective search space: 79856762454
effective search space used: 79856762454
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC144765.9


