

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.8 - phase: 0
(840 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
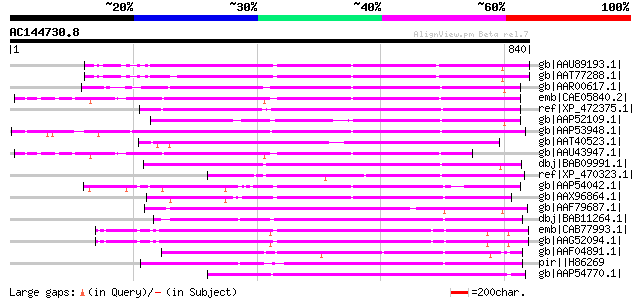
Score E
Sequences producing significant alignments: (bits) Value
gb|AAU89193.1| transposon protein, putative, mutator sub-class [... 386 e-105
gb|AAT77288.1| putative polyprotein [Oryza sativa (japonica cult... 374 e-102
gb|AAR00617.1| hypothetical protein [Oryza sativa (japonica cult... 371 e-101
emb|CAE05840.2| OSJNBa0091C07.2 [Oryza sativa (japonica cultivar... 360 1e-97
ref|XP_472375.1| OSJNBb0118P14.3 [Oryza sativa (japonica cultiva... 350 8e-95
gb|AAP52109.1| putative transposon protein [Oryza sativa (japoni... 325 4e-87
gb|AAP53948.1| putative mutator-like transposase [Oryza sativa (... 322 4e-86
gb|AAT40523.1| putative mutator transposable element [Solanum de... 322 4e-86
gb|AAU43947.1| hypothetical protein [Oryza sativa (japonica cult... 311 6e-83
dbj|BAB09991.1| mutator-like transposase-like [Arabidopsis thali... 302 3e-80
ref|XP_470323.1| putative transposon protein [Oryza sativa (japo... 287 9e-76
gb|AAP54042.1| putative mutator protein [Oryza sativa (japonica ... 280 1e-73
gb|AAX96864.1| transposon protein, putative, mutator sub-class [... 274 8e-72
gb|AAF79687.1| F9C16.9 [Arabidopsis thaliana] gi|25405857|pir||D... 271 9e-71
dbj|BAB11264.1| unnamed protein product [Arabidopsis thaliana] 270 1e-70
emb|CAB77993.1| putative MuDR-A-like transposon protein [Arabido... 265 4e-69
gb|AAG52094.1| putative Mutator-like transposase; 12516-14947 [A... 263 2e-68
gb|AAF04891.1| Mutator-like transposase [Arabidopsis thaliana] g... 256 2e-66
pir||H86269 F21F23.10 protein - Arabidopsis thaliana gi|8920571|... 255 5e-66
gb|AAP54770.1| putative transposon protein [Oryza sativa (japoni... 254 8e-66
>gb|AAU89193.1| transposon protein, putative, mutator sub-class [Oryza sativa
(japonica cultivar-group)]
Length = 1030
Score = 386 bits (992), Expect = e-105
Identities = 236/732 (32%), Positives = 375/732 (50%), Gaps = 55/732 (7%)
Query: 121 DEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANG--NNRVTVNNKTLRI 178
DEED +DD +F DS+ E L+DG +D+ V+ + V K L+
Sbjct: 214 DEEDSGTESDDSDEEFY---DSDYE----LDDGDDDLFVDYVDENVIDEGVAKGKKILKG 266
Query: 179 KKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDS 238
KK + S K N ++++ G DE D D EEL ++D DD
Sbjct: 267 KKA------RGSRLKGN---------LAIVPRG------DESEDTD--EEELNAADSDDE 303
Query: 239 NDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRV 298
+K+ F LN FK+G+ F ++ + ++AI E++V N +IK+ +N+ R+R
Sbjct: 304 GV-RLKFKSFAEEDLNNP-NFKVGLVFPSVEKLRQAITEYSVRNRVEIKMPRNDRRRIRA 361
Query: 299 ECRDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKM 358
C + C + + S+ + F +KT + HTC K S+W+A K + + +KM
Sbjct: 362 HCAEGCPWNLYASEDSRAKAFVVKTCDERHTCQKEWILKRCTSKWLAGKYIEAFRADEKM 421
Query: 359 SVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNV 418
S+ + + + + ++ + RAR+IA I GD +QY ++ Y EL+R ++
Sbjct: 422 SLTSFAKTVQLQWNLTLSRSKLARARRIAMKTIHGDEVEQYKLLWDYGKELRRSNPGSSF 481
Query: 419 KINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDP 478
+ + G F Y D CK+GF++ CRP + +DGCH+KT+YGGQLL AVG DP
Sbjct: 482 FLKLDGSL------FSQCYMSMDACKRGFLNGCRPLICLDGCHIKTKYGGQLLTAVGMDP 535
Query: 479 NDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRY--VFISDQQKGLLSVFDEMFDSI 536
ND +P+AF VVE E TWKWF + L D+G + Y ++D+QKGL+ ++F
Sbjct: 536 NDCIYPIAFAVVEVESLATWKWFLETLKNDLGIENTYPWTIMTDKQKGLIPAVQQVFPES 595
Query: 537 DHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAV 596
+HR C+RHLY+NF+ +F G +++ + +++ Q W + M+ ++ ++ A+EWL +
Sbjct: 596 EHRFCVRHLYSNFQLQF-KGEVLKNQLWACARSSSVQEWNKNMDVMRNLNKSAYEWLEKL 654
Query: 597 EPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATK 656
P W + F+ + KCD+L+NN E FN IL AR+ PI++M E I+ LM R +
Sbjct: 655 PPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIKGQLMTRHFNKQKE 714
Query: 657 L-QKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQ-QFIVDIAKRSCS 714
L ++Q + P RK++ K A + +G G + + R+ Q+IVDI C
Sbjct: 715 LADQFQGLICPKIRKKVLKNADAANTCYALPAGQG----IFQVHEREYQYIVDINAMYCD 770
Query: 715 CNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEA 774
C W++ GIPC H +S L + E + + Y+ ++ Y F I P N W V
Sbjct: 771 CRRWDLTGIPCNHAISCLRHERINAESILPNCYTTDAFSKAYGFNIWPCNDKSKWENVNG 830
Query: 775 PELLPPNYKNGPGRPRKLRIRE----FDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDV 830
PE+ PP Y+ GRP+K R + +NG ++ + GV C + HN CK
Sbjct: 831 PEIKPPVYEKKAGRPKKSRRKAPYEVIGKNGPKLTKHGVMMHYKYCGEENHNSGGCKLKK 890
Query: 831 Q--NPEAAKRKV 840
Q + E AKR V
Sbjct: 891 QGISSEEAKRMV 902
>gb|AAT77288.1| putative polyprotein [Oryza sativa (japonica cultivar-group)]
Length = 1006
Score = 374 bits (959), Expect = e-102
Identities = 234/732 (31%), Positives = 367/732 (49%), Gaps = 64/732 (8%)
Query: 121 DEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANG--NNRVTVNNKTLRI 178
DEED +DD +F DS+ E L+DG +D+ V+ + V K L+
Sbjct: 199 DEEDSGTKSDDSDEEFY---DSDYE----LDDGDDDLFVDYVDENVIDEGVAKGKKILKG 251
Query: 179 KKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDS 238
KK + S K N ++++ G DE D D EEL ++D DD
Sbjct: 252 KKA------RGSRLKGN---------LAIVPRG------DESEDTD--EEELNAADSDDE 288
Query: 239 NDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRV 298
+K+ F LN FK + ++AI E++V N +IK+ +N+ R+R
Sbjct: 289 GV-RLKFKSFAEEDLNNP-NFK---------KLRQAITEYSVRNRVEIKMPRNDRRRIRA 337
Query: 299 ECRDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKM 358
C + C + + S+ + F +KT + HTC K S+W+A K + + +KM
Sbjct: 338 HCEEGCPWNLYTSEDSRAKAFVVKTYDERHTCQKEWILKRCTSKWLAGKYIEAFRADEKM 397
Query: 359 SVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNV 418
S+ + + + + + + RAR+IA I GD +QY ++ Y EL+R +
Sbjct: 398 SLTSFAKTVQLQWNLTPSRSKLARARRIAMKTIHGDEVEQYKLLWDYGKELRRSNPGTSF 457
Query: 419 KINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDP 478
+ + G F Y D CK+GF++ CRP + +DGCH+KT+YGGQLL AVG DP
Sbjct: 458 FLKLDGSL------FSQCYMSMDACKRGFLNGCRPLICLDGCHIKTKYGGQLLTAVGMDP 511
Query: 479 NDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRY--VFISDQQKGLLSVFDEMFDSI 536
ND +P+AF VVE E TWKWF + L D+G + Y ++D+QKGL+ ++F
Sbjct: 512 NDCIYPIAFAVVEVESLATWKWFLETLKNDLGIENTYPWTIMTDKQKGLIPAVQQVFPES 571
Query: 537 DHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAV 596
+HR C+RHLY+NF+ +F G ++ + +++ Q W + M+ ++ ++ A+EWL +
Sbjct: 572 EHRFCVRHLYSNFQLQF-KGEVPKNQLWACARSSSVQEWNKNMDVMRNLNKSAYEWLEKL 630
Query: 597 EPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATK 656
P W + F+ + KCD+L+NN E FN IL AR+ PI++M E I+ LM R +
Sbjct: 631 PPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIKGQLMTRHFNKQKE 690
Query: 657 L-QKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQ-QFIVDIAKRSCS 714
L ++Q + P RK++ K A + +G G + + R+ Q+IVDI C
Sbjct: 691 LADQFQGLICPKIRKKVLKNADAANTCYALPAGQG----IFQVHEREYQYIVDINAMHCD 746
Query: 715 CNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEA 774
C W++ GIPC H +S L + E + + Y+ ++ Y F I P N W +
Sbjct: 747 CRRWDLTGIPCNHAISCLRHERINAESILPNCYTTDAFSKAYGFNIWPCNDKSKWENING 806
Query: 775 PELLPPNYKNGPGRPRKLRIRE----FDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDV 830
PE+ PP Y+ GRP+K R + +NG ++ + GV C C + HN CK
Sbjct: 807 PEIKPPVYEKKAGRPKKSRRKAPYEVIGKNGPKLTKHGVMMHCKYCGEENHNSGGCKLKK 866
Query: 831 Q--NPEAAKRKV 840
Q + E AKR V
Sbjct: 867 QGISSEEAKRMV 878
>gb|AAR00617.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|50901476|ref|XP_463171.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 981
Score = 371 bits (952), Expect = e-101
Identities = 224/720 (31%), Positives = 354/720 (49%), Gaps = 59/720 (8%)
Query: 117 DVGYDEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKTL 176
DV YD + V D F D +D+ DG DV + PA
Sbjct: 182 DVEYDSDGSEVDDSDFVDSDYEFQDDDDDLFEDNVDG--DVVDQGPA------------- 226
Query: 177 RIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPD 236
PKK + KK G++ KGK V+ EE + S+E P+
Sbjct: 227 ---------PKKFNQKKVAGGKL------------KGKRVIREEGSDEESSDEECLELPE 265
Query: 237 DSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRV 296
+ ++ +++ F +N FK+GM F ++ ++ I E+++ N IK+ +N+ R+
Sbjct: 266 NPDEINLRFKSFNPEDMNNPV-FKVGMVFPSVELLRKTITEYSLKNRVDIKMPRNDRTRI 324
Query: 297 RVECRDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTK 356
+ C + C + + S F +KT H C + + W+A K + +
Sbjct: 325 KAHCAEGCPWNMYASLDSRVHGFIVKTYVPQHKCRKEWVLQRCTANWLALKYIESFRADS 384
Query: 357 KMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDN 416
+M++ + + + + + RAR++A I GD QY M+ Y EL+R
Sbjct: 385 RMTLPSFAKTVQKEWNLTPSRSKLARARRLALKEIYGDEVAQYNMLWDYGNELRR----- 439
Query: 417 NVKINVAGPSVTIQLRFGSF---YFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIA 473
+ G S ++L G F YF D CK+GF+S CRP + +DGCH+KT++GGQLL A
Sbjct: 440 ----SNPGSSFYLKLDDGKFSCLYFSLDACKRGFLSGCRPIICLDGCHIKTKFGGQLLTA 495
Query: 474 VGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRY--VFISDQQKGLLSVFDE 531
VG DPND FP+A VVE E TW WF Q L +D+G Y ++D+QKGL+ +
Sbjct: 496 VGIDPNDCIFPIAMAVVEVESFSTWSWFLQTLKDDVGIVNTYPWTIMTDKQKGLIPAVQQ 555
Query: 532 MFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWE 591
+F +HR C+RHLY NF++ F G +++ + +++ Q W K E+K ++ A+
Sbjct: 556 LFPDSEHRFCVRHLYQNFQQSF-KGEILKNQLWACARSSSVQEWNTKFEEMKALNEDAYN 614
Query: 592 WLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMT 651
WL + P W + F+ + KCD+L+NN E FN IL AR+ PI++M E I+ LM R
Sbjct: 615 WLEQMAPNTWVRAFFSDFPKCDILLNNSCEVFNKYILEAREMPILTMLEKIKGQLMTRFF 674
Query: 652 TSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKR 711
+ QKWQ + P RK+L K A +G G FQV ++IVD+ +
Sbjct: 675 NKQKEAQKWQGPICPKIRKKLLKIAEQANICYVLPAGKG-VFQVEE--RGTKYIVDVVTK 731
Query: 712 SCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPE 771
C C W++ GIPC H ++ + + E F+ YS + + Y+ I P N W +
Sbjct: 732 HCDCRRWDLTGIPCCHAIACIREDHLSEEDFLPHCYSINAFKAVYAENIIPCNDKANWEK 791
Query: 772 VEAPELLPPNYKNGPGRPRKLRIREFDE----NGSRMRRQGVAYRCTKCDQFGHNQRRCK 827
+ P++LPP Y+ GRP+K R ++ E NG ++ + GV C+ C + HN++ C+
Sbjct: 792 MNGPQILPPVYEKKVGRPKKSRRKQPQEVQGRNGPKLTKHGVTIHCSYCHEANHNKKGCE 851
>emb|CAE05840.2| OSJNBa0091C07.2 [Oryza sativa (japonica cultivar-group)]
gi|50923321|ref|XP_472021.1| OSJNBa0091C07.2 [Oryza
sativa (japonica cultivar-group)]
Length = 879
Score = 360 bits (923), Expect = 1e-97
Identities = 255/849 (30%), Positives = 407/849 (47%), Gaps = 75/849 (8%)
Query: 8 FHHGGEFVMDK-RIFYRGGVQTVVSGLESNNWGVCDIQNTVASSGYEKNKFRVWSQIEVI 66
FH GGEF D + Y GG QT +S +E + + +I+ GY + + + EVI
Sbjct: 11 FHFGGEFDYDGYSVVYNGG-QTEMSYIERDKVALPEIR------GYLGDHVSLSEEDEVI 63
Query: 67 FEDFFQINEDHIAEEVSGYSVANKVDAHIFVEHNVLDISVKVDVPSFFDLDVGYDEEDVM 126
F F AE +G + +++ + + DV + + ++E+D +
Sbjct: 64 FHWLFPG-----AELNNGLKALSDDKECLYMAQCI----IMGDVAEIYAEILKHNEDDNV 114
Query: 127 VST----------DDDSGKFVGFDDSEDESTT--ALEDGFEDVEVEAPANGNNRVTVNNK 174
V + DD ++ D+ED S T ED E E + A N NK
Sbjct: 115 VLSNGVSDDSDVEDDIEEEYDSHSDAEDSSETDGIAEDDEERSEDDEEAQENREHA--NK 172
Query: 175 TLRIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSD 234
KK+P + + + VS N + +D++ D +L +S
Sbjct: 173 V------------KKNPCATITVTSDNVVLVSDSPNKQDDKGLDDDTDTPFLDSSEEASY 220
Query: 235 PDDSNDDTIKYDQFRMPQLNKDF---KFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKN 291
D + + I+ R P+ + KF+IGM F +FKEA+I++ ++ I+ K+
Sbjct: 221 DDAEDGEAIRRKS-RFPKYDSKAHTPKFEIGMTFAGRAKFKEAVIKYGLVTNRHIRFPKD 279
Query: 292 ESYRVRVECR-DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVP 350
E+ ++R C C + + S + FQ+KT H C +N+ SR +A K
Sbjct: 280 EAKKIRARCSWKDCPWFIFASNGTNCDWFQVKTFNDVHNCPKRRDNRLVTSRRIADKYEH 339
Query: 351 KMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELK 410
++ +Q + + ++ +++ + RA+ I I +Y+ + Y AE+
Sbjct: 340 IIKSNPSWKLQSLKKTVRLDMFADVSISKVKRAKGIVMRRIYDACRGEYSKVFEYQAEIL 399
Query: 411 R--------VCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHL 462
R +C D+ V F Y CFD CKKGF++ CR +G+DGC
Sbjct: 400 RSNPGSTVAICLDHEYNWPV----------FQRMYVCFDACKKGFLAGCRKVIGLDGCFF 449
Query: 463 KTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKR---YVFIS 519
K G+LL A+GRDPN+Q +P+A+ VVE E K+TW WF LL +D+ D +V IS
Sbjct: 450 KGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWFIGLLQKDLNIDPHGAGWVIIS 509
Query: 520 DQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKM 579
DQQKGL+S +E I+HR+C RH+YAN++KK+ + KA+ +
Sbjct: 510 DQQKGLVSAVEEFLPQIEHRMCTRHIYANWRKKY-RDQAFQKPFWKCAKASCRPFFNFCR 568
Query: 580 NELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMA 639
+L ++ + + EP W + F S CD + NN+ ESFN I+ R PIISM
Sbjct: 569 AKLAQLTPAGAKXXXSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMF 628
Query: 640 EWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTY 699
E IR + R+ + +K + W + P K+L+K I L+ + ++ W+G + F+V T
Sbjct: 629 EGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNG-KDGFEV--TD 685
Query: 700 NRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFA 759
+++ VD+ KR+CSC +W++ GIPC H ++AL ++PE ++ D YS Y Y
Sbjct: 686 KDKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCYSVEVYNKIYDHC 745
Query: 760 ISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFDE--NGSRMRRQGVAYRCTKCD 817
+ P+ GM WP P+ PP Y PGRPRK R R+ E G ++ + G RC++C
Sbjct: 746 MMPMEGMMQWPITGHPKPGPPGYVKMPGRPRKERRRDPLEAKKGKKLSKTGTKGRCSQCK 805
Query: 818 QFGHNQRRC 826
Q HN R C
Sbjct: 806 QTTHNIRTC 814
>ref|XP_472375.1| OSJNBb0118P14.3 [Oryza sativa (japonica cultivar-group)]
gi|38346143|emb|CAD40681.2| OSJNBb0118P14.3 [Oryza
sativa (japonica cultivar-group)]
Length = 939
Score = 350 bits (899), Expect = 8e-95
Identities = 202/631 (32%), Positives = 331/631 (52%), Gaps = 28/631 (4%)
Query: 211 GKGKYVVDEEIDGDYLSEE---LGSSDPDDSNDDTIKYDQFRMPQLNKDFK---FKIGME 264
GK + ++E DY+S E L D + ++ IK+ F+ + N D + FK+ +
Sbjct: 204 GKIELAHNKEEGSDYVSSEDERLMMPDISEEEEEKIKFS-FKSFRANVDMETPEFKVSIM 262
Query: 265 FNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQCGYKVLCSKVGDRRTFQIKTL 324
F +VE ++AI ++ + N IK +N R+ +C + C + + S + ++
Sbjct: 263 FADVVELRKAIDQYTIKNQVAIKKTRNTKTRIEAKCAEGCPWMLSASMDNRVKCLVVREY 322
Query: 325 EGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRAR 384
HTC+ E K+ ++++AK+ + + + KM++ ++ + + + RAR
Sbjct: 323 IEKHTCSKQWEIKAVTAKYLAKRYIEEFRDNDKMTLMSFAKKIQKELHLTPSRHKLGRAR 382
Query: 385 KIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSF---YFCFD 441
++A I GD QY + Y EL R +N G S + L FG F Y FD
Sbjct: 383 RMAMRAIYGDEISQYNQLWDYGQEL----RTSN-----PGSSFYLNLHFGCFHTLYMSFD 433
Query: 442 GCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWF 501
CK+GF+ CRP + +DGCH+KT++GG +L AVG DPND FP+A VVE E ++W WF
Sbjct: 434 ACKRGFMFGCRPIICLDGCHIKTKFGGHILTAVGMDPNDCIFPIAIAVVEVESLKSWSWF 493
Query: 502 TQLLMEDIG--EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQI 559
L +D+G + ++D+QKGL+ F + R C+RHLY NF+ G +
Sbjct: 494 LDTLKKDLGIENTSAWTVMTDRQKGLVPAVRREFSDAEQRFCVRHLYQNFQVLHKGETLK 553
Query: 560 RDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNI 619
L A +T + W M ++K + A+++L + P +WC+ F+ + KCD+L+NN
Sbjct: 554 NQLWAIARSSTVPE-WNANMEKMKALSSEAYKYLEEIPPNQWCRAFFSDFPKCDILLNNN 612
Query: 620 SESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQK-WQHNVMPMPRKRLDKEITL 678
SE FN IL AR+ PI+SM E IR+ +M R+ T +L++ W + P +++++K +
Sbjct: 613 SEVFNKYILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPCGICPKIKRKVEKNTEM 672
Query: 679 AAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQR 738
A +G+G FQV Q+IV++ + C C W++ GIPC H +S L + +
Sbjct: 673 ANTCYVFPAGMG-AFQVSDI--GSQYIVELNVKRCDCRRWQLTGIPCNHAISCLRHERIK 729
Query: 739 PEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFD 798
PE V YS Y YS+ I P+ W +++ E+ PP Y+ GRP+K R ++
Sbjct: 730 PEDVVSFCYSTRCYEQAYSYNIMPLRDSIHWEKMQGIEVKPPVYEKKVGRPKKTRRKQPQ 789
Query: 799 --ENGSRMRRQGVAYRCTKCDQFGHNQRRCK 827
E G+++ + GV C+ C GHN+ CK
Sbjct: 790 ELEGGTKISKHGVEMHCSYCKNGGHNKTSCK 820
>gb|AAP52109.1| putative transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37531040|ref|NP_919822.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)] gi|14488302|gb|AAK63883.1| Putative
transposon protein [Oryza sativa]
Length = 841
Score = 325 bits (833), Expect = 4e-87
Identities = 193/606 (31%), Positives = 306/606 (49%), Gaps = 57/606 (9%)
Query: 228 EELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIK 287
E+L D DD + +K+ F M + K+ FK+GM F ++ ++AI E+++ IK
Sbjct: 150 EDLQLPDSDDDGEVRLKFKAF-MAEDVKNLVFKVGMVFPSVEVLRKAITEYSLKARVDIK 208
Query: 288 VLKNESYRVRVECRDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKK 347
+ +NE R+R C + C + + S ++ +KT G H C K ++W+++K
Sbjct: 209 MPRNEQKRLRAHCVEGCPWNLYASFDSRSKSMMVKTYLGEHKCQKEWVLKRCTAKWLSEK 268
Query: 348 AVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAA 407
+ + KM++ G + RAR++A I GD +QY + Y A
Sbjct: 269 YIETFRANDKMTLG------------GFAKSKLARARRLAFKDIYGDEIQQYNQLWNYGA 316
Query: 408 ELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYG 467
EL+R + +N+ + F + Y FD CK+G +S CRP + +DGCH+KT++G
Sbjct: 317 ELRRSNPGSCFFLNL------VDSCFSTCYMSFDACKRGSLSGCRPLICLDGCHIKTKFG 370
Query: 468 GQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRY--VFISDQQKGL 525
GQ+L AVG DPND +P+A VVETE +W+WF Q L ED+G D Y ++D+QK
Sbjct: 371 GQILTAVGIDPNDCIYPIAIVVVETESLRSWRWFLQTLKEDLGIDNTYPWTIMTDKQK-- 428
Query: 526 LSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKI 585
+FK G +++ + +++ W M E+K +
Sbjct: 429 ---------------------VHFK-----GENLKNQLWACARSSSEVEWNANMEEMKSL 462
Query: 586 DLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHY 645
+ A+EWL + PK W K F+ + KCD+L+NN E FN IL AR+ PI+SM E I+
Sbjct: 463 NQDAYEWLQKMPPKTWVKAYFSEFPKCDILLNNNCEVFNKYILEARELPILSMFEKIKSQ 522
Query: 646 LMRRMTTSATKL-QKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQF 704
L+ R + ++ ++W + P RK++ K +A +G G FQV ++
Sbjct: 523 LISRHYSKQKEVAEQWHGPICPKIRKKVLKNADMANTCYVLPAGKG-IFQVED--RNFKY 579
Query: 705 IVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPIN 764
IVD++ + C C W++ GIPC H +S L + E + YS ++ Y+F I P N
Sbjct: 580 IVDLSAKHCDCRRWDLTGIPCNHAISCLRSERISAESILPPCYSLEAFSRAYAFNIWPYN 639
Query: 765 GMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFDE----NGSRMRRQGVAYRCTKCDQFG 820
M W +V PE+ PP Y+ GRP K R + E NG +M + GV C+ C +
Sbjct: 640 DMTKWVQVNGPEVKPPIYEKKVGRPPKSRRKAPHEVQGKNGPKMSKHGVEMHCSFCKEPR 699
Query: 821 HNQRRC 826
HN++ C
Sbjct: 700 HNKKGC 705
>gb|AAP53948.1| putative mutator-like transposase [Oryza sativa (japonica
cultivar-group)] gi|37534718|ref|NP_921661.1| putative
mutator-like transposase [Oryza sativa (japonica
cultivar-group)]
Length = 1005
Score = 322 bits (824), Expect = 4e-86
Identities = 240/881 (27%), Positives = 404/881 (45%), Gaps = 71/881 (8%)
Query: 3 LFDAVFHHGGEFVMDK-RIFYRGGVQTVVSGLESNNWGVCDIQNTVASSGYEKNKFRVW- 60
+ + FH GEFV+D+ ++ Y G VS +E + + +++ + R++
Sbjct: 6 VLEVQFHFNGEFVLDESKMSYCNG-DCGVSHIEKDKISIPELEGHLLDHTTFPQSVRMYW 64
Query: 61 ------------------SQIEVIFE-------DFFQINEDHIAEEVSGYSVANKVDAHI 95
S +++I E D + + D E SG A++ +
Sbjct: 65 LPFGAELNSGMRLLVDDKSCLDMIHELGTATTVDIYTESVDMGGNEESGTQYADEDVFAL 124
Query: 96 FVEHNVLDISVKVDVPSFFDLDVGYDEEDVMVSTDDDSGKFVGFDDS------EDESTTA 149
F + N++D+ + E V T + KFV D+ +
Sbjct: 125 FQDDNIMDLD--------------HAEPIVKEQTIQNRDKFVQGDNMLCLEGPTHGQDSP 170
Query: 150 LEDGFEDVEVEAPANGNNRVTVNNKTLRIKKCGSKTPKKKSPKKNNSGRMNVIAPVSL-- 207
E+ ED+E E+ T +++ +++ +K + S K G + + P+ +
Sbjct: 171 NEEAAEDLEAESDGCDATGFT-SDEDDEVREIRTKYKEFMSEVKKR-GDIPIDNPIEVDG 228
Query: 208 LKNGK---GKYVVDEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDF---KFKI 261
++ G G V E DG + G + D+ +D + R P + +F +
Sbjct: 229 IQGGDIPLGNNQVLEGGDGAEYFDSDGDASYDEDSDGVFTRRKCRFPIFDSFADTPQFAV 288
Query: 262 GMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTFQ 320
M F + K+AI + + I+ +KNE R+R CR + C + + S FQ
Sbjct: 289 DMCFRGKDQLKDAIERYALKKKINIRYVKNEQKRIRAVCRWKGCPWLLYASHNSRSDWFQ 348
Query: 321 IKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRA 380
I T H C P L+NK ++R + + ++ +E+ + + GV +++
Sbjct: 349 IVTYNPNHACCPELKNKRLSTRRICDRYESTIKANPSWKAREMKETVQEDMGVDVSITMI 408
Query: 381 WRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCF 440
RA+ I +Y+ + YA +L+R N+V I + P + F FY CF
Sbjct: 409 KRAKTHVMKKIMDTQTGEYSKLFDYALKLQRSNPGNSVHIAL-DPEEEDHV-FQRFYVCF 466
Query: 441 DGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKW 500
D C++GF+ CR +G+DGC LK G+LL A+GRD N+Q +P+A+ VVE E K++W W
Sbjct: 467 DACRRGFLEGCRRIIGLDGCFLKGPLKGELLSAIGRDANNQLYPIAWAVVEYENKDSWNW 526
Query: 501 FTQLLMEDIG---EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGS 557
F L +DI +VFI+DQQKGLLS+ +F +HR+C RH+YAN++KK
Sbjct: 527 FLGHLQKDINIPVGAAGWVFITDQQKGLLSIVSTLFPFAEHRMCARHIYANWRKKH-RLQ 585
Query: 558 QIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMN 617
+ + KA Q + +L W+ L P W + F S C+ + N
Sbjct: 586 EYQKRFWKIAKAPNEQLFNHYKRKLAAKTPRGWQDLEKTNPIHWSRAWFRLGSNCESVDN 645
Query: 618 NISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEIT 677
N+SESFN I+ +R +PII+M E IR + RR+ + + ++W V P ++ +K
Sbjct: 646 NMSESFNSWIIESRFKPIITMLEDIRIQVTRRIQENRSNSERWTMTVCPNIIRKFNKIRH 705
Query: 678 LAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQ 737
W+G F+V + +F VD+ ++CSC +W++ GIPC+H +AL K Q
Sbjct: 706 RTQFCHVLWNG-DAGFEVRD--KKWRFTVDLTSKTCSCRYWQVSGIPCQHACAALFKMAQ 762
Query: 738 RPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREF 797
P + + +S +Y Y + P+ WP P+ LPP K PGRP+K R ++
Sbjct: 763 EPNNCIHECFSLERYKKTYQHVLQPVEHESAWPVSPNPKPLPPRVKKMPGRPKKNRRKDP 822
Query: 798 D---ENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDVQNPEA 835
++G++ + G RC +C +GHN R C +Q A
Sbjct: 823 SKPVKSGTKSSKVGTKIRCRRCGNYGHNSRTCCVKMQQEHA 863
>gb|AAT40523.1| putative mutator transposable element [Solanum demissum]
Length = 707
Score = 322 bits (824), Expect = 4e-86
Identities = 195/602 (32%), Positives = 313/602 (51%), Gaps = 43/602 (7%)
Query: 209 KNGKGKYVVDEEIDGDYLSEELGSSDPDD-------SNDDTIKYDQFRMPQLNKDF---- 257
KN + V EE+ +EE G SD D ++D +++ P+ N
Sbjct: 124 KNDGNQNYVSEEMHRKMQNEE-GDSDCVDFDDTKSLNSDCDSEFEDCNFPKHNPKIGAFN 182
Query: 258 -KFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECR-DQCGYKVLCSKVG- 314
+ ++GM + EFKEA++ G I+ K++ R R +CR + C +++L S +
Sbjct: 183 PELELGMVLDNKKEFKEAVVANQAKIGKSIRWTKDDRERARAKCRTNACKWRILGSLMQR 242
Query: 315 DRRTFQIKTLEGPHTCAPV-LENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGV 373
D TFQIKT HTC NK+ NS W+A+K V +++ K E + ++
Sbjct: 243 DISTFQIKTFVSEHTCFGWNYNNKTINSSWIARKYVDRVKSNKNWRTSEFRDTLSRELKF 302
Query: 374 GITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQ-LR 432
++M +A RA++ A ++I+GD + Q+ ++ Y E+ R +V + + + + +R
Sbjct: 303 HVSMHQARRAKEKAIAMIDGDINDQFGILWNYCNEIVRTNPGTSVFMKLTPNEILNKPMR 362
Query: 433 FGSFYFCFDGCKKGFISACRPFVGVDGCHLKT-RYGGQLLIAVGRDPNDQYFPLAFGVVE 491
F FY CF CK GF + CR +GV+GC LK YG QLL AV D N+ FP+A+ +VE
Sbjct: 363 FQRFYICFAACKAGFKADCRKIIGVNGCWLKNIMYGAQLLSAVTLDGNNNIFPIAYAIVE 422
Query: 492 TECKETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKK 551
E KE W+WF LM D+ +++Y+F C+RHL+ NFK+
Sbjct: 423 KENKEIWQWFLTYLMNDLEIEEQYLF-----------------------CVRHLHNNFKR 459
Query: 552 KFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSK 611
G +++ + A AT + M +L ++D A+ WL A P +W + F+ K
Sbjct: 460 AGYSGMALKNALWKAASATTIDRFDACMTDLFELDKDAYAWLSAKVPSEWSRSHFSPLPK 519
Query: 612 CDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHN-VMPMPRK 670
CD+L+NN E FN IL ARD+PI+ + E IRH LM R+ + K +KW N + P +K
Sbjct: 520 CDILLNNQCEVFNKFILDARDKPIVKLLETIRHLLMTRINSIREKAEKWNLNDICPTIKK 579
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
+L K + AA++ S + ++V+ + VD+ R+CSC WE+ G+PC+H +S
Sbjct: 580 KLAKTMKKAANYIPQRSNMW-NYEVIGPVEGDNWAVDLYNRTCSCRQWELSGVPCKHAIS 638
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPR 790
++ + +VDD Y Y Y +I P+NG+D+WP+ P PP+Y N + +
Sbjct: 639 SIWLKNDEVLNYVDDCYKVDTYRKIYEASILPMNGLDLWPKSLNPPPFPPSYLNNKKKGK 698
Query: 791 KL 792
K+
Sbjct: 699 KM 700
>gb|AAU43947.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 792
Score = 311 bits (797), Expect = 6e-83
Identities = 226/769 (29%), Positives = 370/769 (47%), Gaps = 73/769 (9%)
Query: 8 FHHGGEFVMDK-RIFYRGGVQTVVSGLESNNWGVCDIQNTVASSGYEKNKFRVWSQIEVI 66
FH GGEF D + Y GG QT +S +E + + +I+ GY + + + EVI
Sbjct: 11 FHFGGEFDYDGYSVVYNGG-QTEMSYIERDKVALPEIR------GYLGDHVSLSEEDEVI 63
Query: 67 FEDFFQINEDHIAEEVSGYSVANKVDAHIFVEHNVLDISVKVDVPSFFDLDVGYDEEDVM 126
F F AE +G + +++ + + DV + + ++E+D +
Sbjct: 64 FHWLFPG-----AELNNGLKALSDDKECLYMAQCI----IMGDVAEIYAEILKHNEDDNV 114
Query: 127 VST----------DDDSGKFVGFDDSEDESTT--ALEDGFEDVEVEAPANGNNRVTVNNK 174
V + DD ++ D+ED S T ED E E + A N NK
Sbjct: 115 VLSNGVSDDSDVEDDIEEEYDSHSDAEDSSETDGIAEDDEERSEDDEEAQENREHA--NK 172
Query: 175 TLRIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSD 234
KK+P + + + VS N + +D++ D +L +S
Sbjct: 173 V------------KKNPCATITVTSDNVVLVSDSPNKQDDKGLDDDTDTPFLDSSEEASY 220
Query: 235 PDDSNDDTIKYDQFRMPQLNKDF---KFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKN 291
D + + I+ R P+ + KF+IGM F +FKEA+I++ ++ I+ K+
Sbjct: 221 DDAEDGEAIRRKS-RFPKYDSKAHTPKFEIGMTFAGRAKFKEAVIKYGLVTNRHIRFPKD 279
Query: 292 ESYRVRVECR-DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVP 350
E+ ++R C C + + S + FQ+KT H C +N+ SR +A K
Sbjct: 280 EAKKIRARCSWKDCPWFIFASNGTNCDWFQVKTFNDVHNCPKRRDNRLVTSRRIADKYEH 339
Query: 351 KMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELK 410
++ +Q + + ++ +++ + RA+ I I +Y+ + Y AE+
Sbjct: 340 IIKSNPSWKLQSLKKTVRLDMFADVSISKVKRAKGIVMRRIYDACRGEYSKVFEYQAEIL 399
Query: 411 R--------VCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHL 462
R +C D+ V F Y CFD CKKGF++ CR +G+DGC
Sbjct: 400 RSNPGSTVAICLDHEYNWPV----------FQRMYVCFDACKKGFLAGCRKVIGLDGCFF 449
Query: 463 KTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKR---YVFIS 519
K G+LL A+GRDPN+Q +P+A+ VVE E K+TW WF LL +D+ D +V IS
Sbjct: 450 KGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWFIGLLQKDLNIDPHGAGWVIIS 509
Query: 520 DQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKM 579
DQQKGL+S +E I+HR+C RH+YAN++KK+ + KA+ +
Sbjct: 510 DQQKGLVSAVEEFLPQIEHRMCTRHIYANWRKKY-RDQAFQKPFWKCAKASCRPFFNFCR 568
Query: 580 NELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMA 639
+L ++ + +M+ EP W + F S CD + NN+ ESFN I+ R PIISM
Sbjct: 569 AKLAQLTPAGAKDMMSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMF 628
Query: 640 EWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTY 699
E IR + R+ + +K + W + P K+L+K I L+ + ++ W+G + F+V T
Sbjct: 629 EGIRTKVYVRIQQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNG-KDGFEV--TD 685
Query: 700 NRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYS 748
+++ VD+ KR+CSC +W++ GIPC H ++AL ++PE ++ D YS
Sbjct: 686 KDKRYTVDLEKRTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCYS 734
>dbj|BAB09991.1| mutator-like transposase-like [Arabidopsis thaliana]
Length = 825
Score = 302 bits (774), Expect = 3e-80
Identities = 185/634 (29%), Positives = 311/634 (48%), Gaps = 29/634 (4%)
Query: 217 VDEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAII 276
V++ + G EE + + N D + ++ + +I F+++ EFKEA+I
Sbjct: 157 VEQNLAGFVDEEEHDNDESTPLNSDCEEGERRYVRCKKGSGDIRIEQVFDSIAEFKEAVI 216
Query: 277 EWNVLNGYQIKVLK--NESYRVRVECRDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVL 334
++ + G IK + +E VR C +++ C+ + +KT H C
Sbjct: 217 DYALKKGVNIKFTRWGSEKSEVRCSIGGNCKFRIYCAYDEKIGVYMVKTFIDEHACTKDG 276
Query: 335 ENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGD 394
K SR + + ++ V+ + + Y + + D+ +AR A +I+ +
Sbjct: 277 FCKVLKSRIIVDMFLNDIRQDPTFKVKAMQKAIEERYSLIASSDQCRKARGKALKMIQDE 336
Query: 395 ADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQ--LRFGSFYFCFDGCKKGFISACR 452
D+Q+A I Y +L + N+ V ++TI ++F FY CFD +K +++ CR
Sbjct: 337 YDEQFARIHDYKEQLL----ETNLGSTVEVETITIDGIVKFDKFYVCFDALRKTWLAYCR 392
Query: 453 PFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIG-- 510
P +G+DGC LK GQLL AVGRD N+Q++P+A+ VVETE ++W WF + L D+
Sbjct: 393 PIIGIDGCFLKNNMKGQLLAAVGRDANNQFYPVAWAVVETENIDSWLWFIRKLKSDLKLQ 452
Query: 511 EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKAT 570
+ + ISD+QKGLL+ D+ ++HR+C RH+Y N ++ + G ++L K+
Sbjct: 453 DGDGFTLISDRQKGLLNTVDQELPKVEHRMCARHIYGNLRRVYPGKDLSKNLFCAVAKSF 512
Query: 571 YNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSA 630
+ + ++ELK+ D G ++ +M P+ + F S C+ + NN SES+N I A
Sbjct: 513 NDYEYNRAIDELKQFDQGVYDAVMMRNPRNCSRAFFGCKSSCEDVSNNFSESYNNAINKA 572
Query: 631 RDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIG 690
R+ P++ M E IR M R+ K K Q + +E + + G G
Sbjct: 573 REMPLVEMLETIRRQTMIRIDMRLRKAIKHQGKFTLKVANTIKEETIYRKYCQVVPCGNG 632
Query: 691 EQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVD-DYYSR 749
QF+V+ N F VD+ ++C+C W + GIPCRHV+ + +K PE V+ D+Y
Sbjct: 633 -QFEVLE--NNHGFRVDMNAKTCACRRWNMTGIPCRHVLRIILNKKVNPEELVNSDWYLA 689
Query: 750 SKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLR---------------I 794
SK YS +IS +NG+ W P + PP G + K + +
Sbjct: 690 SKNLKIYSESISGVNGIGFWIRSGEPRIQPPPRPTGTKKKEKRKKGKNESPKKKKKGKGV 749
Query: 795 REFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKS 828
+E E +++ ++G + RC +C GHN R C S
Sbjct: 750 QEPPEKRTKVSKEGRSIRCGRCSTEGHNTRSCPS 783
>ref|XP_470323.1| putative transposon protein [Oryza sativa (japonica
cultivar-group)] gi|40714668|gb|AAR88574.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 839
Score = 287 bits (735), Expect = 9e-76
Identities = 169/524 (32%), Positives = 271/524 (51%), Gaps = 22/524 (4%)
Query: 320 QIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDR 379
+I T G H+C P L+NK ++ + K ++ +E+ + GV ++M
Sbjct: 233 KIVTYNGNHSCCPDLKNKRLSTSRICDKYESVIKANPSWKARELKETVQEEMGVDVSMTM 292
Query: 380 AWRAR-KIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFYF 438
RA+ ++ K +++ + +Y+ + YA EL R + ++VA + F FY
Sbjct: 293 VKRAKSRVIKKVMDAHSG-EYSRLFDYALELMR--SNPGSSVHVALDPDENEHVFQRFYV 349
Query: 439 CFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETW 498
C D C++GF+ CR +G DGC LK G+LL AVGRD N+Q +P+A+ VVE E +W
Sbjct: 350 CLDACRRGFLDGCRRVIGFDGCFLKGVVKGELLSAVGRDANNQIYPIAWAVVEYENASSW 409
Query: 499 KWFTQLLMEDI----GEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKF- 553
WF L +D+ G D +VF++DQQKGLLSV + +F +HR+C RH+YAN++K+
Sbjct: 410 NWFLGHLQKDLNIPYGGD-GWVFLTDQQKGLLSVIEHLFPKAEHRMCARHIYANWRKRHR 468
Query: 554 --GGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSK 611
+ + + +N K K +G WE L P WC+ F S
Sbjct: 469 LQEYQKRFWKIARSSNAVLFNH---YKSKLANKTPMG-WEDLEKTNPIHWCRAWFKLGSN 524
Query: 612 CDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKR 671
CD + NNI ESFN I+ AR +PII+M E IR + RR+ + T ++W + P K+
Sbjct: 525 CDSVENNICESFNNWIIEARFKPIITMLEDIRMKVTRRIQENKTNSERWTMGICPNILKK 584
Query: 672 LDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSA 731
++K W+G F+V + +F VD++ +CSC +W+I GIPC+H +A
Sbjct: 585 INKIRHATQFCHVLWNG-SSGFEVRE--KKWRFTVDLSANTCSCRYWQISGIPCQHACAA 641
Query: 732 LGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRK 791
K + P V+ +S +Y Y + P+ +WP P LPP K PG P++
Sbjct: 642 YFKMAEEPNNHVNMCFSIDQYRNTYQDVLQPVEHESVWPLSTNPRPLPPRVKKMPGSPKR 701
Query: 792 LRIREFDE---NGSRMRRQGVAYRCTKCDQFGHNQRRCKSDVQN 832
R ++ E + ++ ++G + +C C + GHN R CK +++
Sbjct: 702 ARRKDPTEAAGSSTKSSKRGGSVKCGFCHEKGHNSRGCKKKMEH 745
>gb|AAP54042.1| putative mutator protein [Oryza sativa (japonica cultivar-group)]
gi|37534906|ref|NP_921755.1| putative mutator protein
[Oryza sativa (japonica cultivar-group)]
Length = 929
Score = 280 bits (716), Expect = 1e-73
Identities = 227/785 (28%), Positives = 346/785 (43%), Gaps = 119/785 (15%)
Query: 120 YDEEDVMV----STDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKT 175
Y EE +V S+DDDS + + E E G E E G T N+
Sbjct: 100 YAEEPTIVDLCDSSDDDSSYEADESEDDSEEMEVAEGGMEGTENAVALEGGEIRTKTNEV 159
Query: 176 LRIKKCGSKTPK------------------------------------------------ 187
+ K S+ P+
Sbjct: 160 RKGKAISSEIPENRLVVYNCDNDPLASQAPSESESNSEYIPGDDAGSDDDEEIIEIEKHY 219
Query: 188 KKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDSNDDTIKYD- 246
K +K +G+++ + V L G+G ++ E G+ + + + P+D + + I D
Sbjct: 220 KDMKRKVKAGQLDNLDDVYL--GGQGWQNMNNEAGGE--GDGIAAESPEDESMEEIGTDG 275
Query: 247 -----QFRMPQLNKDFK---FKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRV 298
+ + P+ K F++GM+F++ +FK+A+ ++ + I +K++ RVR
Sbjct: 276 EVSTKENKFPRYKKKVGTPCFELGMKFSSKKQFKKAVTKYALAERKVINFIKDDLKRVRA 335
Query: 299 ECR-DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKK------AVPK 351
+C C + L SK ++QI TLE H C P +NK + +A+K A P
Sbjct: 336 KCDWASCPWVCLLSKNTRSDSWQIVTLESLHACPPRRDNKMVTATRIAEKYGKIILANPG 395
Query: 352 MQVTK-KMSVQE-VFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADK-QYAMIRRYAAE 408
+ K +VQE +F E +V + RA +A + K++ DA K QY + Y E
Sbjct: 396 WNLAHMKATVQEEMFGEASVP-----KLKRA-KAMVMKKAM---DATKGQYQKLYNYQLE 446
Query: 409 LKRVCRDNNVKINV---AGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTR 465
L R + V +N P V F Y C D CK+GF+ CR VG+DGC K
Sbjct: 447 LLRSNPGSTVVVNREIGTDPPV-----FKRIYICLDACKRGFVDGCRKVVGLDGCFFKGA 501
Query: 466 YGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIG--EDKRYVFISDQQK 523
G+LL A+GRD N+Q +P+A+ VV E E W WF LL D+ + +VFISDQQK
Sbjct: 502 TNGELLCAIGRDANNQMYPIAWAVVHKENNEEWDWFLDLLCSDLKVLDGTGWVFISDQQK 561
Query: 524 GLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELK 583
G+++ ++ +HR C RH+YAN+K+ F + + KA + +L
Sbjct: 562 GIINAVEKWAPQAEHRNCARHIYANWKRHF-SDKEFQKKFWRCAKAPCRMLFNLARAKLA 620
Query: 584 KIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIR 643
++ + ++ P+ W + F S CD + NN+ ESFN IL AR PII+M E IR
Sbjct: 621 QVTPAGAQAILNTHPEHWSRAWFKLGSNCDSVDNNMCESFNKWILEARFFPIITMLETIR 680
Query: 644 HYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQ 703
+M R++ +W V P K+L+ IT +A+ + +G + F+V H NR
Sbjct: 681 RKVMVRISEQRKVSARWNTVVCPGILKKLNVYITESAYCHAICNG-ADSFEVKHHTNR-- 737
Query: 704 FIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPI 763
F V + K+ CSC R + F D Y + Y + P+
Sbjct: 738 FTVQLDKKECSC-------------------RTNTLDEFTADCYKVDAFKSTYKHCLLPV 778
Query: 764 NGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFDE--NGSRMRRQGVAYRCTKCDQFGH 821
GM+ WPE + L P Y PGRPR R RE E S+ + G RC C Q GH
Sbjct: 779 EGMNAWPEDDREPLTAPGYIKMPGRPRTERRREAHEPAKPSKASKFGTKVRCRTCKQVGH 838
Query: 822 NQRRC 826
N+ C
Sbjct: 839 NKSSC 843
>gb|AAX96864.1| transposon protein, putative, mutator sub-class [Oryza sativa
(japonica cultivar-group)]
Length = 868
Score = 274 bits (701), Expect = 8e-72
Identities = 192/613 (31%), Positives = 293/613 (47%), Gaps = 38/613 (6%)
Query: 222 DGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFK-------FKIGMEFNTLVEFKEA 274
+GD E SD ++S ++ + N FK F++GM+F+ +FK+A
Sbjct: 196 EGDGNDTEYIDSDDEESVEEGSDGELRVRGSNNARFKKKPGYPTFELGMKFSCKSQFKKA 255
Query: 275 IIEWNVLNGYQIKVLKNESYRVRVECR-DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPV 333
I + + I +K++ RVR +C C + L S +QI T E H C P
Sbjct: 256 ITAYALAERKVINFVKDDPKRVRAKCDWHSCPWVCLLSNNSRSDGWQIVTFENFHACPPR 315
Query: 334 LENKSANSRWVAKK------AVPKMQVTK-KMSVQEVFNEMAVNYGVGITMDRAWRARKI 386
+NK + +A K A P + K ++QE EM N +++ + RA+ I
Sbjct: 316 RDNKLVTATRIADKYGKFIIANPSWPLAHMKATIQE---EMFAN----VSVSKLKRAKSI 368
Query: 387 AKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVA---GPSVTIQLRFGSFYFCFDGC 443
QY + Y EL R + V +N P V F Y C D C
Sbjct: 369 VMKKAMDATKGQYQKLYNYQKELLRSNPGSTVVVNREVDMDPPV-----FKRIYICLDAC 423
Query: 444 KKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQ 503
K+GFIS CR +G+DGC K G+LL A+GRD N+Q + +A+ V+ E E W WF
Sbjct: 424 KRGFISGCRKVIGLDGCFFKGATNGKLLCAIGRDANNQMYLVAWAVIHKENNEEWDWFLD 483
Query: 504 LLMED--IGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRD 561
LL D +G+ +VFISDQQKG+++ ++ +HR C RH+YA++K+ F ++
Sbjct: 484 LLCGDLKVGDGSGWVFISDQQKGIINAVEKWAPEAEHRNCARHIYADWKRHF-NEKILQK 542
Query: 562 LMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISE 621
KA + +L ++ + +M P+ W + F S CD + NN+ E
Sbjct: 543 KFWRCAKAPCILLFNLARAKLAQLTPPGAQAIMNTHPQHWSRAWFRLGSNCDSVDNNLCE 602
Query: 622 SFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAH 681
SFN IL AR PII+M E IR +M R+ T +W + P K+L+ IT +A
Sbjct: 603 SFNKWILEARFFPIITMLETIRRKVMVRIHDQITTSARWNTAICPGILKKLNAYITKSAF 662
Query: 682 WKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEM 741
+ +G ++V H NR F V + K +CS +W++ G+PC H +S + + +
Sbjct: 663 SHAICNG-ASSYEVKHHDNR--FTVQLDKMACSYRYWQLSGLPCPHAISCIFFKTNSLDG 719
Query: 742 FVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKLRIREFDE-- 799
++ D YS +++ YS + P GM+ WP + L P Y PGRPR R RE E
Sbjct: 720 YISDCYSVTEFKKTYSHCLEPFEGMNNWPYDDRQPLNAPGYIKMPGRPRTERRREPTEAP 779
Query: 800 NGSRMRRQGVAYR 812
+RM + G R
Sbjct: 780 KATRMSKMGTKIR 792
>gb|AAF79687.1| F9C16.9 [Arabidopsis thaliana] gi|25405857|pir||D96503 protein
F9C16.9 [imported] - Arabidopsis thaliana
Length = 946
Score = 271 bits (692), Expect = 9e-71
Identities = 172/641 (26%), Positives = 301/641 (46%), Gaps = 34/641 (5%)
Query: 218 DEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIE 277
D+ D + + + L SD + NDD Q P+ + ++ FN +FK A++
Sbjct: 200 DDIWDDERIPDPLVHSDDEGDNDDEDGAPQTDDPEES----LRLAKTFNNPEDFKIAVLR 255
Query: 278 WNVLNGYQIKVLKNESYRVRVECRD---QCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVL 334
+++ Y IK+ +++S RV +C D C ++ CS ++ Q+K + H+C
Sbjct: 256 YSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDGHSCVRSG 315
Query: 335 ENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGD 394
+K R +A +++ K++ QE+ +E+ Y + +T D+ +A+ + +
Sbjct: 316 YSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTEDQCSKAKTMVMRERQAT 375
Query: 395 ADKQYAMIRRYAAELKRVCRDNNVKIN-VAGPSVTIQLRFGSFYFCFDGCKKGFISACRP 453
+ +A I Y A++ R +I + GP++ RF + CF K+ + CRP
Sbjct: 376 HQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQRFLRLFICFKSQKESWKQTCRP 435
Query: 454 FVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLME--DIGE 511
+G+DG LK G LL A GRD +++ P+A+ VVE E + W WF ++L D+ +
Sbjct: 436 IIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDDNWDWFVRMLSRTLDLQD 495
Query: 512 DKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATY 571
+ ISD+Q GL+ + +HR C RH+ N+K+ +++ L ++
Sbjct: 496 GRNVAIISDKQSGLVKAIHSVIPQAEHRQCARHIMENWKRN-SHDMELQRLFWKIVRSYT 554
Query: 572 NQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSAR 631
+ M LK + A+E L+ P W + F S C+ +NN+SESFN TI AR
Sbjct: 555 EGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREAR 614
Query: 632 DQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGE 691
+P++ M E IR M R + ++ + R R K S + +
Sbjct: 615 RKPLLDMLEDIRRQCMVR--------NEKRYVIADRLRTRFTKRAHAEIEKMIAGSQVCQ 666
Query: 692 QFQVMHTYNR------QQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDD 745
++ H + F VD+ +C C W++ GIPC H S + ++Q+ E +V D
Sbjct: 667 RWMARHNKHEIKVGPVDSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVEDYVSD 726
Query: 746 YYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNG-PGRPRKLRIRE-------- 796
+Y+ S + Y+ I P+ G +WP V +LPP ++ G PGRP+ R+
Sbjct: 727 WYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTA 786
Query: 797 FDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDVQNPEAAK 837
+ + + R C+ C GHN++ CK++ P A +
Sbjct: 787 SSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKR 827
>dbj|BAB11264.1| unnamed protein product [Arabidopsis thaliana]
Length = 733
Score = 270 bits (690), Expect = 1e-70
Identities = 166/605 (27%), Positives = 302/605 (49%), Gaps = 30/605 (4%)
Query: 234 DPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNES 293
+ DDS+DD + D F +G EF + + K+ I ++ + I ++E
Sbjct: 92 ESDDSDDDGARID------------FYVGQEFVSKEKCKDTIEKYAIREKVNIHFKRSER 139
Query: 294 YRVRVEC-RDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKM 352
++ C +D C +++ S +++ +G H+C P+ ++ +A + +
Sbjct: 140 NKIEGVCVQDCCKWRIYASITSRSDKMVVQSYKGIHSCYPIGVVDLYSAPKIAADFINEF 199
Query: 353 QVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRV 412
+ +S ++ + +N G+ +T + AR+I K II + +Q+ + Y EL++
Sbjct: 200 RTNSNLSAGQIMQRLYLN-GLRVTKTKCQSARQIMKHIISDEYVEQFTRMYDYVEELRKT 258
Query: 413 CRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLI 472
+ + + T F FY CF+ K G+ +ACR + + G LK R GQLL
Sbjct: 259 NPGSTIILG------TKDRIFEKFYTCFEAQKTGWKTACRRVIHLGGTFLKGRMKGQLLT 312
Query: 473 AVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGED--KRYVFISDQQKGLLSVFD 530
AVGRDPND + +A+ +V E K W+WF +LL ED+ + SDQQKGL+
Sbjct: 313 AVGRDPNDAMYIIAWAIVPVENKVYWQWFMELLGEDLRLELGNGLALSSDQQKGLIYAIK 372
Query: 531 EMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAW 590
+ +HR+C RH++AN +K++ + + +A + +++ ++K I A+
Sbjct: 373 NVLPYAEHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKMKTIKFEAY 432
Query: 591 EWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRM 650
+ + W + F+ +K + NNISES+N + AR++P++++ E IR ++M
Sbjct: 433 DEVKRSVGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDIRRHIMASN 492
Query: 651 TTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIA- 709
++Q + P ++K + W +S ++V H N+ ++V +
Sbjct: 493 LVKLKEMQNVTGLITPKAIAIMEKR-KKSLKWCYPFSNGRGIYEVDHGKNK--YVVHVRD 549
Query: 710 KRSCSCNFWEIVGIPCRHVVSALG---KRKQRPEMFVDDYYSRSKYALCYSFAISPINGM 766
K SC+C +++ GIPC H++SA+ K + PE + D+YS K+ LCY+ + P+NGM
Sbjct: 550 KTSCTCREYDVSGIPCCHIMSAMWAEYKETKLPETAILDWYSVEKWKLCYNSLLFPVNGM 609
Query: 767 DMWPEVEAPELLPPNYKNGPGRPRKL-RIREFDENGSRMRRQGVAYRCTKCDQFGHNQRR 825
++W ++PP + PGRP+K RI++ E S Q C+ C Q GHN+R
Sbjct: 610 ELWETHSDVVVMPPPDRIMPGRPKKNDRIKDPSEEASSENSQKALVTCSNCGQTGHNKRT 669
Query: 826 CKSDV 830
C+ ++
Sbjct: 670 CQIEL 674
>emb|CAB77993.1| putative MuDR-A-like transposon protein [Arabidopsis thaliana]
gi|2565011|gb|AAB81881.1| putative MuDR-A-like
transposon protein [Arabidopsis thaliana]
gi|7487609|pir||T00940 hypothetical protein T3F12.2 -
Arabidopsis thaliana
Length = 761
Score = 265 bits (678), Expect = 4e-69
Identities = 205/726 (28%), Positives = 332/726 (45%), Gaps = 46/726 (6%)
Query: 139 FDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKTLRIKKCGSKTPKKKSPKKNNSGR 198
FDD D DG DVE + + + +R KK K K+K K
Sbjct: 2 FDDDVD-MFYKYSDG--DVEYNGGNESESEERIEEEIVRSKKNTKKNKKEKKKKDEEDPL 58
Query: 199 MNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFK 258
V +G + + I D + E SSD D+ D I R+ + D K
Sbjct: 59 ERFDFEVD---DGVANFEDEIPIGRDIIPE---SSDEDE---DPILVRDRRIRRAMGD-K 108
Query: 259 FKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECR--DQCGYKVLCSKVGDR 316
F IG F T +EFKEAI+E + +G+ + + E ++ +C +C ++V CS DR
Sbjct: 109 FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPDR 168
Query: 317 RTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGIT 376
+ F +KT H+C+P + + S +A+ + K+++ +K ++ + + + T
Sbjct: 169 QLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVST 228
Query: 377 MDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKI----NVAGPSVTIQLR 432
+ + R R +A +++ + ++Q+A IR Y E+ + I N G V
Sbjct: 229 IPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDV----- 283
Query: 433 FGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVET 492
F FY CF+ + + +CRP +G+DG LK G LL AVG DPN+Q +P+A+ VV++
Sbjct: 284 FNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQS 343
Query: 493 ECKETWKWFTQLLMEDIG--EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFK 550
E E W WF Q + +D+ + R+V +SD+ KGLLS + + +HR+C++H+ N K
Sbjct: 344 ENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSAVKQELPNAEHRMCVKHIVENLK 403
Query: 551 KKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYS 610
K ++ L+ + + + + +N L+ D + ++ EP W + + S
Sbjct: 404 KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLNEEPHTWSRCFYKLGS 463
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
C+ + NN +ESFN TI AR + +I M E IR M R+ K + + K
Sbjct: 464 CCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNKKSLRHEGRFTKYALK 523
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
L E T A K + F+V N + VDI K CSC W+I GIPC H
Sbjct: 524 MLALEKTDADRSK-VYRCTHGVFEVYIDENGHR--VDIPKTQCSCGKWQISGIPCEHSYG 580
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPE-----VEAP--ELLPPNYK 783
A+ + E ++ +++S + Y A P+ G W V AP +LP K
Sbjct: 581 AMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKYWLNSSYGLVTAPPEPILPGRKK 640
Query: 784 NGPGRPRKLRIREFDENGSRMRR----------QGVAYRCTKCDQFGHNQRRCKSDVQNP 833
+ + RI+ +E+ + +R +G C C + GHN RCK +
Sbjct: 641 EKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKIIHCKSCGEAGHNALRCKKFPKEK 700
Query: 834 EAAKRK 839
KRK
Sbjct: 701 VPRKRK 706
>gb|AAG52094.1| putative Mutator-like transposase; 12516-14947 [Arabidopsis
thaliana] gi|25406536|pir||A96810 probable Mutator-like
transposase, 12516-14947 [imported] - Arabidopsis
thaliana
Length = 761
Score = 263 bits (671), Expect = 2e-68
Identities = 204/726 (28%), Positives = 331/726 (45%), Gaps = 46/726 (6%)
Query: 139 FDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKTLRIKKCGSKTPKKKSPKKNNSGR 198
FDD D DG DVE + + + +R KK K K+K K
Sbjct: 2 FDDDVD-MFYKYSDG--DVEYNGGNESESEERIEEEIVRRKKNTKKNKKEKKKKDEEDPL 58
Query: 199 MNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFK 258
V +G + + I D + E SSD D+ D I R+ + D K
Sbjct: 59 ERFDFEVD---DGVANFEDEIPIGRDIIPE---SSDEDE---DPILVRDRRIRRAMGD-K 108
Query: 259 FKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVEC--RDQCGYKVLCSKVGDR 316
F IG F T +EFKEAI+E + +G+ + + E ++ +C +C ++V CS DR
Sbjct: 109 FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMSGKCKWRVYCSYDPDR 168
Query: 317 RTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGIT 376
+ F +KT H+C+P + + S +A+ + K+++ +K ++ + + + T
Sbjct: 169 QLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVST 228
Query: 377 MDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKI----NVAGPSVTIQLR 432
+ + R R +A +++ + ++Q+A IR Y E+ + I N G V
Sbjct: 229 IPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDV----- 283
Query: 433 FGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVET 492
F FY CF+ + + +CRP +G+DG LK G LL AVG DPN+Q +P+A+ VV++
Sbjct: 284 FNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQS 343
Query: 493 ECKETWKWFTQLLMEDIG--EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFK 550
E E W WF Q + +D+ + R+V +SD+ KGLLS + + +HR+C++H+ N K
Sbjct: 344 ENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSAVKQELPNAEHRMCVKHIVENLK 403
Query: 551 KKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYS 610
K ++ L+ + + + + +N L+ D + ++ EP W + + S
Sbjct: 404 KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLNEEPHTWSRCFYKLGS 463
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
C+ + NN +ESFN TI AR + +I M E IR M R+ K + + K
Sbjct: 464 CCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNKKSLRHEGRFTKYALK 523
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
L E T A K + F+V N + VDI K CSC W+I GIPC H
Sbjct: 524 MLALEKTDADRSK-VYRCTHGVFEVYIDGNGHR--VDIPKTQCSCGKWQISGIPCEHAYG 580
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPE-----VEAP--ELLPPNYK 783
A+ + E ++ +++S + Y A P+ G W V AP +LP K
Sbjct: 581 AMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKYWLNSSYGLVTAPPEPILPGRKK 640
Query: 784 NGPGRPRKLRIREFDENGSRMRR----------QGVAYRCTKCDQFGHNQRRCKSDVQNP 833
+ + RI+ +E+ + +R +G C C + GHN CK +
Sbjct: 641 EKSKKGKFARIKGKNESPKKKKRKKNEVEKLGKKGKIIHCKSCGEAGHNALGCKKFPKEK 700
Query: 834 EAAKRK 839
KRK
Sbjct: 701 VPRKRK 706
>gb|AAF04891.1| Mutator-like transposase [Arabidopsis thaliana]
gi|20465953|gb|AAM20162.1| putative mutator transposase
[Arabidopsis thaliana] gi|18389286|gb|AAL67086.1|
putative Mutator transposase [Arabidopsis thaliana]
Length = 757
Score = 256 bits (654), Expect = 2e-66
Identities = 164/602 (27%), Positives = 287/602 (47%), Gaps = 32/602 (5%)
Query: 247 QFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECR-DQCG 305
Q R + ++ +G EF + + A+ + + ++++ +K++ R +C D C
Sbjct: 170 QARALTADMTYQLTVGQEFPDVKSCRRALRDMAIALHFEMQTIKSDKTRFTAKCSSDGCP 229
Query: 306 YKVLCSKVGDRRTFQIKTLEGPHTCAPV--LENKSANSRWVAKKAVPKMQVTKKMSVQEV 363
++V +K+ TF I+T+ H+C + L ++ A+ +WVA +++ +E+
Sbjct: 230 WRVHAAKLPGVPTFTIRTIHESHSCGGINHLGHQQASVQWVASSVEQRLRENPNCKPKEI 289
Query: 364 FNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVA 423
E+ +G+ ++ +AWR ++ + + G ++ Y ++ +Y ++KR ++
Sbjct: 290 LEEIHRVHGITLSYKQAWRGKERIMATMRGSFEEGYRLLPQYCEQVKRT-NPGSIASVYG 348
Query: 424 GPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYF 483
P+ F + F GF++ACRP +G+D +LK++Y G LL+A G D + F
Sbjct: 349 SPADNC---FQRLFISFQASIYGFLNACRPLLGLDRTYLKSKYLGTLLLATGFDGDGALF 405
Query: 484 PLAFGVVETECKETWKWFT----QLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHR 539
PLAFG+V+ E E W WF LL + R +SD+QKG++ ++ F + H
Sbjct: 406 PLAFGIVDEENDENWMWFLCELHNLLETNTENMPRLTILSDRQKGIVEGVEQNFPTAFHG 465
Query: 540 LCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPK 599
C+RHL +F+K+F + L A T + + K+ E+++I A W+ + P+
Sbjct: 466 FCMRHLSESFRKEFNNTLLVNYLWEAAQALTVIE-FEAKILEIEEISQDAAYWIRRIPPR 524
Query: 600 KWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQK 659
W A+ + L NI ES N I A PII M E IR LM +
Sbjct: 525 LWAT-AYFEGQRFGHLTANIVESLNSWIAEASGLPIIQMMECIRRQLMTWFNERRETSMQ 583
Query: 660 WQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWE 719
W ++P +R+ + + LA ++ + E + H N IVDI R C C W+
Sbjct: 584 WTSILVPTAERRVAEALELARTYQVLRANEAEFEVISHEGNN---IVDIRNRCCLCRGWQ 640
Query: 720 IVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE--- 776
+ G+PC H V+AL +Q F + ++ + Y YS I PI W E+ +
Sbjct: 641 LYGLPCAHAVAALLSCRQNVHRFTESCFTVATYRKTYSQTIHPIPDKSHWRELSEGDPNV 700
Query: 777 --------LLPPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKS 828
+ PP PGRPRK R+R E+ R++R C++C+Q GH + C +
Sbjct: 701 NKAALDAIINPPKSLRPPGRPRKRRVRA--EDRGRVKR---VVHCSRCNQTGHFRTTCAA 755
Query: 829 DV 830
+
Sbjct: 756 PI 757
>pir||H86269 F21F23.10 protein - Arabidopsis thaliana gi|8920571|gb|AAF81293.1|
Strong similarity to a mutator-like transposase from
Arabidopsis thaliana gb|AC006067. It contains a zinc
finger, CCHC class domain PF|00098
Length = 753
Score = 255 bits (651), Expect = 5e-66
Identities = 167/633 (26%), Positives = 303/633 (47%), Gaps = 43/633 (6%)
Query: 212 KGKYVV---DEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKD---FKFKIGMEF 265
K YVV D +D + E +P+ D + D + D F +G EF
Sbjct: 91 KNLYVVRADDHYLDDVLIGEREEEEEPESEQLDFYRDDYVESDDSDDDGARIDFYVGQEF 150
Query: 266 NTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVEC-RDQCGYKVLCSKVGDRRTFQIKTL 324
+ + K+ I ++ + I ++E ++ C +D C +++ S +++
Sbjct: 151 VSKEKCKDTIEKYAIREKVNIHFKRSERNKIEGVCVQDCCKWRIYASITSRSDKMVVQSY 210
Query: 325 EGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRAR 384
+G H+C P+ ++ +A + + + +S ++ + +N G+ +T + AR
Sbjct: 211 KGIHSCYPIGVVDLYSAPKIAADFINEFRTNSNLSAGQIMQRLYLN-GLRVTKTKCQSAR 269
Query: 385 KIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCK 444
+I K II + +Q+ + Y EL++ P T+ K
Sbjct: 270 QIIKHIISDEYAEQFTRMYDYVEELRKT-----------NPGSTL--------------K 304
Query: 445 KGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQL 504
G+ +ACR + + G LK R GQLL AVGRDPND + +A+ +V E K W+WF +L
Sbjct: 305 TGWKTACRRVIHLGGTFLKGRMKGQLLTAVGRDPNDAMYIIAWAIVPVENKVYWQWFMKL 364
Query: 505 LMEDIGED--KRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDL 562
L ED+ + SDQQKGL+ + +HR+C RH++AN +K++ + +
Sbjct: 365 LGEDLRLELGNGLALSSDQQKGLIYAIKNVLPYAEHRMCARHIFANLQKRYKQMGPLHKV 424
Query: 563 MMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISES 622
+A + +++ ++K I A++ + W + F+ +K + NNISES
Sbjct: 425 FWKCARAYNETVFWKQLEKMKTIKFEAYDEVKRSVGSNWSRAFFSDITKSAAVENNISES 484
Query: 623 FNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHW 682
+N + AR++P++++ E IR ++M ++Q + P ++K + W
Sbjct: 485 YNAVLKDAREKPVVALLEDIRRHIMASNLVKIKEMQNVTGLITPKAIAIMEKR-KKSLKW 543
Query: 683 KSTWSGIGEQFQVMHTYNRQQFIVDIA-KRSCSCNFWEIVGIPCRHVVSALG---KRKQR 738
+S ++V H N+ ++V + K SC+C +++ GIPC H++SA+ K +
Sbjct: 544 CYPFSNGRGIYEVDHGKNK--YVVHVRDKTSCTCREYDVSGIPCCHIMSAMWAEYKETKL 601
Query: 739 PEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNGPGRPRKL-RIREF 797
PE + D+YS K+ LCY+ + P+NGM++W ++PP + PGRP+K RI++
Sbjct: 602 PETAILDWYSVEKWKLCYNSLLFPVNGMELWETHSDVVVMPPPDRIMPGRPKKNDRIKDP 661
Query: 798 DENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDV 830
E S Q C+ C Q GHN+R C+ ++
Sbjct: 662 SEEASSENSQKALVTCSNCGQTGHNKRTCQIEL 694
>gb|AAP54770.1| putative transposon protein [Oryza sativa (japonica
cultivar-group)] gi|22213194|gb|AAM94534.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)] gi|37536362|ref|NP_922483.1| putative
transposon protein [Oryza sativa (japonica
cultivar-group)]
Length = 835
Score = 254 bits (649), Expect = 8e-66
Identities = 158/522 (30%), Positives = 252/522 (48%), Gaps = 14/522 (2%)
Query: 321 IKTLEGPHTCAPVL---ENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITM 377
IK L PH CA + N A + WV + + K++ + + + Y + I+
Sbjct: 289 IKKLPYPHNCASIAGVENNCMATNSWVRDRVINKLRQEPTIGATALKKVLEEMYKIKISY 348
Query: 378 DRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFY 437
W R++A I G + + + AEL+R+ + V+++ SV + F +
Sbjct: 349 YVVWDDRQMALDEILGKWEDSFDAAYSFKAELERISPRSIVEVDHV--SVNGKNHFSRMF 406
Query: 438 FCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKET 497
C GF++ CRP++G+D L ++ GQL A+G D ++ FP+A+GV E+E E
Sbjct: 407 VALKPCVDGFLNGCRPYLGIDSTVLTGKWRGQLASAIGVDGHNWMFPVAYGVFESESTEN 466
Query: 498 WKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDS-IDHRLCLRHLYANFKKKFGGG 556
W WF L IG V +D KG+ + +F++ ++HR C+RHL NF+K+F G
Sbjct: 467 WAWFMDKLHMAIGSPVGLVLSTDAGKGIDTTVTRVFNNGVEHRECMRHLVKNFQKRFSGE 526
Query: 557 SQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLM 616
R+L A++A + NE+K+ A EW+ W + F+ SKCD +
Sbjct: 527 VFERNL-WSASRAYRQDIYEGHYNEMKEACPRATEWIDDFHKHIWTRSKFSPVSKCDYVT 585
Query: 617 NNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL-DKE 675
NNI+E+FN I + P++ + + IRH +M R++ +K ++P K L +
Sbjct: 586 NNIAETFNSWIRHEKSLPVVDLMDKIRHMIMERISIRKRLAEKLTGQILPSVMKTLYARS 645
Query: 676 ITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVV-SALGK 734
L S +GE + VD+ R CSC W+I GIPC HV+ + +
Sbjct: 646 RDLGYKLYSAHKHLGEIGGTGRDLKTWRHTVDLNTRECSCRQWQITGIPCTHVIFLIISR 705
Query: 735 RKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVE-APELLPPNYKNGPGRPRKLR 793
R E FVD+YY + + Y+ + P+ W + +L PP K GRPR R
Sbjct: 706 RGLELEQFVDEYYYVAAFKRAYAGHVVPMTDKAQWAKGNIGLKLHPPLLKRSAGRPRSRR 765
Query: 794 IREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDVQNPEA 835
I+ +E G R+ YRC +C QFGH ++ C V +P A
Sbjct: 766 IKGVEEGGVGKRK----YRCKRCGQFGHTKKTCNEPVHDPNA 803
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,502,863,104
Number of Sequences: 2540612
Number of extensions: 66924359
Number of successful extensions: 164500
Number of sequences better than 10.0: 523
Number of HSP's better than 10.0 without gapping: 358
Number of HSP's successfully gapped in prelim test: 166
Number of HSP's that attempted gapping in prelim test: 162509
Number of HSP's gapped (non-prelim): 1086
length of query: 840
length of database: 863,360,394
effective HSP length: 137
effective length of query: 703
effective length of database: 515,296,550
effective search space: 362253474650
effective search space used: 362253474650
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC144730.8


