

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.19 - phase: 0
(146 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
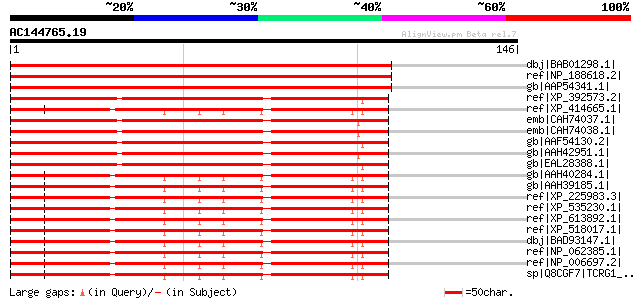
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB01298.1| unnamed protein product [Arabidopsis thaliana] 185 2e-46
ref|NP_188618.2| FF domain-containing protein / WW domain-contai... 185 2e-46
gb|AAP54341.1| hypothetical protein [Oryza sativa (japonica cult... 175 2e-43
ref|XP_392573.2| PREDICTED: similar to ENSANGP00000027066 [Apis ... 95 4e-19
ref|XP_414665.1| PREDICTED: similar to transcription elongation ... 94 7e-19
emb|CAH74037.1| transcription elongation regulator 1-like [Homo ... 93 2e-18
emb|CAH74038.1| transcription elongation regulator 1-like [Homo ... 93 2e-18
gb|AAF54130.2| CG33097-PA [Drosophila melanogaster] gi|17861548|... 92 2e-18
gb|AAH42951.1| Transcription elongation regulator 1-like [Homo s... 91 6e-18
gb|EAL28388.1| GA17277-PA [Drosophila pseudoobscura] 91 6e-18
gb|AAH40284.1| Tcerg1 protein [Mus musculus] 91 8e-18
gb|AAH39185.1| Tcerg1 protein [Mus musculus] 91 8e-18
ref|XP_225983.3| PREDICTED: similar to Transcription elongation ... 91 8e-18
ref|XP_535230.1| PREDICTED: similar to transcription elongation ... 91 8e-18
ref|XP_613892.1| PREDICTED: similar to transcription factor CA15... 91 8e-18
ref|XP_518017.1| PREDICTED: similar to transcription elongation ... 91 8e-18
dbj|BAD93147.1| transcription elongation regulator 1 variant [Ho... 91 8e-18
ref|NP_062385.1| transcription elongation regulator 1 (CA150) [M... 91 8e-18
ref|NP_006697.2| transcription elongation regulator 1 [Homo sapi... 91 8e-18
sp|Q8CGF7|TCRG1_MOUSE Transcription elongation regulator 1 (TATA... 91 8e-18
>dbj|BAB01298.1| unnamed protein product [Arabidopsis thaliana]
Length = 835
Score = 185 bits (470), Expect = 2e-46
Identities = 89/110 (80%), Positives = 98/110 (88%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
MLKERG+APFSKWEKELPKI+FDPRFKAIPS+S RRSLFE YVK RAEEER+EKRAA KA
Sbjct: 465 MLKERGIAPFSKWEKELPKIIFDPRFKAIPSHSVRRSLFEQYVKTRAEEERREKRAAHKA 524
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNER 110
AIEGF+QLLD+AS DID TD F+KKWGND RFEA++RKERE LLNER
Sbjct: 525 AIEGFRQLLDDASTDIDQHTDYRAFKKKWGNDLRFEAIERKEREGLLNER 574
>ref|NP_188618.2| FF domain-containing protein / WW domain-containing protein
[Arabidopsis thaliana]
Length = 743
Score = 185 bits (470), Expect = 2e-46
Identities = 89/110 (80%), Positives = 98/110 (88%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
MLKERG+APFSKWEKELPKI+FDPRFKAIPS+S RRSLFE YVK RAEEER+EKRAA KA
Sbjct: 373 MLKERGIAPFSKWEKELPKIIFDPRFKAIPSHSVRRSLFEQYVKTRAEEERREKRAAHKA 432
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNER 110
AIEGF+QLLD+AS DID TD F+KKWGND RFEA++RKERE LLNER
Sbjct: 433 AIEGFRQLLDDASTDIDQHTDYRAFKKKWGNDLRFEAIERKEREGLLNER 482
>gb|AAP54341.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|37535504|ref|NP_922054.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|18087875|gb|AAL59029.1| hypothetical protein [Oryza
sativa]
Length = 1099
Score = 175 bits (443), Expect = 2e-43
Identities = 83/110 (75%), Positives = 95/110 (85%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
MLKERGV PFSKWEKELPKIVFDPRFKAIPS+S RRS FE YV+ RA+EERKEKRAAQ+A
Sbjct: 721 MLKERGVLPFSKWEKELPKIVFDPRFKAIPSHSRRRSTFEQYVRTRADEERKEKRAAQRA 780
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKEREHLLNER 110
A+E +KQLL+EASEDI+ D F++KWG DPRFEALDRKER+ L NE+
Sbjct: 781 AVEAYKQLLEEASEDINSNKDYKEFKRKWGTDPRFEALDRKERDALFNEK 830
Score = 38.9 bits (89), Expect = 0.026
Identities = 28/87 (32%), Positives = 47/87 (53%), Gaps = 6/87 (6%)
Query: 23 DPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKAAIEGFKQLLDEASEDIDDKTDS 82
DPRF+A+ R +LF VK+ E+ + + + A I FK +L E S+DI +
Sbjct: 812 DPRFEALDR-KERDALFNEKVKSIEEKVQ----SVRNAVIAEFKSMLRE-SKDITSTSRW 865
Query: 83 HTFRKKWGNDPRFEALDRKEREHLLNE 109
++ + +D R++A+ +ERE NE
Sbjct: 866 TKVKENFRSDARYKAMKHEEREVAFNE 892
>ref|XP_392573.2| PREDICTED: similar to ENSANGP00000027066 [Apis mellifera]
Length = 1153
Score = 94.7 bits (234), Expect = 4e-19
Identities = 54/110 (49%), Positives = 72/110 (65%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML E+ V+ FS WEKEL KIVFDPR+ + S R+ +FE YVK RAEEER+EKR K
Sbjct: 640 MLAEKDVSAFSTWEKELHKIVFDPRYLLLTS-KERKQVFEKYVKERAEEERREKRNKMKE 698
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
E F++LL+EA + K+ F +K G D RF+ +++ +ERE L NE
Sbjct: 699 RKEQFQKLLEEAG--LHGKSSFSDFAQKHGRDERFKNVEKMRERESLFNE 746
Score = 39.7 bits (91), Expect = 0.015
Identities = 28/99 (28%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
+L+E G+ S + K D RFK + R SLF Y+ ++E++EK A ++
Sbjct: 706 LLEEAGLHGKSSFSDFAQKHGRDERFKNVEKMRERESLFNEYLLEVRKKEKEEKTAKREQ 765
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD 99
+ F +L E +DID + +KK +D R+ ++
Sbjct: 766 VKKEFIAMLRE-HKDIDRHSHWSDCKKKLESDWRYRVVE 803
>ref|XP_414665.1| PREDICTED: similar to transcription elongation regulator 1;
transcription factor CA150; TATA box binding protein
(TBP)-associated factor, RNA polymerase II, S, 150kD;
TATA box-binding protein-associated factor 2S [Gallus
gallus]
Length = 1125
Score = 94.0 bits (232), Expect = 7e-19
Identities = 53/110 (48%), Positives = 73/110 (66%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEERKEK+
Sbjct: 696 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERKEKKNKIMQ 754
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++E+ I+ +T F K D RF+A+++ K+RE L NE
Sbjct: 755 AKEDFKKMMEESK--INPRTTFSEFAAKHAKDSRFKAIEKMKDREALFNE 802
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 839 SRWSKVKDKVETDPRYKAVDSSSQREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 898
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 899 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 954
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 955 DHRWESGSLLEREEKEKLFNE 975
Score = 43.5 bits (101), Expect = 0.001
Identities = 28/110 (25%), Positives = 55/110 (49%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E + P + + + K D RFKAI R +LF ++ ++E+++ + +
Sbjct: 762 MMEESKINPRTTFSEFAAKHAKDSRFKAIEKMKDREALFNEFITAARKKEKEDSKTRGEK 821
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKEREHLLNE 109
F +LL A+ +D ++ + K DPR++A+D +RE L +
Sbjct: 822 IKMDFFELL--ANHHLDSQSRWSKVKDKVETDPRYKAVDSSSQREDLFKQ 869
>emb|CAH74037.1| transcription elongation regulator 1-like [Homo sapiens]
Length = 225
Score = 92.8 bits (229), Expect = 2e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + S R+ +FE +VK R +EE KEK++
Sbjct: 99 MLLERGVSAFSTWEKELHKIVFDPRYLLLNS-EERKQIFEQFVKTRIKEEYKEKKSKLLL 157
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKEREHLLNE 109
A E FK+LL+E+ + +T F +K+G D RF + RK++EH N+
Sbjct: 158 AKEEFKKLLEESK--VSPRTTFKEFAEKYGRDQRFRLVQKRKDQEHFFNQ 205
>emb|CAH74038.1| transcription elongation regulator 1-like [Homo sapiens]
gi|62739680|gb|AAH93639.1| Transcription elongation
regulator 1-like [Homo sapiens]
Length = 545
Score = 92.8 bits (229), Expect = 2e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + S R+ +FE +VK R +EE KEK++
Sbjct: 419 MLLERGVSAFSTWEKELHKIVFDPRYLLLNS-EERKQIFEQFVKTRIKEEYKEKKSKLLL 477
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKEREHLLNE 109
A E FK+LL+E+ + +T F +K+G D RF + RK++EH N+
Sbjct: 478 AKEEFKKLLEESK--VSPRTTFKEFAEKYGRDQRFRLVQKRKDQEHFFNQ 525
>gb|AAF54130.2| CG33097-PA [Drosophila melanogaster] gi|17861548|gb|AAL39251.1|
GH12404p [Drosophila melanogaster]
gi|28573212|ref|NP_788585.1| CG33097-PA [Drosophila
melanogaster]
Length = 763
Score = 92.4 bits (228), Expect = 2e-18
Identities = 51/110 (46%), Positives = 72/110 (65%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML+E+ V+ FS WEKEL KIVFDPR+ + S R+ +FE YVK+RAEEERKEKR +
Sbjct: 594 MLREKDVSAFSTWEKELHKIVFDPRYLLLTS-KERKQVFEKYVKDRAEEERKEKRNKMRQ 652
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
E F+ L++EA + K+ F ++ D R+ A+++ +ERE L NE
Sbjct: 653 KREDFRSLMEEAR--LHGKSSFSEFSQRNAKDERYRAIEKVRERESLFNE 700
>gb|AAH42951.1| Transcription elongation regulator 1-like [Homo sapiens]
gi|28372559|ref|NP_777597.1| transcription elongation
regulator 1-like [Homo sapiens]
Length = 545
Score = 90.9 bits (224), Expect = 6e-18
Identities = 50/110 (45%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + S R+ +FE +VK R +EE KEK++
Sbjct: 419 MLLERGVSAFSTWEKELHKIVFDPRYLLLNS-EERKQIFEQFVKTRIKEEYKEKKSKLLL 477
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALD-RKEREHLLNE 109
A E FK+LL+++ + +T F +K+G D RF + RK++EH N+
Sbjct: 478 AKEEFKKLLEKSK--VSPRTTFKEFAEKYGRDQRFRLVQKRKDQEHFFNK 525
>gb|EAL28388.1| GA17277-PA [Drosophila pseudoobscura]
Length = 676
Score = 90.9 bits (224), Expect = 6e-18
Identities = 50/110 (45%), Positives = 72/110 (65%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML+E+ V+ FS WEKEL KIVFDPR+ + S R+ +FE YVK+RAEEERKEKR +
Sbjct: 551 MLREKDVSAFSTWEKELHKIVFDPRYLLLTS-KERKQVFEKYVKDRAEEERKEKRNKMRQ 609
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
+ F+ L++EA + K+ F +K + R+ A+++ +ERE L NE
Sbjct: 610 KRDDFRSLMEEAR--LHGKSSFSEFSQKNAKEERYRAIEKVRERESLFNE 657
>gb|AAH40284.1| Tcerg1 protein [Mus musculus]
Length = 1079
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 650 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 708
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 709 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 756
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 793 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 852
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 853 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 908
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 909 DHRWESGSLLEREEKEKLFNE 929
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 716 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 775
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 776 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 823
>gb|AAH39185.1| Tcerg1 protein [Mus musculus]
Length = 1057
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 628 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 686
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 687 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 734
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 771 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 830
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 831 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 886
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 887 DHRWESGSLLEREEKEKLFNE 907
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 694 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 753
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 754 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 801
>ref|XP_225983.3| PREDICTED: similar to Transcription elongation regulator 1 (TATA
box-binding protein-associated factor 2S) (Transcription
factor CA150) (p144) (Formin-binding protein 28) (FBP
28) [Rattus norvegicus]
Length = 1081
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 652 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 710
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 711 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 758
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 795 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 854
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 855 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 910
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 911 DHRWESGSLLEREEKEKLFNE 931
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 718 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 777
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 778 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 825
>ref|XP_535230.1| PREDICTED: similar to transcription elongation regulator 1 [Canis
familiaris]
Length = 1129
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 700 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 758
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 759 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 806
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 843 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 902
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 903 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 958
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 959 DHRWESGSLLEREEKEKLFNE 979
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 766 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 825
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 826 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 873
>ref|XP_613892.1| PREDICTED: similar to transcription factor CA150b [Bos taurus]
Length = 389
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 26 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 84
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 85 AKEDFKKMMEEAK--FNPRVTFSEFAAKHAKDSRFKAIEKMKDREALFNE 132
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 169 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 228
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 229 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 284
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 285 DHRWESGSLLEREEKEKLFNE 305
Score = 40.4 bits (93), Expect = 0.009
Identities = 28/110 (25%), Positives = 53/110 (47%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 92 MMEEAKFNPRVTFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 151
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 152 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 199
>ref|XP_518017.1| PREDICTED: similar to transcription elongation regulator 1;
transcription factor CA150; TATA box binding protein
(TBP)-associated factor, RNA polymerase II, S, 150kD;
TATA box-binding protein-associated factor 2S [Pan
troglodytes]
Length = 1032
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 603 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 661
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 662 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 709
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 746 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 805
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 806 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 861
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 862 DHRWESGSLLEREEKEKLFNE 882
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 669 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 728
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 729 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 776
>dbj|BAD93147.1| transcription elongation regulator 1 variant [Homo sapiens]
Length = 1081
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 652 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 710
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 711 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 758
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 795 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 854
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 855 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 910
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 911 DHRWESGSLLEREEKEKLFNE 931
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 718 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 777
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 778 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 825
>ref|NP_062385.1| transcription elongation regulator 1 (CA150) [Mus musculus]
gi|6329166|dbj|BAA86392.1| transcription factor CA150b
[Mus musculus]
Length = 1034
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 671 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 729
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 730 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 777
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 814 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 873
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 874 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 929
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 930 DHRWESGSLLEREEKEKLFNE 950
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 737 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 796
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 797 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 844
>ref|NP_006697.2| transcription elongation regulator 1 [Homo sapiens]
Length = 1098
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 669 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 727
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 728 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 775
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 812 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 871
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 872 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 927
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 928 DHRWESGSLLEREEKEKLFNE 948
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 735 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 794
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 795 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 842
>sp|Q8CGF7|TCRG1_MOUSE Transcription elongation regulator 1 (TATA box-binding
protein-associated factor 2S) (Transcription factor
CA150) (p144) (Formin-binding protein 28) (FBP 28)
Length = 1100
Score = 90.5 bits (223), Expect = 8e-18
Identities = 51/110 (46%), Positives = 71/110 (64%), Gaps = 4/110 (3%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
ML ERGV+ FS WEKEL KIVFDPR+ + + R+ +F+ YVK RAEEER+EK+
Sbjct: 671 MLLERGVSAFSTWEKELHKIVFDPRYLLL-NPKERKQVFDQYVKTRAEEERREKKNKIMQ 729
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDR-KEREHLLNE 109
A E FK++++EA + + F K D RF+A+++ K+RE L NE
Sbjct: 730 AKEDFKKMMEEAK--FNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNE 777
Score = 50.1 bits (118), Expect = 1e-05
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 46/141 (32%)
Query: 11 SKWEKELPKIVFDPRFKAIPSYSARRSLFEHYV----KNRAEEERKE------------- 53
S+W K K+ DPR+KA+ S S R LF+ Y+ KN E+ KE
Sbjct: 814 SRWSKVKDKVESDPRYKAVDSSSMREDLFKQYIEKIAKNLDSEKEKELERQARIEASLRE 873
Query: 54 -KRAAQKA-------------------AIEGFKQLLDE--ASEDIDDKTDSHTFRKKWGN 91
+R QKA AI+ FK LL + S D+ T RK
Sbjct: 874 REREVQKARSEQTKEIDREREQHKREEAIQNFKALLSDMVRSSDVSWSDTRRTLRK---- 929
Query: 92 DPRFEA---LDRKEREHLLNE 109
D R+E+ L+R+E+E L NE
Sbjct: 930 DHRWESGSLLEREEKEKLFNE 950
Score = 41.6 bits (96), Expect = 0.004
Identities = 28/110 (25%), Positives = 54/110 (48%), Gaps = 3/110 (2%)
Query: 1 MLKERGVAPFSKWEKELPKIVFDPRFKAIPSYSARRSLFEHYVKNRAEEERKEKRAAQKA 60
M++E P + + + K D RFKAI R +LF +V ++E+++ + +
Sbjct: 737 MMEEAKFNPRATFSEFAAKHAKDSRFKAIEKMKDREALFNEFVAAARKKEKEDSKTRGEK 796
Query: 61 AIEGFKQLLDEASEDIDDKTDSHTFRKKWGNDPRFEALDRKE-REHLLNE 109
F +LL ++ +D ++ + K +DPR++A+D RE L +
Sbjct: 797 IKSDFFELL--SNHHLDSQSRWSKVKDKVESDPRYKAVDSSSMREDLFKQ 844
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.137 0.437
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 261,103,333
Number of Sequences: 2540612
Number of extensions: 10052047
Number of successful extensions: 39611
Number of sequences better than 10.0: 240
Number of HSP's better than 10.0 without gapping: 133
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 39162
Number of HSP's gapped (non-prelim): 476
length of query: 146
length of database: 863,360,394
effective HSP length: 122
effective length of query: 24
effective length of database: 553,405,730
effective search space: 13281737520
effective search space used: 13281737520
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144765.19


