

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.8 - phase: 0
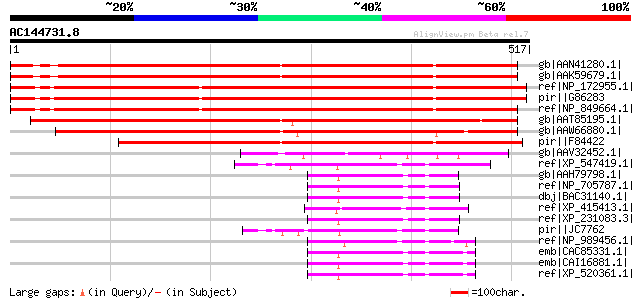
(517 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN41280.1| unknown protein [Arabidopsis thaliana] gi|2192804... 711 0.0
gb|AAK59679.1| unknown protein [Arabidopsis thaliana] 708 0.0
ref|NP_172955.1| thioredoxin family protein [Arabidopsis thalian... 693 0.0
pir||G86283 hypothetical protein T15D22.7 - Arabidopsis thaliana... 693 0.0
ref|NP_849664.1| thioredoxin family protein [Arabidopsis thaliana] 686 0.0
gb|AAT85195.1| unknown protein [Oryza sativa (japonica cultivar-... 627 e-178
gb|AAW66880.1| thiol oxidoreductase [Zea mays] 624 e-177
pir||F84422 hypothetical protein At2g01270 [imported] - Arabidop... 593 e-168
gb|AAV32452.1| thioredoxin-like protein [Chlamydomonas reinhardtii] 135 4e-30
ref|XP_547419.1| PREDICTED: similar to quiescin Q6 isoform a [Ca... 119 3e-25
gb|AAH79798.1| MGC86371 protein [Xenopus laevis] 118 4e-25
ref|NP_705787.1| quiescin Q6-like 1 [Mus musculus] gi|22658418|g... 115 3e-24
dbj|BAC31140.1| unnamed protein product [Mus musculus] 115 3e-24
ref|XP_415413.1| PREDICTED: similar to quiescin Q6-like 1; putat... 115 4e-24
ref|XP_231083.3| PREDICTED: LIM homeobox protein 3 [Rattus norve... 114 1e-23
pir||JC7762 SOX-3 protein - guinea pig gi|12597921|gb|AAB58401.2... 113 1e-23
ref|NP_989456.1| quiescin Q6 [Gallus gallus] gi|21314229|gb|AAM4... 112 3e-23
emb|CAC85331.1| putative sulfhydryl oxidase precursor [Homo sapi... 110 8e-23
emb|CAI16881.1| RP11-83N9.4 [Homo sapiens] 110 8e-23
ref|XP_520361.1| PREDICTED: similar to quiescin Q6-like 1; putat... 110 8e-23
>gb|AAN41280.1| unknown protein [Arabidopsis thaliana] gi|21928041|gb|AAM78049.1|
At2g01270/F10A8.15 [Arabidopsis thaliana]
gi|20197588|gb|AAD14527.2| expressed protein
[Arabidopsis thaliana] gi|19699314|gb|AAL91267.1|
At2g01270/F10A8.15 [Arabidopsis thaliana]
gi|18379190|ref|NP_565258.1| thioredoxin family protein
[Arabidopsis thaliana]
Length = 495
Score = 711 bits (1834), Expect = 0.0
Identities = 343/507 (67%), Positives = 409/507 (80%), Gaps = 17/507 (3%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ LF + +SS++ S R ILRE+ D D AVELN TNFD+
Sbjct: 1 MSLVHLLLFAGLVIAASSSSPGS------RLILREISDQK-------DKAVELNTTNFDS 47
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
VLKDTPA +AVVEFFA+WCPACRNYKPHYEKVA+LFNGPDA+HPGI+L+TRVDCA K N
Sbjct: 48 VLKDTPAKYAVVEFFAHWCPACRNYKPHYEKVARLFNGPDAIHPGIVLMTRVDCAMKTNT 107
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFSV HYPMLFWGPP KFV GSWEPK++KS+I VIDD RTA+RLLNWINKQ+ SS+
Sbjct: 108 KLCDKFSVSHYPMLFWGPPTKFVSGSWEPKKDKSEILVIDDGRTAERLLNWINKQIGSSY 167
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIK-PETRASLVKFLQ 239
GLDDQKF+NEH SN++D QI++A+YDVEEAT+ AFDIIL +K IK ET AS ++F+Q
Sbjct: 168 GLDDQKFKNEHALSNLTDYNQISQAVYDVEEATAEAFDIILAHKAIKSSETSASFIRFIQ 227
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHH SRRCRKGA ++LV++ DL P+ S+++ ++ NF ICGK+VPRGY+MF
Sbjct: 228 LLAAHHLSRRCRKGAAEILVNYDDLCPSGN-CSYEKSGGNDTLGNFPICGKDVPRGYYMF 286
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKN+TRGFSCGLWVL+HSLSVRIEDGES F F ICDFV+NFF+C+ECR HF DMC
Sbjct: 287 CRGSKNDTRGFSCGLWVLMHSLSVRIEDGESHFAFTTICDFVNNFFMCDECRLHFNDMCL 346
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARDFVLW+WS+HNKVNERL K+EASLGTGDPKFPK IWP K+LCP CYL +
Sbjct: 347 SVKTPFKKARDFVLWVWSTHNKVNERLLKDEASLGTGDPKFPKIIWPPKELCPLCYLSSN 406
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
QKS IEW+ + VYK LKNYY LVSLYKEK ++ + +A EDL V TNA+VVP+G
Sbjct: 407 QKS--IEWDHEHVYKFLKNYYGPKLVSLYKEKSVSRSKEETVSATEDLTVATNALVVPIG 464
Query: 480 AALAIAVASCAFGALACYWRSQQKSRK 506
AALAIA+ASCAFGALACYWR+QQK+RK
Sbjct: 465 AALAIAIASCAFGALACYWRTQQKNRK 491
>gb|AAK59679.1| unknown protein [Arabidopsis thaliana]
Length = 495
Score = 708 bits (1827), Expect = 0.0
Identities = 342/507 (67%), Positives = 408/507 (80%), Gaps = 17/507 (3%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ LF + +SS++ S R ILRE+ D D AVELN TNFD+
Sbjct: 1 MSLVHLLLFAGLVIAASSSSPGS------RLILREISDQK-------DKAVELNTTNFDS 47
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
VLKDTPA +AVVEFFA+WCPACRNYKPHYEKVA+LFN PDA+HPGI+L+TRVDCA K N
Sbjct: 48 VLKDTPAKYAVVEFFAHWCPACRNYKPHYEKVARLFNDPDAIHPGIVLMTRVDCAMKTNT 107
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFSV HYPMLFWGPP KFV GSWEPK++KS+I VIDD RTA+RLLNWINKQ+ SS+
Sbjct: 108 KLCDKFSVSHYPMLFWGPPTKFVSGSWEPKKDKSEILVIDDGRTAERLLNWINKQIGSSY 167
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIK-PETRASLVKFLQ 239
GLDDQKF+NEH SN++D QI++A+YDVEEAT+ AFDIIL +K IK ET AS ++F+Q
Sbjct: 168 GLDDQKFKNEHALSNLTDYNQISQAVYDVEEATAEAFDIILAHKAIKSSETSASFIRFIQ 227
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHH SRRCRKGA ++LV++ DL P+ S+++ ++ NF ICGK+VPRGY+MF
Sbjct: 228 LLAAHHLSRRCRKGAAEILVNYDDLCPSGN-CSYEKSGGNDTLGNFPICGKDVPRGYYMF 286
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKN+TRGFSCGLWVL+HSLSVRIEDGES F F ICDFV+NFF+C+ECR HF DMC
Sbjct: 287 CRGSKNDTRGFSCGLWVLMHSLSVRIEDGESHFAFTTICDFVNNFFMCDECRLHFNDMCL 346
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARDFVLW+WS+HNKVNERL K+EASLGTGDPKFPK IWP K+LCP CYL +
Sbjct: 347 SVKTPFKKARDFVLWVWSTHNKVNERLLKDEASLGTGDPKFPKIIWPPKELCPLCYLSSN 406
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
QKS IEW+ + VYK LKNYY LVSLYKEK ++ + +A EDL V TNA+VVP+G
Sbjct: 407 QKS--IEWDHEHVYKFLKNYYGPKLVSLYKEKSVSRSKEETVSATEDLTVATNALVVPIG 464
Query: 480 AALAIAVASCAFGALACYWRSQQKSRK 506
AALAIA+ASCAFGALACYWR+QQK+RK
Sbjct: 465 AALAIAIASCAFGALACYWRTQQKNRK 491
>ref|NP_172955.1| thioredoxin family protein [Arabidopsis thaliana]
gi|17064904|gb|AAL32606.1| Unknown protein [Arabidopsis
thaliana] gi|24899781|gb|AAN65105.1| Unknown protein
[Arabidopsis thaliana]
Length = 528
Score = 693 bits (1788), Expect = 0.0
Identities = 332/516 (64%), Positives = 414/516 (79%), Gaps = 12/516 (2%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ L L +S+ ++A+ S S RSILR++ N D A+ELNATNFD+
Sbjct: 1 MSLIHLFLLLGLLSLEAAASFSPGS----RSILRDIGSNV---ADQKDNAIELNATNFDS 53
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
V +D+PA +AV+EFFA+WCPACRNYKPHYEKVA+LFNG DAV+PG++L+TRVDCA K+N+
Sbjct: 54 VFQDSPAKYAVLEFFAHWCPACRNYKPHYEKVARLFNGADAVYPGVVLMTRVDCAIKMNV 113
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFS+ HYPMLFW PP +FVGGSW PKQEK++I V+++ RTAD LLNWINKQ+ SS+
Sbjct: 114 KLCDKFSINHYPMLFWAPPKRFVGGSWGPKQEKNEISVVNEWRTADLLLNWINKQIGSSY 173
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP-ETRASLVKFLQ 239
GLDDQK N L SN+SD EQI++AI+D+EEAT AFDIIL +K IK ET AS ++FLQ
Sbjct: 174 GLDDQKLGN--LLSNISDQEQISQAIFDIEEATEEAFDIILAHKAIKSSETSASFIRFLQ 231
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHHPSRRCR G+ ++LV+F D+ P+ S ++ K S++NF ICGK+VPRGY+ F
Sbjct: 232 LLVAHHPSRRCRTGSAEILVNFDDICPSGECSYDQESGAKDSLRNFHICGKDVPRGYYRF 291
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKNETRGFSCGLWVL+HSLSVRIEDGESQF F AICDF++NFF+C++CR+HF+DMC
Sbjct: 292 CRGSKNETRGFSCGLWVLMHSLSVRIEDGESQFAFTAICDFINNFFMCDDCRRHFHDMCL 351
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARD LWLWS+HNKVNERL K+E SLGTGDPKFPK IWP KQLCPSCYL
Sbjct: 352 SVKTPFKKARDIALWLWSTHNKVNERLKKDEDSLGTGDPKFPKMIWPPKQLCPSCYLSST 411
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
+K+ I+W+ D+VYK LK YY + LVS+YK+ + + AA E++ V TNA+VVPVG
Sbjct: 412 EKN--IDWDHDQVYKFLKKYYGQKLVSVYKKNGESVSKEEVIAAAEEMAVPTNALVVPVG 469
Query: 480 AALAIAVASCAFGALACYWRSQQKSRKYFHHLHSLK 515
AALAIA+ASCAFGALACYWR+QQK+RKY ++ H LK
Sbjct: 470 AALAIALASCAFGALACYWRTQQKNRKYNYNPHYLK 505
>pir||G86283 hypothetical protein T15D22.7 - Arabidopsis thaliana
gi|6899648|gb|AAF31025.1| Contains Thioredoxin domain
PF|00085. ESTs gb|T42351, gb|AA042405 come from this
gene. [Arabidopsis thaliana]
Length = 536
Score = 693 bits (1788), Expect = 0.0
Identities = 332/516 (64%), Positives = 414/516 (79%), Gaps = 12/516 (2%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ L L +S+ ++A+ S S RSILR++ N D A+ELNATNFD+
Sbjct: 1 MSLIHLFLLLGLLSLEAAASFSPGS----RSILRDIGSNV---ADQKDNAIELNATNFDS 53
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
V +D+PA +AV+EFFA+WCPACRNYKPHYEKVA+LFNG DAV+PG++L+TRVDCA K+N+
Sbjct: 54 VFQDSPAKYAVLEFFAHWCPACRNYKPHYEKVARLFNGADAVYPGVVLMTRVDCAIKMNV 113
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFS+ HYPMLFW PP +FVGGSW PKQEK++I V+++ RTAD LLNWINKQ+ SS+
Sbjct: 114 KLCDKFSINHYPMLFWAPPKRFVGGSWGPKQEKNEISVVNEWRTADLLLNWINKQIGSSY 173
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP-ETRASLVKFLQ 239
GLDDQK N L SN+SD EQI++AI+D+EEAT AFDIIL +K IK ET AS ++FLQ
Sbjct: 174 GLDDQKLGN--LLSNISDQEQISQAIFDIEEATEEAFDIILAHKAIKSSETSASFIRFLQ 231
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHHPSRRCR G+ ++LV+F D+ P+ S ++ K S++NF ICGK+VPRGY+ F
Sbjct: 232 LLVAHHPSRRCRTGSAEILVNFDDICPSGECSYDQESGAKDSLRNFHICGKDVPRGYYRF 291
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKNETRGFSCGLWVL+HSLSVRIEDGESQF F AICDF++NFF+C++CR+HF+DMC
Sbjct: 292 CRGSKNETRGFSCGLWVLMHSLSVRIEDGESQFAFTAICDFINNFFMCDDCRRHFHDMCL 351
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARD LWLWS+HNKVNERL K+E SLGTGDPKFPK IWP KQLCPSCYL
Sbjct: 352 SVKTPFKKARDIALWLWSTHNKVNERLKKDEDSLGTGDPKFPKMIWPPKQLCPSCYLSST 411
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
+K+ I+W+ D+VYK LK YY + LVS+YK+ + + AA E++ V TNA+VVPVG
Sbjct: 412 EKN--IDWDHDQVYKFLKKYYGQKLVSVYKKNGESVSKEEVIAAAEEMAVPTNALVVPVG 469
Query: 480 AALAIAVASCAFGALACYWRSQQKSRKYFHHLHSLK 515
AALAIA+ASCAFGALACYWR+QQK+RKY ++ H LK
Sbjct: 470 AALAIALASCAFGALACYWRTQQKNRKYNYNPHYLK 505
>ref|NP_849664.1| thioredoxin family protein [Arabidopsis thaliana]
Length = 502
Score = 686 bits (1769), Expect = 0.0
Identities = 328/507 (64%), Positives = 408/507 (79%), Gaps = 12/507 (2%)
Query: 1 MNLLYFALFLCTISVSSSATSSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDA 60
M+L++ L L +S+ ++A+ S S RSILR++ N D A+ELNATNFD+
Sbjct: 1 MSLIHLFLLLGLLSLEAAASFSPGS----RSILRDIGSNV---ADQKDNAIELNATNFDS 53
Query: 61 VLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINI 120
V +D+PA +AV+EFFA+WCPACRNYKPHYEKVA+LFNG DAV+PG++L+TRVDCA K+N+
Sbjct: 54 VFQDSPAKYAVLEFFAHWCPACRNYKPHYEKVARLFNGADAVYPGVVLMTRVDCAIKMNV 113
Query: 121 KLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSF 180
KLCDKFS+ HYPMLFW PP +FVGGSW PKQEK++I V+++ RTAD LLNWINKQ+ SS+
Sbjct: 114 KLCDKFSINHYPMLFWAPPKRFVGGSWGPKQEKNEISVVNEWRTADLLLNWINKQIGSSY 173
Query: 181 GLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP-ETRASLVKFLQ 239
GLDDQK N L SN+SD EQI++AI+D+EEAT AFDIIL +K IK ET AS ++FLQ
Sbjct: 174 GLDDQKLGN--LLSNISDQEQISQAIFDIEEATEEAFDIILAHKAIKSSETSASFIRFLQ 231
Query: 240 VLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGKEVPRGYWMF 299
+L AHHPSRRCR G+ ++LV+F D+ P+ S ++ K S++NF ICGK+VPRGY+ F
Sbjct: 232 LLVAHHPSRRCRTGSAEILVNFDDICPSGECSYDQESGAKDSLRNFHICGKDVPRGYYRF 291
Query: 300 CRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCS 359
CRGSKNETRGFSCGLWVL+HSLSVRIEDGESQF F AICDF++NFF+C++CR+HF+DMC
Sbjct: 292 CRGSKNETRGFSCGLWVLMHSLSVRIEDGESQFAFTAICDFINNFFMCDDCRRHFHDMCL 351
Query: 360 SVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHD 419
SV TPF KARD LWLWS+HNKVNERL K+E SLGTGDPKFPK IWP KQLCPSCYL
Sbjct: 352 SVKTPFKKARDIALWLWSTHNKVNERLKKDEDSLGTGDPKFPKMIWPPKQLCPSCYLSST 411
Query: 420 QKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDLIVGTNAVVVPVG 479
+K+ I+W+ D+VYK LK YY + LVS+YK+ + + AA E++ V TNA+VVPVG
Sbjct: 412 EKN--IDWDHDQVYKFLKKYYGQKLVSVYKKNGESVSKEEVIAAAEEMAVPTNALVVPVG 469
Query: 480 AALAIAVASCAFGALACYWRSQQKSRK 506
AALAIA+ASCAFGALACYWR+QQK+RK
Sbjct: 470 AALAIALASCAFGALACYWRTQQKNRK 496
>gb|AAT85195.1| unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 513
Score = 627 bits (1618), Expect = e-178
Identities = 299/492 (60%), Positives = 367/492 (73%), Gaps = 8/492 (1%)
Query: 21 SSSSSSFPGRSILREVHDNDQTGTTDHDYAVELNATNFDAVLKDTPATFAVVEFFANWCP 80
++S ++ P + R + + G D D AV+LNATNFDA LK + +AVVEFFA+WCP
Sbjct: 18 AASLAAAPRGAAARSLGGREGPGEVDADAAVDLNATNFDAFLKASLEPWAVVEFFAHWCP 77
Query: 81 ACRNYKPHYEKVAKLFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPP 140
ACRNYKPHYEKVAKLFNG DA HPG+IL+ RVDCASK+NI LC++FSV HYP L WGPP
Sbjct: 78 ACRNYKPHYEKVAKLFNGRDAAHPGLILMARVDCASKVNIDLCNRFSVDHYPFLLWGPPT 137
Query: 141 KFVGGSWEPKQEKSDIHVIDDARTADRLLNWINKQMSSSFGLDDQKFQNEH-LSSNVSDP 199
KF W+PKQE ++I +IDD RTA+RLL WIN QM SSF L+D+K++NE+ L N SDP
Sbjct: 138 KFASAKWDPKQENNEIKLIDDGRTAERLLKWINNQMKSSFSLEDKKYENENMLPKNASDP 197
Query: 200 EQIARAIYDVEEATSLAFDIILENKMIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLV 259
EQI +AIYDVEEAT+ A IILE K IKP+ R SL++FLQ+L A HPS+RCR+G+ +LL+
Sbjct: 198 EQIVQAIYDVEEATAQALQIILERKTIKPKNRDSLIRFLQILVARHPSKRCRRGSAELLI 257
Query: 260 SFADLYPTDFWSSHKQEDDKS----SVKNFQICGKEVPRGYWMFCRGSKNETRGFSCGLW 315
+F D + ++ S QE K + +N ICGKEVPRGYW+FCRGSK+ETRGFSCGLW
Sbjct: 258 NFDDHWSSNL-SLSSQEGSKLLESVAEENHWICGKEVPRGYWLFCRGSKSETRGFSCGLW 316
Query: 316 VLLHSLSVRIEDGESQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWL 375
VL+HSL+VRI DGESQ TF +ICDF+HNFF+CEECR+HFY+MCSSVS PF AR+ LWL
Sbjct: 317 VLMHSLTVRIGDGESQSTFTSICDFIHNFFICEECRKHFYEMCSSVSAPFRTARELSLWL 376
Query: 376 WSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKA 435
WS+HNKVN RL KEE +GTGDP FPK WP QLCPSCY ++WN+D VY+
Sbjct: 377 WSTHNKVNMRLMKEEKDMGTGDPLFPKVTWPPNQLCPSCYRSSKVTDGAVDWNEDAVYQF 436
Query: 436 LKNYYEKTLVSLYKEKDIAG-NDGTKGAALEDLIVGTNAVVVPVGAALAIAVASCAFGAL 494
L NYY K LVS YKE + K ED + +NA VP+GAAL +A+ASC FGAL
Sbjct: 437 LVNYYGKKLVSSYKETYMESLQQQEKKIVSEDSSI-SNAASVPIGAALGVAIASCTFGAL 495
Query: 495 ACYWRSQQKSRK 506
AC+WR+QQK+RK
Sbjct: 496 ACFWRAQQKNRK 507
>gb|AAW66880.1| thiol oxidoreductase [Zea mays]
Length = 511
Score = 624 bits (1610), Expect = e-177
Identities = 294/467 (62%), Positives = 359/467 (75%), Gaps = 11/467 (2%)
Query: 46 DHDYAVELNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPG 105
D D AV+L+A+NF A L+ +P +FAVVEFFA+WCPACRNYKPHYE+VAKLFNGPDA HPG
Sbjct: 44 DGDAAVDLDASNFTAFLQTSPESFAVVEFFAHWCPACRNYKPHYERVAKLFNGPDAAHPG 103
Query: 106 IILLTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTA 165
+++ RVDCASK+N+ LC+KFSV HYP L WGPP KF W+PKQE S++ +IDD RTA
Sbjct: 104 TVVMARVDCASKVNVDLCNKFSVDHYPYLVWGPPTKFNLAQWKPKQENSELELIDDGRTA 163
Query: 166 DRLLNWINKQMSSSFGLDDQKFQNEHLS-SNVSDPEQIARAIYDVEEATSLAFDIILENK 224
DRLL WINK+M SSF LDD+K++NE + N SDPEQI RAIYDVEEATS A IILE+K
Sbjct: 164 DRLLKWINKKMGSSFNLDDKKYENESMHPKNTSDPEQIVRAIYDVEEATSHALQIILEHK 223
Query: 225 MIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKN 284
MIKP+TR SL+ FLQ+L AHHPS+RCR+G+ +LL+ F D + T+ S ED + +K
Sbjct: 224 MIKPDTRDSLISFLQILVAHHPSKRCRRGSAELLIDFDDHWHTNL--SLSLEDSTTLLKG 281
Query: 285 F--QICGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVH 342
++CG VPRGYW+FCRGSK ETRGFSCGLWVLLHSL+VRI DGESQ TF +ICDF+H
Sbjct: 282 AGEKVCGNGVPRGYWIFCRGSKKETRGFSCGLWVLLHSLTVRIGDGESQTTFTSICDFIH 341
Query: 343 NFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPK 402
NFF+CEECR HFY+MCSSVS PF ARD LWLW++HNKVNERL KEE L DP FPK
Sbjct: 342 NFFICEECRTHFYEMCSSVSVPFKSARDLALWLWTAHNKVNERLMKEEKELDNADPSFPK 401
Query: 403 TIWPTKQLCPSCYLGHDQKSN---KIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGT 459
IWP KQLCP CY + ++ ++EWN+DEV+ L NYY K LVS Y+E +
Sbjct: 402 IIWPPKQLCPLCYRSSSRTADGAMQVEWNEDEVFNFLVNYYGKMLVSSYRETSM---QSF 458
Query: 460 KGAALEDLIVGTNAVVVPVGAALAIAVASCAFGALACYWRSQQKSRK 506
+ A++ D ++A VP+GAAL IA+ASC FGALAC+WR+QQK+RK
Sbjct: 459 QVASISDDSSASSAATVPIGAALGIALASCTFGALACFWRTQQKNRK 505
>pir||F84422 hypothetical protein At2g01270 [imported] - Arabidopsis thaliana
Length = 403
Score = 593 bits (1529), Expect = e-168
Identities = 281/404 (69%), Positives = 334/404 (82%), Gaps = 4/404 (0%)
Query: 109 LTRVDCASKINIKLCDKFSVGHYPMLFWGPPPKFVGGSWEPKQEKSDIHVIDDARTADRL 168
+TRVDCA K N KLCDKFSV HYPMLFWGPP KFV GSWEPK++KS+I VIDD RTA+RL
Sbjct: 1 MTRVDCAMKTNTKLCDKFSVSHYPMLFWGPPTKFVSGSWEPKKDKSEILVIDDGRTAERL 60
Query: 169 LNWINKQMSSSFGLDDQKFQNEHLSSNVSDPEQIARAIYDVEEATSLAFDIILENKMIKP 228
LNWINKQ+ SS+GLDDQKF+NEH SN++D QI++A+YDVEEAT+ AFDIIL +K IK
Sbjct: 61 LNWINKQIGSSYGLDDQKFKNEHALSNLTDYNQISQAVYDVEEATAEAFDIILAHKAIKS 120
Query: 229 -ETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQI 287
ET AS ++F+Q+L AHH SRRCRKGA ++LV++ DL P+ S+++ ++ NF I
Sbjct: 121 SETSASFIRFIQLLAAHHLSRRCRKGAAEILVNYDDLCPSGN-CSYEKSGGNDTLGNFPI 179
Query: 288 CGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVC 347
CGK+VPRGY+MFCRGSKN+TRGFSCGLWVL+HSLSVRIEDGES F F ICDFV+NFF+C
Sbjct: 180 CGKDVPRGYYMFCRGSKNDTRGFSCGLWVLMHSLSVRIEDGESHFAFTTICDFVNNFFMC 239
Query: 348 EECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPT 407
+ECR HF DMC SV TPF KARDFVLW+WS+HNKVNERL K+EASLGTGDPKFPK IWP
Sbjct: 240 DECRLHFNDMCLSVKTPFKKARDFVLWVWSTHNKVNERLLKDEASLGTGDPKFPKIIWPP 299
Query: 408 KQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDIAGNDGTKGAALEDL 467
K+LCP CYL +QKS IEW+ + VYK LKNYY LVSLYKEK ++ + +A EDL
Sbjct: 300 KELCPLCYLSSNQKS--IEWDHEHVYKFLKNYYGPKLVSLYKEKSVSRSKEETVSATEDL 357
Query: 468 IVGTNAVVVPVGAALAIAVASCAFGALACYWRSQQKSRKYFHHL 511
V TNA+VVP+GAALAIA+ASCAFGALACYWR+QQK+RKY+ HL
Sbjct: 358 TVATNALVVPIGAALAIAIASCAFGALACYWRTQQKNRKYYFHL 401
>gb|AAV32452.1| thioredoxin-like protein [Chlamydomonas reinhardtii]
Length = 438
Score = 135 bits (339), Expect = 4e-30
Identities = 90/288 (31%), Positives = 132/288 (45%), Gaps = 29/288 (10%)
Query: 231 RASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDDKSSVKNFQICGK 290
R +L L HPS C+ G+ LL + +P ED + +CG
Sbjct: 19 REALQLLLAAWAEAHPSPSCKAGSARLLEGYEAAWPA------AAEDAPPGLTRAALCGP 72
Query: 291 E---VPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQFTFNAICDFVHNFFVC 347
RG W C GS+ ++RGFSCGLW L+H+L+ R+ S + + F +FF+C
Sbjct: 73 PGGLAWRGLWSACAGSQPDSRGFSCGLWFLIHTLAARMPSPTSVLEY--LRAFNTHFFLC 130
Query: 348 EECRQHFYDMCSSVSTPFNKA--RDFVLWLWSSHNKVNERLSKEEASLG---TGDPKFPK 402
E C++HF + +S A RD VLWLW +HN+VNERL E G TGDP++PK
Sbjct: 131 EPCQKHFGRILASPEAAAATASRRDLVLWLWRTHNEVNERLRGIETRYGHSTTGDPEWPK 190
Query: 403 TIWPTKQLCPSCYLGHDQKSNK--------IEWNQDEVYKALKNYYEKTLV----SLYKE 450
+WP + CP+C + + S W+++ VY+ L Y + KE
Sbjct: 191 EVWPAPEACPACRVQYKPGSGSGSGSGALPGSWDEEAVYQYLTAAYGPGSAPGADGIVKE 250
Query: 451 KDIAGNDGTKGAALEDLIVGTNAVVVPVGAALA-IAVASCAFGALACY 497
+ G G G L + + + V P + + VA AL Y
Sbjct: 251 AAVGGGGGGGGVLLSEKGLYSRRVPPPTSTLYSMLPVAGVLAAALMLY 298
>ref|XP_547419.1| PREDICTED: similar to quiescin Q6 isoform a [Canis familiaris]
Length = 1236
Score = 119 bits (297), Expect = 3e-25
Identities = 79/271 (29%), Positives = 123/271 (45%), Gaps = 32/271 (11%)
Query: 225 MIKPETRASLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFWSSHKQEDD------ 278
+++ + +L KF+ VL H P + L+ +F L+ + W +Q
Sbjct: 815 VLEGQRLVALKKFMAVLAQHFPGQ-------PLVQNF--LHSVNDWLKRQQRKKIPYSFF 865
Query: 279 KSSVKNFQICGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRI---------EDGE 329
K+++ N + + +W+ C+GS+ RGF C LW+L H L+V+ E E
Sbjct: 866 KAALDNRKEGTMIAKKVHWVGCQGSEPHFRGFPCSLWILFHFLTVQAARHNLDHSQETAE 925
Query: 330 SQFTFNAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKE 389
+Q AI +V FF C +C HF M ++ + VLWLWSSHNKVN RL
Sbjct: 926 AQEVLQAIRSYVRFFFGCRDCANHFEQMAAASMNRVDSLNSAVLWLWSSHNKVNARL--- 982
Query: 390 EASLGTGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSLYK 449
A + DP FPK WP + LC +C H+ W+ D + LK ++ + V L
Sbjct: 983 -AGAASEDPHFPKVQWPPRGLCSAC---HNDLRGSPVWDLDNTLRFLKTHFSPSNVVLDM 1038
Query: 450 EKDIAGND-GTKGAALEDLIVGTNAVVVPVG 479
+ G G A ++ +G + VG
Sbjct: 1039 PSAVPGPQMGALRMAAGNVTLGPKKAGIAVG 1069
Score = 35.8 bits (81), Expect = 3.4
Identities = 19/62 (30%), Positives = 35/62 (55%), Gaps = 6/62 (9%)
Query: 72 VEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVGH 130
VEFFA+WC C + P ++ +A+ + + A++ L +DCA + N +C F++
Sbjct: 363 VEFFASWCGHCIAFAPTWKALAQDVKDWRPALN-----LAALDCADETNNAVCRDFNIPG 417
Query: 131 YP 132
+P
Sbjct: 418 FP 419
>gb|AAH79798.1| MGC86371 protein [Xenopus laevis]
Length = 661
Score = 118 bits (296), Expect = 4e-25
Identities = 61/162 (37%), Positives = 77/162 (46%), Gaps = 18/162 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ CRGSK+ RG+ C LW L HSL+V+ + E + + ++ FF
Sbjct: 394 WVGCRGSKSNLRGYPCSLWKLFHSLTVQAAVKPDALANTAFEAEPRAVLQTMRRYIREFF 453
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C EC +HF M +LWLW HN VN RLS + DPKFPK W
Sbjct: 454 GCRECAKHFEAMAKETVDSVKTPDQAILWLWRKHNVVNNRLSGAPSE----DPKFPKVQW 509
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSL 447
PT LC +C H Q WN+DEV LK YY +SL
Sbjct: 510 PTSDLCSAC---HSQTGGVHSWNEDEVLAFLKRYYGNQEISL 548
Score = 40.0 bits (92), Expect = 0.18
Identities = 20/63 (31%), Positives = 35/63 (54%), Gaps = 6/63 (9%)
Query: 71 VVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGIILLTRVDCASKINIKLCDKFSVG 129
++EF+++WC C NY P ++ +A+ + + A+ G++ DCA + N C F V
Sbjct: 59 LLEFYSSWCGHCINYAPTWKALARDVRDWEPAIKIGVL-----DCAEEDNYGACKDFGVT 113
Query: 130 HYP 132
YP
Sbjct: 114 LYP 116
>ref|NP_705787.1| quiescin Q6-like 1 [Mus musculus] gi|22658418|gb|AAH30934.1|
Quiescin Q6-like 1 [Mus musculus]
Length = 639
Score = 115 bits (289), Expect = 3e-24
Identities = 60/164 (36%), Positives = 82/164 (49%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G Q AI ++ FF
Sbjct: 409 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALAGTGFEGHPQAVLQAIRRYIRTFF 468
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M VLWLW HN VN RL+ + DPKFPK W
Sbjct: 469 GCKECGEHFEEMAKESMDSVKTPDQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 524
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP+C H++ WN+ +V LK +Y + LV Y
Sbjct: 525 PTPDLCPAC---HEEIKGLDSWNEGQVLLFLKQHYSRDNLVDAY 565
>dbj|BAC31140.1| unnamed protein product [Mus musculus]
Length = 527
Score = 115 bits (289), Expect = 3e-24
Identities = 60/164 (36%), Positives = 82/164 (49%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G Q AI ++ FF
Sbjct: 244 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALAGTGFEGHPQAVLQAIRRYIRTFF 303
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M VLWLW HN VN RL+ + DPKFPK W
Sbjct: 304 GCKECGEHFEEMAKESMDSVKTPDQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 359
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP+C H++ WN+ +V LK +Y + LV Y
Sbjct: 360 PTPDLCPAC---HEEIKGLDSWNEGQVLLFLKQHYSRDNLVDAY 400
>ref|XP_415413.1| PREDICTED: similar to quiescin Q6-like 1; putative sulfhydryl
oxidase [Gallus gallus]
Length = 808
Score = 115 bits (287), Expect = 4e-24
Identities = 60/177 (33%), Positives = 91/177 (50%), Gaps = 21/177 (11%)
Query: 294 RGYWMFCRGSKNETRGFSCGLWVLLHSLSVR------------IEDGESQFTFNAICDFV 341
R W+ C+GS+ E RG++C LW L H+L+V+ +ED + + ++
Sbjct: 521 RTEWVGCQGSRPELRGYTCSLWKLFHTLTVQAALRPTALISTGLEDNP-RIVLEVMRRYI 579
Query: 342 HNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFP 401
H+FF C+ C QHF +M VLWLW HN VN RL+ + T DPKFP
Sbjct: 580 HHFFGCKACAQHFEEMAKESMDSVQTLDKAVLWLWEKHNVVNNRLAGDL----TEDPKFP 635
Query: 402 KTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYY-EKTLVSLYKEKDIAGND 457
K WPT LCP+C H++ WN+ +V + +K++Y + ++ Y E +D
Sbjct: 636 KVQWPTPDLCPAC---HEEIKGLHSWNEAQVLQFMKHHYNSENILYRYTESQTDSSD 689
>ref|XP_231083.3| PREDICTED: LIM homeobox protein 3 [Rattus norvegicus]
Length = 1457
Score = 114 bits (284), Expect = 1e-23
Identities = 58/164 (35%), Positives = 83/164 (50%), Gaps = 19/164 (11%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS+ E RG+ C LW L H+L+V+ +G+ Q + ++ FF
Sbjct: 475 WVGCQGSRLELRGYPCSLWKLFHTLTVQASTHPEALVGTGFEGDPQAVLQTMRRYIRTFF 534
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M A VLWLW HN VN RL+ + DPKFPK W
Sbjct: 535 GCKECGEHFEEMAKESMDSVKTADQAVLWLWRKHNMVNSRLAGHLSE----DPKFPKVPW 590
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLY 448
PT LCP C H++ W++++V LK +Y + LV Y
Sbjct: 591 PTPDLCPVC---HEEIKGLDSWDEEQVLVFLKQHYSRDNLVDTY 631
>pir||JC7762 SOX-3 protein - guinea pig gi|12597921|gb|AAB58401.2| GEC-3 [Cavia
porcellus]
Length = 613
Score = 113 bits (283), Expect = 1e-23
Identities = 75/233 (32%), Positives = 111/233 (47%), Gaps = 37/233 (15%)
Query: 233 SLVKFLQVLTAHHPSRRCRKGAGDLLVSFADLYPTDFW--SSHKQEDDKSSVKNFQ---- 286
+L KF+ VLT + P + L+ +F L T+ W HK++ S K
Sbjct: 330 ALKKFVTVLTKYFPGQ-------PLVRNF--LQSTNEWLKRQHKKKMPYSFFKTAMDSRN 380
Query: 287 ---ICGKEVPRGYWMFCRGSKNETRGFSCGLWVLLHSLSVRIED---------GESQFTF 334
+ KEV W+ C+GS++ RGF C LW+L H L+V+ Q
Sbjct: 381 EEAVITKEVN---WVGCQGSESHFRGFPCSLWILFHFLTVQASQKNAESSQKPANGQEVL 437
Query: 335 NAICDFVHNFFVCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLG 394
AI ++V FF C +C HF M + D VLWLW+SHN+VN RL A
Sbjct: 438 QAIRNYVRFFFGCRDCANHFEQMAAGSMHRVKSPNDAVLWLWTSHNRVNARL----AGAP 493
Query: 395 TGDPKFPKTIWPTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSL 447
+ DP+FPK WP +LC +C H++ S + W+ D + LK ++ + + L
Sbjct: 494 SEDPQFPKVQWPPPELCSAC---HNELSGEPVWDVDATLRFLKTHFSPSNIVL 543
Score = 39.3 bits (90), Expect = 0.30
Identities = 23/81 (28%), Positives = 45/81 (55%), Gaps = 7/81 (8%)
Query: 53 LNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAK-LFNGPDAVHPGIILLTR 111
L A + + ++P+ +AV EFFA+WC C + P ++ +AK + + A++ L
Sbjct: 46 LQADTVRSTVLNSPSAWAV-EFFASWCGHCIAFAPTWKALAKDIKDWRPALN-----LAA 99
Query: 112 VDCASKINIKLCDKFSVGHYP 132
++CA + N +C F++ +P
Sbjct: 100 LNCADETNNAVCRDFNIAGFP 120
>ref|NP_989456.1| quiescin Q6 [Gallus gallus] gi|21314229|gb|AAM44079.1|
quiescin/sulfhydryl oxidase [Gallus gallus]
Length = 743
Score = 112 bits (280), Expect = 3e-23
Identities = 64/177 (36%), Positives = 89/177 (50%), Gaps = 18/177 (10%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIEDGESQF-----TFNAICDFVHNFFVCEECR 351
W+ CRGS+ RG+ CGLW + H L+V+ G N + +V +FF C+EC
Sbjct: 404 WVGCRGSEPHFRGYPCGLWTIFHLLTVQAAQGGPDEELPLEVLNTMRCYVKHFFGCQECA 463
Query: 352 QHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIWPTKQLC 411
QHF M + R+ VLWLWS HN+VN RL A T DP+FPK WP +C
Sbjct: 464 QHFEAMAAKSMDQVKSRREAVLWLWSHHNEVNARL----AGGDTEDPQFPKLQWPPPDMC 519
Query: 412 PSCYLGHDQKSNKIEWNQDEVYKALKNYYEKTLVSLYKEKDI----AGNDGTKGAAL 464
P C H ++ W++ V LK ++ +L +LY + I AG + A L
Sbjct: 520 PQC---HREERGVHTWDEAAVLSFLKEHF--SLGNLYLDHAIPIPMAGEEAAASARL 571
Score = 41.6 bits (96), Expect = 0.061
Identities = 24/82 (29%), Positives = 43/82 (52%), Gaps = 5/82 (6%)
Query: 53 LNATNFDAVLKDTPATFAVVEFFANWCPACRNYKPHYEKVAKLFNGPDAVHPGIILLTRV 112
L A + L +P+ +AV EFFA+WC C ++ P + +A+ P +++ +
Sbjct: 55 LGADTAERRLLGSPSAWAV-EFFASWCGHCIHFAPTWRALAE---DVREWRPA-VMIAAL 109
Query: 113 DCASKINIKLCDKFSVGHYPML 134
DCA + N ++C F + +P L
Sbjct: 110 DCADEANQQVCADFGITGFPTL 131
>emb|CAC85331.1| putative sulfhydryl oxidase precursor [Homo sapiens]
Length = 698
Score = 110 bits (276), Expect = 8e-23
Identities = 58/180 (32%), Positives = 86/180 (47%), Gaps = 22/180 (12%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS++E RG+ C LW L H+L+V + + Q + +VH FF
Sbjct: 415 WVGCQGSRSELRGYPCSLWKLFHTLTVEASTHPDALVGTGFEDDPQAVLQTMRRYVHTFF 474
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M +LWLW HN VN RL+ + DP+FPK W
Sbjct: 475 GCKECGEHFEEMAKESMDSVKTPDQAILWLWKKHNMVNGRLAGHLSE----DPRFPKLQW 530
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLYKEKDIAGNDGTKGAAL 464
PT LCP+C H++ W++ V LK +Y + L+ Y D ++G L
Sbjct: 531 PTPDLCPAC---HEEIKGLASWDEGHVLTFLKQHYGRDNLLDTYSADQ---GDSSEGGTL 584
>emb|CAI16881.1| RP11-83N9.4 [Homo sapiens]
Length = 588
Score = 110 bits (276), Expect = 8e-23
Identities = 58/180 (32%), Positives = 86/180 (47%), Gaps = 22/180 (12%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS++E RG+ C LW L H+L+V + + Q + +VH FF
Sbjct: 305 WVGCQGSRSELRGYPCSLWKLFHTLTVEASTHPDALVGTGFEDDPQAVLQTMRRYVHTFF 364
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M +LWLW HN VN RL+ + DP+FPK W
Sbjct: 365 GCKECGEHFEEMAKESMDSVKTPDQAILWLWKKHNMVNGRLAGHLSE----DPRFPKLQW 420
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLYKEKDIAGNDGTKGAAL 464
PT LCP+C H++ W++ V LK +Y + L+ Y D ++G L
Sbjct: 421 PTPDLCPAC---HEEIKGLASWDEGHVLTFLKQHYGRDNLLDTYSADQ---GDSSEGGTL 474
>ref|XP_520361.1| PREDICTED: similar to quiescin Q6-like 1; putative sulfhydryl
oxidase [Pan troglodytes]
Length = 312
Score = 110 bits (276), Expect = 8e-23
Identities = 57/180 (31%), Positives = 86/180 (47%), Gaps = 22/180 (12%)
Query: 297 WMFCRGSKNETRGFSCGLWVLLHSLSVRIE-----------DGESQFTFNAICDFVHNFF 345
W+ C+GS++E RG+ C LW L H+L+V + + Q + ++H FF
Sbjct: 40 WVGCQGSRSELRGYPCSLWKLFHTLTVEASTHPDALVGTGFEDDPQAVLQTMRRYIHTFF 99
Query: 346 VCEECRQHFYDMCSSVSTPFNKARDFVLWLWSSHNKVNERLSKEEASLGTGDPKFPKTIW 405
C+EC +HF +M +LWLW HN VN RL+ + DP+FPK W
Sbjct: 100 GCKECGEHFEEMAKESMDSVKTPDQAILWLWKKHNMVNGRLAGHLSE----DPRFPKLQW 155
Query: 406 PTKQLCPSCYLGHDQKSNKIEWNQDEVYKALKNYYEK-TLVSLYKEKDIAGNDGTKGAAL 464
PT LCP+C H++ W++ V LK +Y + L+ Y D ++G L
Sbjct: 156 PTPDLCPAC---HEEIKGLASWDEGHVLTLLKQHYGRDNLLDTYSADQ---GDSSEGGTL 209
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 933,653,471
Number of Sequences: 2540612
Number of extensions: 39944269
Number of successful extensions: 104071
Number of sequences better than 10.0: 744
Number of HSP's better than 10.0 without gapping: 306
Number of HSP's successfully gapped in prelim test: 438
Number of HSP's that attempted gapping in prelim test: 102946
Number of HSP's gapped (non-prelim): 1220
length of query: 517
length of database: 863,360,394
effective HSP length: 133
effective length of query: 384
effective length of database: 525,458,998
effective search space: 201776255232
effective search space used: 201776255232
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144731.8


