

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
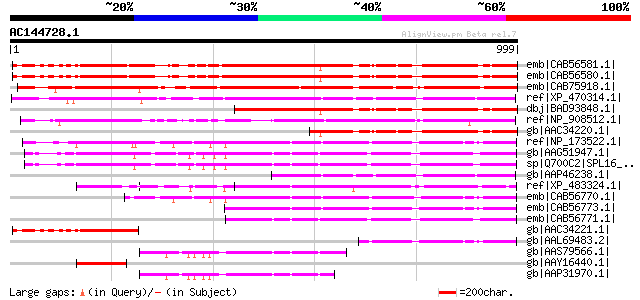
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabid... 1000 0.0
emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabid... 999 0.0
emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabi... 983 0.0
ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japo... 734 0.0
dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabid... 650 0.0
ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica... 489 e-136
gb|AAC34220.1| unknown protein [Arabidopsis thaliana] 439 e-121
ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis tha... 426 e-117
gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana] 411 e-113
sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 ... 411 e-113
gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japoni... 363 2e-98
ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (j... 327 9e-88
emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana] 294 1e-77
emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana] 293 3e-77
emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana] 283 2e-74
gb|AAC34221.1| putative squamosa-promoter binding protein [Arabi... 255 5e-66
gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana] 162 5e-38
gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|... 146 4e-33
gb|AAY16440.1| squamosa promoter binding-like protein [Betula pl... 133 3e-29
gb|AAP31970.1| At1g76580 [Arabidopsis thaliana] gi|22330667|ref|... 130 3e-28
>emb|CAB56581.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
gi|3650274|emb|CAA09698.1| squamosa-promoter binding
protein-like 1 [Arabidopsis thaliana]
gi|67461551|sp|Q9SMX9|SPL1_ARATH Squamosa-promoter
binding-like protein 1 gi|30690591|ref|NP_850468.1|
squamosa promoter-binding protein-like 1 (SPL1)
[Arabidopsis thaliana]
Length = 881
Score = 1000 bits (2586), Expect = 0.0
Identities = 566/1002 (56%), Positives = 684/1002 (67%), Gaps = 136/1002 (13%)
Query: 6 GAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPL 65
G E FYG +GKRS EW+LNDW+WDGDLF+A++ + G+QFFPL
Sbjct: 8 GGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR--------GRQFFPL 51
Query: 66 GSGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 125
G+ SSN+SSSCS+EG+ +KKRR + ++ D GAL+LNL G
Sbjct: 52 GN-------SSNSSSSCSDEGN--------DKKRRAVAIQ----GDTNGALTLNLNG--- 89
Query: 126 PVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQ 185
E DG A T + AVCQVE+C ADLS+ KDYHRRHKVCEMHSKA+ A VG +Q
Sbjct: 90 ---ESDGLFP--AKKTKSGAVCQVENCEADLSKVKDYHRRHKVCEMHSKATSATVGGILQ 144
Query: 186 RFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISL 245
RFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRKTN E NG+P+ DD +S+YLLI+L
Sbjct: 145 RFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNPS-DDHSSNYLLITL 203
Query: 246 LKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVS 305
LKILSNMH DQDL++HLL+SL S EQ KNL LL + GG
Sbjct: 204 LKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQ-------GG--------- 247
Query: 306 ALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSET 365
GSQGS I + + Q QE +K + +E
Sbjct: 248 -----GSQGSLN-IGNSALLGIEQAPQE----------------ELKQFSARQDGTATEN 285
Query: 366 RDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYP-WTQQDSHQSSPAQT 424
R S Q KMN+FDLNDIY+DSDD D+ER P TN ATSS+DYP W HQSSP QT
Sbjct: 286 R-SEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPSWI----HQSSPPQT 338
Query: 425 SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESY 484
S NSDSAS QSPSSSS +AQ RT RIVFKLFGKEPNEFP+VLR QILDWLS SPTD+ESY
Sbjct: 339 SRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESY 398
Query: 485 IRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFN 544
IRPGCIVLTIYLRQAE WEEL DL SL KLLD+SDD W TGW+++RVQ+Q+AF++N
Sbjct: 399 IRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYN 458
Query: 545 GQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVC 604
GQVV+DTSL +S +YS I +V P+A+ A+++AQF+VKG+NL + TRL+C++EGKYL+
Sbjct: 459 GQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEGKYLIQ 518
Query: 605 EDAHEST----DQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAE-ED 659
E H+ST D + + + ++C+ FSC +P+ +GRGF+EIEDQGLSSSFFPF+V E +D
Sbjct: 519 ETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDD 578
Query: 660 VCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGV 719
VC+EIR+LE LE + TD + QAMDFIHE+GWLLHRS+L + N GV
Sbjct: 579 VCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLHRSKLGES--DPNPGV 626
Query: 720 DLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRR 779
FPL RF WL+EFSMD +WCAV++KLLN+ D V + + LSE+ LLHRAVR+
Sbjct: 627 --FPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NATLSELCLLHRAVRK 683
Query: 780 NSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGS 839
NSK +VE+LLRY+P + + LFRPDA GPAGLTPLHIAAGKDGS
Sbjct: 684 NSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPAGLTPLHIAAGKDGS 727
Query: 840 EDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAA-HV 898
EDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+Q+KINK HV
Sbjct: 728 EDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHV 787
Query: 899 VVEIPSNMTE-SNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRP 957
VV IP + ++ K PK ++LEI +Q CKLCD K+ T RS+ YRP
Sbjct: 788 VVNIPVSFSDREQKEPKSGPMASALEI-------TQIPCKLCDHKLVYGT-TRRSVAYRP 839
Query: 958 AMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
AMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE LD+GTS
Sbjct: 840 AMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>emb|CAB56580.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 881
Score = 999 bits (2582), Expect = 0.0
Identities = 566/1002 (56%), Positives = 683/1002 (67%), Gaps = 136/1002 (13%)
Query: 6 GAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPL 65
G E FYG +GKRS EW+LNDW+WDGDLF+A++ + G+QFFPL
Sbjct: 8 GGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR--------GRQFFPL 51
Query: 66 GSGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 125
G+ SSN+SSSCS+EG+ +KKRR + ++ D GAL+LNL G
Sbjct: 52 GN-------SSNSSSSCSDEGN--------DKKRRAVAIQ----GDTNGALTLNLNG--- 89
Query: 126 PVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQ 185
E DG A T + AVCQVE+C ADLS+ KDYHRRHKVCEMHSKA+ A V +Q
Sbjct: 90 ---ESDGLFP--AKKTKSGAVCQVENCEADLSKVKDYHRRHKVCEMHSKATSATVEGILQ 144
Query: 186 RFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISL 245
RFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRKTN E NG+P+ DD +S+YLLI+L
Sbjct: 145 RFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNPS-DDHSSNYLLITL 203
Query: 246 LKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVS 305
LKILSNMH DQDL++HLL+SL S EQ KNL LL + GG
Sbjct: 204 LKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQ-------GG--------- 247
Query: 306 ALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSET 365
GSQGS I + + Q QE +K + +E
Sbjct: 248 -----GSQGSLN-IGNSALLGIEQAPQE----------------ELKQFSARQDGTATEN 285
Query: 366 RDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYP-WTQQDSHQSSPAQT 424
R S Q KMN+FDLNDIY+DSDD D+ER P TN ATSS+DYP W HQSSP QT
Sbjct: 286 R-SEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPSWI----HQSSPPQT 338
Query: 425 SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESY 484
S NSDSAS QSPSSSS +AQ RT RIVFKLFGKEPNEFP+VLR QILDWLS SPTD+ESY
Sbjct: 339 SRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESY 398
Query: 485 IRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFN 544
IRPGCIVLTIYLRQAE WEEL DL SL KLLD+SDD W TGW+++RVQ+Q+AF++N
Sbjct: 399 IRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYN 458
Query: 545 GQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVC 604
GQVV+DTSL +S +YS I +V P+A+ A+++AQF+VKG+NL R TRL+C++EGKYL+
Sbjct: 459 GQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRRRGTRLLCSVEGKYLIQ 518
Query: 605 EDAHEST----DQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAE-ED 659
E H+ST D + + + ++C+ FSC +P+ +GRGF+EIEDQGLSSSFFPF+V E +D
Sbjct: 519 ETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDD 578
Query: 660 VCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGV 719
VC+EIR+LE LE + TD + QAMDFIHE+GWLLHRS+L + N GV
Sbjct: 579 VCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLHRSKLGES--DPNPGV 626
Query: 720 DLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRR 779
FPL RF WL+EFSMD +WCAV++KLLN+ D V + + LSE+ LLHRAVR+
Sbjct: 627 --FPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NATLSELCLLHRAVRK 683
Query: 780 NSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGS 839
NSK +VE+LLRY+P + + LFRPDA GPAGLTPLHIAAGKDGS
Sbjct: 684 NSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPAGLTPLHIAAGKDGS 727
Query: 840 EDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAA-HV 898
EDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+Q+KINK HV
Sbjct: 728 EDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHV 787
Query: 899 VVEIPSNMTE-SNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRP 957
VV IP + ++ K PK ++LEI +Q CKLCD K+ T RS+ YRP
Sbjct: 788 VVNIPVSFSDREQKEPKSGPMASALEI-------TQIPCKLCDHKLVYGT-TRRSVAYRP 839
Query: 958 AMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
AMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE LD+GTS
Sbjct: 840 AMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>emb|CAB75918.1| squamosa promoter binding protein-like 12 [Arabidopsis thaliana]
gi|6006403|emb|CAB56769.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|6006395|emb|CAB56768.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|28392866|gb|AAO41870.1| putative squamosa promoter
binding protein 12 [Arabidopsis thaliana]
gi|67461584|sp|Q9S7P5|SPL12_ARATH Squamosa-promoter
binding-like protein 12 gi|15232241|ref|NP_191562.1|
squamosa promoter-binding protein-like 12 (SPL12)
[Arabidopsis thaliana]
Length = 927
Score = 983 bits (2542), Expect = 0.0
Identities = 559/1008 (55%), Positives = 704/1008 (69%), Gaps = 115/1008 (11%)
Query: 16 GGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGS 75
G S + GKRS EW+LNDW+W+GDLF+A+++N GS
Sbjct: 11 GHSLEYGFSGKRSVEWDLNDWKWNGDLFVATQLNH-----------------------GS 47
Query: 76 SNTSSSCSEEGDLE-----KGNKEGEKKRR---VIVLEDDGL-NDKAGALSLNLAGHVSP 126
SN+SS+CS+EG++E + E +KKRR V+ +E+D L +D A L+LNL G+
Sbjct: 48 SNSSSTCSDEGNVEIMERRRIEMEKKKKRRAVTVVAMEEDNLKDDDAHRLTLNLGGNN-- 105
Query: 127 VVERDG-KKSRGAGGTSNRAVC-QVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAM 184
+E +G KK++ GG +RA+C QV++CGADLS+ KDYHRRHKVCE+HSKA+ ALVG M
Sbjct: 106 -IEGNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCEIHSKATTALVGGIM 164
Query: 185 QRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLIS 244
QRFCQQCSRFH+LEEFDEGKRSCRRRLAGHNKRRRK N + + NG+ +DDQTS+Y+LI+
Sbjct: 165 QRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGTSMSDDQTSNYMLIT 224
Query: 245 LLKILSNMHY----QPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRN 300
LLKILSN+H Q DQDLL+HLL+SL SQ E +NL LL+ L +S+N
Sbjct: 225 LLKILSNIHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNLVGLLQGGGGLQ----ASQN 280
Query: 301 SGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPP 360
G +SAL +S+ Q +E + H V + Q + +NS
Sbjct: 281 IGNLSAL-----------------LSLEQAPREDIKHHSVSETPWQEV------YANSAQ 317
Query: 361 AYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSS 420
S Q K+N+FDLNDIY+DSDD T+ P TN ATSS+DY QDS QSS
Sbjct: 318 ERVAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSPPPTNPATSSLDY---HQDSRQSS 374
Query: 421 PAQTSG-NSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPT 479
P QTS NSDSAS QSPSSSSG+AQSRTDRIVFKLFGKEPN+FP+ LR QIL+WL+ +PT
Sbjct: 375 PPQTSRRNSDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDFPVALRGQILNWLAHTPT 434
Query: 480 DIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQM 539
D+ESYIRPGCIVLTIYLRQ EA WEELCCDL+ SL +LLD+SDD W GW+++RVQ+Q+
Sbjct: 435 DMESYIRPGCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSDDPLWTDGWLYLRVQNQL 494
Query: 540 AFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEG 599
AF FNGQVV+DTSLP RS++YS+I TV P+AV +K+AQF+VKG+NL RP TRL+C +EG
Sbjct: 495 AFAFNGQVVLDTSLPLRSHDYSQIITVRPLAV--TKKAQFTVKGINLRRPGTRLLCTVEG 552
Query: 600 KYLVCEDAHESTDQYSE--ELDELQCIQFSCSVPVSNGRGFIEIEDQ-GLSSSFFPFIVA 656
+LV E ++ + E +E+ + FSC +P+++GRGF+EIEDQ GLSSSFFPFIV+
Sbjct: 553 THLVQEATQGGMEERDDLKENNEIDFVNFSCEMPIASGRGFMEIEDQGGLSSSFFPFIVS 612
Query: 657 E-EDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRM-VN 714
E ED+C+EIR LE LE + TD + QAMDFIHE+GWLLHRS+LK R+ +
Sbjct: 613 EDEDICSEIRRLESTLEFTGTD----------SAMQAMDFIHEIGWLLHRSELKSRLAAS 662
Query: 715 LNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDE-TVNKGDHPTLYQALSEMGLL 773
++ DLF L RF +L+EFSMD +WC V+KKLLN+L +E TV+ P+ ALSE+ LL
Sbjct: 663 DHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNILFEEGTVD----PSPDAALSELCLL 718
Query: 774 HRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIA 833
HRAVR+NSK +VE+LLR+ P +++ L G LFRPDA GP GLTPLHIA
Sbjct: 719 HRAVRKNSKPMVEMLLRFSPKK-------KNQTLAG------LFRPDAAGPGGLTPLHIA 765
Query: 834 AGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKIN-KT 892
AGKDGSEDVLDALT DP M GI+AWKN+RD+TG TPEDYARLRGH++YIHLVQ+K++ K
Sbjct: 766 AGKDGSEDVLDALTEDPGMTGIQAWKNSRDNTGFTPEDYARLRGHFSYIHLVQRKLSRKP 825
Query: 893 QGAAHVVVEIPSNMTESNKNPKQNE-SFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGR 951
HVVV IP + +K K++ +SLEI + CKLCD K T +
Sbjct: 826 IAKEHVVVNIPESFNIEHKQEKRSPMDSSSLEITQI------NQCKLCDHKRVFVTTHHK 879
Query: 952 SMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
S+ YRPAMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE L++GTS
Sbjct: 880 SVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLEYGTS 927
>ref|XP_470314.1| putative SBP-domain protein [Oryza sativa (japonica
cultivar-group)] gi|40714694|gb|AAR88600.1| putative
SBP-domain protein [Oryza sativa (japonica
cultivar-group)]
Length = 969
Score = 734 bits (1894), Expect = 0.0
Identities = 440/1026 (42%), Positives = 603/1026 (57%), Gaps = 95/1026 (9%)
Query: 4 RLGAENYHFYGVG-GSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQF 62
R+GA++ H YG G G D+ KR W+LNDWRWD D F+A+ V +A L +
Sbjct: 5 RVGAQSRHLYGGGLGEPDMDRRDKRLFGWDLNDWRWDSDRFVATPVPAAEASGLAL---- 60
Query: 63 FPLGSGIPVVGGSSNTSSSCSEEG---DLEKGNKEGE--KKRRVIVLEDDGLNDK----- 112
N+S S SEE + N G+ K++RV+V++DD + D
Sbjct: 61 --------------NSSPSSSEEAGAASVRNVNARGDSDKRKRVVVIDDDDVEDDELVEN 106
Query: 113 -AGALSLNLAGHV----------SPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKD 161
G+LSL + G + +R+GKK R GG+ + CQVE C ADL+ +D
Sbjct: 107 GGGSLSLRIGGDAVAHGAGVGGGADEEDRNGKKIRVQGGSPSGPACQVEGCTADLTGVRD 166
Query: 162 YHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKT 221
YHRRHKVCEMH+KA+ A+VGN +QRFCQQCSRFH L+EFDEGKRSCRRRLAGHN+RRRKT
Sbjct: 167 YHRRHKVCEMHAKATTAVVGNTVQRFCQQCSRFHPLQEFDEGKRSCRRRLAGHNRRRRKT 226
Query: 222 NQEAVPNGSPTNDDQTSSYLLISLLKILSNMHYQPTD----QDLLTHLLRSLASQNDEQG 277
E GS +D+ SSYLL+ LL + +N++ + Q+L++ LLR+L +
Sbjct: 227 RPEVAVGGSAFTEDKISSYLLLGLLGVCANLNADNAEHLRGQELISGLLRNLGAVAKSLD 286
Query: 278 SKNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHT 337
K L LL +++ ++G ++ S +AL + T + + S ++M
Sbjct: 287 PKELCKLLEACQSM-QDGSNAGTSETANALVN-------TAVAEAAGPSNSKMPFVNGDQ 338
Query: 338 HDVRTSDHQLISSIKPSISN-SPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERL 396
+ +S + S P+++ PPA K +FDLND Y + + E
Sbjct: 339 CGLASSSVVPVQSKSPTVATPDPPA----------CKFKDFDLNDTYGGMEGFEDGYEGS 388
Query: 397 PVSTNLATSSVDYP-WTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLF 455
P T S + P W QDS QS P QTSGNSDS SAQS SSS+G+AQ RTD+IVFKLF
Sbjct: 389 PTPAFKTTDSPNCPSWMHQDSTQSPP-QTSGNSDSTSAQSLSSSNGDAQCRTDKIVFKLF 447
Query: 456 GKEPNEFPLVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLI 515
K P++ P VLR+QIL WLS SPTDIESYIRPGCI+LT+YLR E+ W+EL +++S L
Sbjct: 448 EKVPSDLPPVLRSQILGWLSSSPTDIESYIRPGCIILTVYLRLVESAWKELSDNMSSYLD 507
Query: 516 KLLDVSDDTFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASK 575
KLL+ S FW +G V + V+HQ+AF+ NGQ+++D L +++Y KI V PIA P S
Sbjct: 508 KLLNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRPLANSAHHYCKILCVRPIAAPFST 567
Query: 576 RAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNG 635
+ F V+G+NL+ ++RL+C+ EG + ED D E D+++ + F C +P S G
Sbjct: 568 KVNFRVEGLNLVSDSSRLICSFEGSCIFQEDTDNIVDDV--EHDDIEYLNFCCPLPSSRG 625
Query: 636 RGFIEIEDQGLSSSFFPFIVAEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDF 695
RGF+E+ED G S+ FFPFI+AE+D+C+E+ LE + ESS E A++QA++F
Sbjct: 626 RGFVEVEDGGFSNGFFPFIIAEQDICSEVCELESIFESSSH----EQADDDNARNQALEF 681
Query: 696 IHEMGWLLHRSQL--KYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDE 753
++E+GWLLHR+ + K V L S F + RF L F+M+ +WCAV K LL+ L
Sbjct: 682 LNELGWLLHRANIISKQDKVPLAS----FNIWRFRNLGIFAMEREWCAVTKLLLDFLFTG 737
Query: 754 TVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNH 813
V+ G LSE LLH AVR S Q+V LL Y P+ +
Sbjct: 738 LVDIGSQSPEEVVLSE-NLLHAAVRMKSAQMVRFLLGYKPNESLKRTA-----------E 785
Query: 814 SYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYA 873
++LFRPDA GP+ TPLHIAA D +EDVLDALTNDP +VGI W+NARD G TPEDYA
Sbjct: 786 TFLFRPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLVGINTWRNARDGAGFTPEDYA 845
Query: 874 RLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNM-TESNKNPKQNESFTSLEIGKAEVRRS 932
R RG+ Y+++V+KKINK G HVV+ +PS++ K E SLEIG V
Sbjct: 846 RQRGNDAYLNMVEKKINKHLGKGHVVLGVPSSIHPVITDGVKPGE--VSLEIGMT-VPPP 902
Query: 933 QGNCKLCDTK-ISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRW 991
+C C + + + R+ +YRPAML+++ IA +CVCV LL + P+V Y FRW
Sbjct: 903 APSCNACSRQALMYPNSTARTFLYRPAMLTVMGIAVICVCVGLLLHTCPKV-YAAPTFRW 961
Query: 992 ESLDFG 997
E L+ G
Sbjct: 962 ELLERG 967
>dbj|BAD93848.1| squamosa promoter binding protein-like 1 [Arabidopsis thaliana]
Length = 528
Score = 650 bits (1678), Expect = 0.0
Identities = 341/564 (60%), Positives = 417/564 (73%), Gaps = 46/564 (8%)
Query: 443 AQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAV 502
+Q RT RIVFKLFGKEPNEFP+VLR QILDWLS SPTD+ESYIRPGCIVLTIYLRQAE
Sbjct: 4 SQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESYIRPGCIVLTIYLRQAETA 63
Query: 503 WEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSK 562
WEEL DL SL KLLD+SDD W TGW+++RVQ+Q+AF++NGQVV+DTSL +S +YS
Sbjct: 64 WEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYNGQVVVDTSLSLKSRDYSH 123
Query: 563 IWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHEST----DQYSEEL 618
I +V P+A+ A+++AQF+VKG+NL + TRL+C++EGKYL+ E H+ST D + +
Sbjct: 124 IISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEGKYLIQETTHDSTTREDDDFKDNS 183
Query: 619 DELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAE-EDVCTEIRVLEPLLESSETD 677
+ ++C+ FSC +P+ +GRGF+EIEDQGLSSSFFPF+V E +DVC+EIR+LE LE + TD
Sbjct: 184 EIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDDVCSEIRILETTLEFTGTD 243
Query: 678 PDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDH 737
+ QAMDFIHE+GWLLHRS+L + N GV FPL RF WL+EFSMD
Sbjct: 244 ----------SAKQAMDFIHEIGWLLHRSKLGES--DPNPGV--FPLIRFQWLIEFSMDR 289
Query: 738 DWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTS 797
+WCAV++KLLN+ D V + + LSE+ LLHRAVR+NSK +VE+LLRY+P
Sbjct: 290 EWCAVIRKLLNMFFDGAVGEFSSSS-NATLSELCLLHRAVRKNSKPMVEMLLRYIPK--- 345
Query: 798 DELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEA 857
+ + LFRPDA GPAGLTPLHIAAGKDGSEDVLDALT DP MVGIEA
Sbjct: 346 -------------QQRNSLFRPDAAGPAGLTPLHIAAGKDGSEDVLDALTEDPAMVGIEA 392
Query: 858 WKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAA-HVVVEIPSNMTE-SNKNPKQ 915
WK RDSTG TPEDYARLRGH++YIHL+Q+KINK HVVV IP + ++ K PK
Sbjct: 393 WKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVNIPVSFSDREQKEPKS 452
Query: 916 NESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALL 975
++LEI +Q CKLCD K+ T RS+ YRPAMLSMVAIAAVCVCVALL
Sbjct: 453 GPMASALEI-------TQIPCKLCDHKLVYGT-TRRSVAYRPAMLSMVAIAAVCVCVALL 504
Query: 976 FKSSPEVLYMFRPFRWESLDFGTS 999
FKS PEVLY+F+PFRWE LD+GTS
Sbjct: 505 FKSCPEVLYVFQPFRWELLDYGTS 528
>ref|NP_908512.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|8468036|dbj|BAA96636.1| putative squamosa promoter
binding protein-like 1 [Oryza sativa (japonica
cultivar-group)]
Length = 862
Score = 489 bits (1260), Expect = e-136
Identities = 338/993 (34%), Positives = 487/993 (49%), Gaps = 152/993 (15%)
Query: 22 SGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSS 81
SG+ K+ EW+LNDWRWD +LF+A+ N + S
Sbjct: 3 SGLKKKGLEWDLNDWRWDSNLFLATPSNA---------------------------SPSK 35
Query: 82 CSEEGDLEKGNKEGE-------KKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKK 134
CS E G EGE K+RRV +DDG A + G +S R +
Sbjct: 36 CSRR---ELGRAEGEIDFGVVDKRRRVSPEDDDGEECINAATTNGDDGQISGQRGRSSED 92
Query: 135 SRGAGGT--SNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
GT S+ CQV+ C +LS +DY++RHKVCE+H+K+ + N RFCQQCS
Sbjct: 93 EMPRQGTCSSSGPCCQVDGCTVNLSSARDYNKRHKVCEVHTKSGVVRIKNVEHRFCQQCS 152
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSNM 252
RFH L+EFDEGK+SCR RLA HN+RRRK +A + + +++ + S L+ LLK LS +
Sbjct: 153 RFHFLQEFDEGKKSCRSRLAQHNRRRRKVQVQAGVDVNSLHENHSLSNTLLLLLKQLSGL 212
Query: 253 HYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSALFSNGS 312
+ + G L+NL++ NL G+ RN M+ N +
Sbjct: 213 DSSGPSEQI--------------NGPNYLTNLVK---NLAALAGTQRNQDMLK----NAN 251
Query: 313 QGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQT 372
+ T + N + HD R P I + +E + + +
Sbjct: 252 SAAIASHTGNYVAKGNSL-------HDSR-----------PHIPVGTESTAE--EPTVER 291
Query: 373 KMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTSGNSDSAS 432
++ NFDLND YV+ D+ D V D+P + + S
Sbjct: 292 RVQNFDLNDAYVEGDENRTD---KIVFKLFGKEPNDFP-------------SDLRAQILS 335
Query: 433 AQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESYIRPGCIVL 492
S S E+ R I+ ++ + PN L A W+ +
Sbjct: 336 WLSNCPSDIESYIRPGCIILTIYMRLPNWMWDKLAADPAHWIQK---------------- 379
Query: 493 TIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFNGQVVIDTS 552
LI L S DT W+TGW++ RVQ + NG +++ +
Sbjct: 380 ---------------------LISL---STDTLWRTGWMYARVQDYLTLSCNGNLMLASP 415
Query: 553 LPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHESTD 612
N +I ++PIAV S A FSVKG+N+ +P T+L+C GKYL+ E + D
Sbjct: 416 WQPAIGNKHQILFITPIAVACSSTANFSVKGLNIAQPTTKLLCIFGGKYLIQEATEKLLD 475
Query: 613 QYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAEEDVCTEIRVLEPLLE 672
+ QC+ FSCS P ++GRGFIE+ED SS FPF+VAEEDVC+EIR LE LL
Sbjct: 476 DTKMQRGP-QCLTFSCSFPSTSGRGFIEVEDLDQSSLSFPFVVAEEDVCSEIRTLEHLLN 534
Query: 673 -SSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLM 731
S D +E + ++ +A++F+HE GW L RS ++ + FP RF WL+
Sbjct: 535 LVSFDDTLVEKNDLLASRDRALNFLHEFGWFLQRSHIRATSETPKDCTEGFPAARFRWLL 594
Query: 732 EFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRY 791
F++D ++CAV+KKLL+ L V+ T+ L + L+ AV + SK L++ LL Y
Sbjct: 595 SFAVDREFCAVIKKLLDTLFQGGVDLDVQSTVEFVLKQ-DLVFVAVNKRSKPLIDFLLTY 653
Query: 792 VPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPC 851
+ + G E A +LF PD GP+ +TPLHIAA + VLDALT+DP
Sbjct: 654 TTSSAPMD-GTESAAPA-----QFLFTPDIAGPSDITPLHIAATYSDTAGVLDALTDDPQ 707
Query: 852 MVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPS-----NM 906
+GI+AWKNARD+TG TPEDYAR RGH +YI +VQ KI+ AHV V I S +
Sbjct: 708 QLGIKAWKNARDATGLTPEDYARKRGHESYIEMVQNKIDSRLPKAHVSVTISSTTSTTDF 767
Query: 907 TESNKNPKQNESFTSLEIGKAEVRRSQG--NCKLCDTKISCRTAVGRSMVYRPAMLSMVA 964
TE + + + T+ ++ K + ++ +C+ C +++ R + R + RPA+LS+VA
Sbjct: 768 TEKHASQSKTTDQTAFDVEKGQQISTKPPLSCRQCLPELAYRHHLNRFLSTRPAVLSLVA 827
Query: 965 IAAVCVCVALLFKSSPEVLYMFRPFRWESLDFG 997
IAAVCVCV L+ + P + M PFRW SL G
Sbjct: 828 IAAVCVCVGLIMQGPPHIGGMRGPFRWNSLRSG 860
>gb|AAC34220.1| unknown protein [Arabidopsis thaliana]
Length = 383
Score = 439 bits (1128), Expect = e-121
Identities = 239/415 (57%), Positives = 292/415 (69%), Gaps = 46/415 (11%)
Query: 592 RLMCALEGKYLVCEDAHEST----DQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLS 647
RL+C++EGKYL+ E H+ST D + + + ++C+ FSC +P+ +GRGF+EIEDQGLS
Sbjct: 8 RLLCSVEGKYLIQETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLS 67
Query: 648 SSFFPFIVAEED-VCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRS 706
SSFFPF+V E+D VC+EIR+LE LE + TD + QAMDFIHE+GWLLHRS
Sbjct: 68 SSFFPFLVVEDDDVCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLHRS 117
Query: 707 QLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQA 766
+L + N GV FPL RF WL+EFSMD +WCAV++KLLN+ D V + +
Sbjct: 118 KLGES--DPNPGV--FPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NAT 172
Query: 767 LSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAG 826
LSE+ LLHRAVR+NSK +VE+LLRY+P + + LFRPDA GPAG
Sbjct: 173 LSELCLLHRAVRKNSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPAG 216
Query: 827 LTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQ 886
LTPLHIAAGKDGSEDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+Q
Sbjct: 217 LTPLHIAAGKDGSEDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQ 276
Query: 887 KKINKTQGAA-HVVVEIPSNMTE-SNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKIS 944
+KINK HVVV IP + ++ K PK ++LEI +Q CKLCD K+
Sbjct: 277 RKINKKSTTEDHVVVNIPVSFSDREQKEPKSGPMASALEI-------TQIPCKLCDHKLV 329
Query: 945 CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
T RS+ YRPAMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE LD+GTS
Sbjct: 330 YGT-TRRSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 383
>ref|NP_173522.1| SPL1-Related2 protein (SPL1R2) [Arabidopsis thaliana]
gi|67461574|sp|Q8RY95|SPL14_ARATH Squamosa-promoter
binding-like protein 14 (SPL1-related protein 2)
gi|4836890|gb|AAD30593.1| Unknown protein [Arabidopsis
thaliana]
Length = 1035
Score = 426 bits (1095), Expect = e-117
Identities = 331/1087 (30%), Positives = 498/1087 (45%), Gaps = 204/1087 (18%)
Query: 26 KRSREWNLNDWRWDGDLFIASRVN-QVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE 84
+R EWN W WD F A V+ +VQ
Sbjct: 38 QRRDEWNSKMWDWDSRRFEAKPVDVEVQ-------------------------------- 65
Query: 85 EGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVE--------RDGKKSR 136
E DL N+ GE++ L LNL ++ V E R KK R
Sbjct: 66 EFDLTLRNRSGEER----------------GLDLNLGSGLTAVEETTTTTQNVRPNKKVR 109
Query: 137 GAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHI 196
N +CQV++C DLS KDYHRRHKVCE+HSKA++ALVG MQRFCQQCSRFH+
Sbjct: 110 SGSPGGNYPMCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHL 169
Query: 197 LEEFDEGKRSCRRRLAGHNKRRRKTNQ-EAVPNGS--PTNDDQTSSY---------LLIS 244
L EFDEGKRSCRRRLAGHN+RRRKT Q E V +G P N D T++ LL +
Sbjct: 170 LSEFDEGKRSCRRRLAGHNRRRRKTTQPEEVASGVVVPGNHDTTNNTANANMDLMALLTA 229
Query: 245 L------------------------LKILSNMHYQPTDQDLLTHL--LRSLASQNDEQGS 278
L L+IL+ ++ P DL++ L + SLA +N + +
Sbjct: 230 LACAQGKNAVKPPVGSPAVPDREQLLQILNKINALPLPMDLVSKLNNIGSLARKNMDHPT 289
Query: 279 KNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVIT----------QHQPVSMN 328
N N + G S +++ L + SP + + ++
Sbjct: 290 VNPQNDMN--------GASPSTMDLLAVLSTTLGSSSPDALAILSQGGFGNKDSEKTKLS 341
Query: 329 QMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDD 388
+ + + RT + + S SN P S+ DS GQ ++ L +D
Sbjct: 342 SYENGVTTNLEKRTFGFSSVGGERSSSSNQSP--SQDSDSRGQDTRSSLSLQLFTSSPED 399
Query: 389 GTEDL---------------------ERLPVSTNLATSSVDYPWTQQDSHQSSPAQT--- 424
+ PV L + +H++S +T
Sbjct: 400 ESRPTVASSRKYYSSASSNPVEDRSPSSSPVMQELFPLQASPETMRSKNHKNSSPRTGCL 459
Query: 425 -----------------------SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNE 461
SG + S S SP S + +AQ RT +IVFKL K+P++
Sbjct: 460 PLELFGASNRGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQ 519
Query: 462 FPLVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVS 521
P LR++I +WLS P+++ESYIRPGC+VL++Y+ + A WE+L L L LL S
Sbjct: 520 LPGTLRSEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNS 579
Query: 522 DDTFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSV 581
FW+ + Q+A NG+V S +R+ N ++ +VSP+AV A + V
Sbjct: 580 PSDFWRNARFIVNTGRQLASHKNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVV 637
Query: 582 KGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCS--VPVSNGRGFI 639
+G +L + C G Y+ E Q DEL F P GR FI
Sbjct: 638 RGRSLTNDGISIRCTHMGSYMAMEVTRAVCRQ--TIFDELNVNSFKVQNVHPGFLGRCFI 695
Query: 640 EIEDQGLSSSFFPFIVAEEDVCTEI-RVLEPLLESSE--TDPDIEGTGK-IKAKSQAMDF 695
E+E+ G FP I+A +C E+ R+ E S+ T+ + + + ++ + + F
Sbjct: 696 EVEN-GFRGDSFPLIIANASICKELNRLGEEFHPKSQDMTEEQAQSSNRGPTSREEVLCF 754
Query: 696 IHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDET- 754
++E+GWL ++Q L D F L RF +L+ S++ D+CA+++ LL++L++
Sbjct: 755 LNELGWLFQKNQTS----ELREQSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNL 809
Query: 755 VNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHS 814
VN + L+E+ LL+RAV+R S ++VELL+ Y+ + L +
Sbjct: 810 VNDELNREALDMLAEIQLLNRAVKRKSTKMVELLIHYLVN-----------PLTLSSSRK 858
Query: 815 YLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYAR 874
++F P+ GP G+TPLH+AA GS+D++D LTNDP +G+ +W RD+TG TP YA
Sbjct: 859 FVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAA 918
Query: 875 LRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQG 934
+R ++ Y LV +K+ + V + I + + K+ SLE+ K S
Sbjct: 919 IRNNHNYNSLVARKLADKRN-KQVSLNIEHEVVDQTGLSKR----LSLEMNK-----SSS 968
Query: 935 NCKLCDT---KISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRW 991
+C C T K R + + + P + SM+A+A VCVCV + + P ++ F W
Sbjct: 969 SCASCATVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSW 1027
Query: 992 ESLDFGT 998
LD+G+
Sbjct: 1028 GGLDYGS 1034
>gb|AAG51947.1| unknown protein; 70902-74753 [Arabidopsis thaliana]
Length = 1020
Score = 411 bits (1057), Expect = e-113
Identities = 314/1074 (29%), Positives = 488/1074 (45%), Gaps = 199/1074 (18%)
Query: 30 EWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSEEGDLE 89
+W +N W+WDG F A ++Q ESL++
Sbjct: 40 DWQMNRWKWDGQRFEAI---ELQGESLQLS------------------------------ 66
Query: 90 KGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKKSRGAGGTSNRAVCQV 149
NK+G G ND G ++L V R G G GG N CQV
Sbjct: 67 --NKKGLDLNLPC-----GFNDVEGT-PVDLTRPSKKV--RSGSPGSGGGGGGNYPKCQV 116
Query: 150 EDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRR 209
++C DLS KDYHRRHKVCE+HSKA++ALVG MQRFCQQCSRFH+L EFDEGKRSCRR
Sbjct: 117 DNCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRR 176
Query: 210 RLAGHNKRRRKTNQEAVPNG--SPTNDDQTSSYLLIS----------------------- 244
RL GHN+RRRKT +A+ + + N D TS+ +
Sbjct: 177 RLDGHNRRRRKTQPDAITSQVVALENRDNTSNNTNMDVMALLTALVCAQGRNEATTNGSP 236
Query: 245 -------LLKILSNMHYQPTDQDLLTHL--LRSLASQNDEQGSKNLSNLLREQENLLREG 295
LL+IL+ + P +L + L + LA +N EQ S +N +
Sbjct: 237 GVPQREQLLQILNKIKALPLPMNLTSKLNNIGILARKNPEQPSP------MNPQNSMNGA 290
Query: 296 GSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKP-- 353
S +++AL ++ +P I +E + +SDH +S++
Sbjct: 291 SSPSTMDLLAALSASLGSSAPEAIAFLSQGGFGN--KESNDRTKLTSSDHSATTSLEKKT 348
Query: 354 -------------SISNSPPAYSETRDSSGQTKMNNFDL-----------------NDIY 383
S ++SP YS DS GQ ++ L + Y
Sbjct: 349 LEFPSFGGGERTSSTNHSPSQYS---DSRGQDTRSSLSLQLFTSSPEEESRPKVASSTKY 405
Query: 384 VDSDDGTEDLERLPVSTN-------LATSSVDYPWTQQDSHQSSPAQT------------ 424
S +R P S+ L TS + +SP +
Sbjct: 406 YSSASSNPVEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNR 465
Query: 425 --------------SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQI 470
SG + S S SP S + AQ RT +I FKLF K+P++ P LR +I
Sbjct: 466 GATANPNYNVLRHQSGYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEI 525
Query: 471 LDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGW 530
WLS P+D+ES+IRPGC++L++Y+ + + WE+L +L + L V D FW
Sbjct: 526 FRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSR 583
Query: 531 VHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPA 590
+ Q+A +G++ + S +R+ N ++ TVSP+AV A + V+G NL
Sbjct: 584 FLVNAGRQLASHKHGEIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDG 641
Query: 591 TRLMCALEGKYLVCEDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIEIEDQGLSS 648
RL CA G Y E + ++DEL Q + VS GR FIE+E+ GL
Sbjct: 642 MRLRCAHMGNYASMEVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIELEN-GLRG 698
Query: 649 SFFPFIVAEEDVCTEIRVLEPLLESSET-DPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQ 707
FP I+A +C E+ LE + + I+ + +++ + + F++E+GWL R
Sbjct: 699 DNFPLIIANATICKELNRLEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQR-- 756
Query: 708 LKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGD--HPTLYQ 765
+ + G F L RF +L+ S++ D+C++++ +L+++++ + K +
Sbjct: 757 ---KWTSDIHGEPDFSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKESLD 813
Query: 766 ALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPA 825
L+++ LL+RA++R + ++ E L+ Y V +++F P GP
Sbjct: 814 MLADIQLLNRAIKRRNTKMAETLIHY---------------SVNPSTRNFIFLPSIAGPG 858
Query: 826 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 885
+TPLH+AA S+D++DALTNDP +G+ W D+TG TP YA +R +++Y LV
Sbjct: 859 DITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYNTLV 918
Query: 886 QKKI-NKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKIS 944
+K+ +K G + +E + +K +E++RS C K
Sbjct: 919 ARKLADKRNGQISLNIENGIDQIGLSKRL------------SSELKRSCNTCASVALKYQ 966
Query: 945 CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+ + R + P + SM+A+A VCVCV + + P ++ F W LD+G+
Sbjct: 967 RKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 1019
>sp|Q700C2|SPL16_ARATH Squamosa-promoter binding-like protein 16 (SPL1-related protein 3)
Length = 988
Score = 411 bits (1056), Expect = e-113
Identities = 314/1074 (29%), Positives = 488/1074 (45%), Gaps = 199/1074 (18%)
Query: 30 EWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSEEGDLE 89
+W +N W+WDG F A ++Q ESL++
Sbjct: 8 DWQMNRWKWDGQRFEAI---ELQGESLQLS------------------------------ 34
Query: 90 KGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVERDGKKSRGAGGTSNRAVCQV 149
NK+G G ND G ++L V R G G GG N CQV
Sbjct: 35 --NKKGLDLNLPC-----GFNDVEGT-PVDLTRPSKKV--RSGSPGSGGGGGGNYPKCQV 84
Query: 150 EDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRR 209
++C DLS KDYHRRHKVCE+HSKA++ALVG MQRFCQQCSRFH+L EFDEGKRSCRR
Sbjct: 85 DNCKEDLSIAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRR 144
Query: 210 RLAGHNKRRRKTNQEAVPNG--SPTNDDQTSSYLLIS----------------------- 244
RL GHN+RRRKT +A+ + + N D TS+ +
Sbjct: 145 RLDGHNRRRRKTQPDAITSQVVALENRDNTSNNTNMDVMALLTALVCAQGRNEATTNGSP 204
Query: 245 -------LLKILSNMHYQPTDQDLLTHL--LRSLASQNDEQGSKNLSNLLREQENLLREG 295
LL+IL+ + P +L + L + LA +N EQ S +N +
Sbjct: 205 GVPQREQLLQILNKIKALPLPMNLTSKLNNIGILARKNPEQPSP------MNPQNSMNGA 258
Query: 296 GSSRNSGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKP-- 353
S +++AL ++ +P I +E + +SDH +S++
Sbjct: 259 SSPSTMDLLAALSASLGSSAPEAIAFLSQGGFGN--KESNDRTKLTSSDHSATTSLEKKT 316
Query: 354 -------------SISNSPPAYSETRDSSGQTKMNNFDL-----------------NDIY 383
S ++SP YS DS GQ ++ L + Y
Sbjct: 317 LEFPSFGGGERTSSTNHSPSQYS---DSRGQDTRSSLSLQLFTSSPEEESRPKVASSTKY 373
Query: 384 VDSDDGTEDLERLPVSTN-------LATSSVDYPWTQQDSHQSSPAQT------------ 424
S +R P S+ L TS + +SP +
Sbjct: 374 YSSASSNPVEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNR 433
Query: 425 --------------SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQI 470
SG + S S SP S + AQ RT +I FKLF K+P++ P LR +I
Sbjct: 434 GATANPNYNVLRHQSGYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEI 493
Query: 471 LDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGW 530
WLS P+D+ES+IRPGC++L++Y+ + + WE+L +L + L V D FW
Sbjct: 494 FRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSR 551
Query: 531 VHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPA 590
+ Q+A +G++ + S +R+ N ++ TVSP+AV A + V+G NL
Sbjct: 552 FLVNAGRQLASHKHGRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDG 609
Query: 591 TRLMCALEGKYLVCEDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIEIEDQGLSS 648
RL CA G Y E + ++DEL Q + VS GR FIE+E+ GL
Sbjct: 610 MRLRCAHMGNYASMEVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIELEN-GLRG 666
Query: 649 SFFPFIVAEEDVCTEIRVLEPLLESSET-DPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQ 707
FP I+A +C E+ LE + + I+ + +++ + + F++E+GWL R
Sbjct: 667 DNFPLIIANATICKELNRLEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQR-- 724
Query: 708 LKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGD--HPTLYQ 765
+ + G F L RF +L+ S++ D+C++++ +L+++++ + K +
Sbjct: 725 ---KWTSDIHGEPDFSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKESLD 781
Query: 766 ALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPA 825
L+++ LL+RA++R + ++ E L+ Y V +++F P GP
Sbjct: 782 MLADIQLLNRAIKRRNTKMAETLIHY---------------SVNPSTRNFIFLPSIAGPG 826
Query: 826 GLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLV 885
+TPLH+AA S+D++DALTNDP +G+ W D+TG TP YA +R +++Y LV
Sbjct: 827 DITPLHLAASTSSSDDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYNTLV 886
Query: 886 QKKI-NKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKIS 944
+K+ +K G + +E + +K +E++RS C K
Sbjct: 887 ARKLADKRNGQISLNIENGIDQIGLSKRL------------SSELKRSCNTCASVALKYQ 934
Query: 945 CRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+ + R + P + SM+A+A VCVCV + + P ++ F W LD+G+
Sbjct: 935 RKVSGSRRLFPTPIIHSMLAVATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 987
>gb|AAP46238.1| unknown protein, 5'-partial [Oryza sativa (japonica
cultivar-group)] gi|50919619|ref|XP_470170.1| unknown
protein, 5'-partial [Oryza sativa (japonica
cultivar-group)]
Length = 462
Score = 363 bits (931), Expect = 2e-98
Identities = 210/486 (43%), Positives = 291/486 (59%), Gaps = 30/486 (6%)
Query: 516 KLLDVSDDTFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASK 575
KLL+ S FW +G V + V+HQ+AF+ NGQ+++D L +++Y KI V PIA P S
Sbjct: 1 KLLNSSTGNFWASGLVFVMVRHQIAFMHNGQLMLDRPLANSAHHYCKILCVRPIAAPFST 60
Query: 576 RAQFSVKGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNG 635
+ F V+G+NL+ ++RL+C+ EG + ED D E D+++ + F C +P S G
Sbjct: 61 KVNFRVEGLNLVSDSSRLICSFEGSCIFQEDTDNIVDDV--EHDDIEYLNFCCPLPSSRG 118
Query: 636 RGFIEIEDQGLSSSFFPFIVAEEDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDF 695
RGF+E+ED G S+ FFPFI+AE+D+C+E+ LE + ESS E A++QA++F
Sbjct: 119 RGFVEVEDGGFSNGFFPFIIAEQDICSEVCELESIFESSSH----EQADDDNARNQALEF 174
Query: 696 IHEMGWLLHRSQL--KYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDE 753
++E+GWLLHR+ + K V L S F + RF L F+M+ +WCAV K LL+ L
Sbjct: 175 LNELGWLLHRANIISKQDKVPLAS----FNIWRFRNLGIFAMEREWCAVTKLLLDFLFTG 230
Query: 754 TVNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNH 813
V+ G LSE LLH AVR S Q+V LL Y P+ +
Sbjct: 231 LVDIGSQSPEEVVLSE-NLLHAAVRMKSAQMVRFLLGYKPNESLKRTA-----------E 278
Query: 814 SYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYA 873
++LFRPDA GP+ TPLHIAA D +EDVLDALTNDP +VGI W+NARD G TPEDYA
Sbjct: 279 TFLFRPDAQGPSKFTPLHIAAATDDAEDVLDALTNDPGLVGINTWRNARDGAGFTPEDYA 338
Query: 874 RLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNM-TESNKNPKQNESFTSLEIGKAEVRRS 932
R RG+ Y+++V+KKINK G HVV+ +PS++ K E SLEIG V
Sbjct: 339 RQRGNDAYLNMVEKKINKHLGKGHVVLGVPSSIHPVITDGVKPGE--VSLEIGMT-VPPP 395
Query: 933 QGNCKLCDTK-ISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRW 991
+C C + + + R+ +YRPAML+++ IA +CVCV LL + P+V Y FRW
Sbjct: 396 APSCNACSRQALMYPNSTARTFLYRPAMLTVMGIAVICVCVGLLLHTCPKV-YAAPTFRW 454
Query: 992 ESLDFG 997
E L+ G
Sbjct: 455 ELLERG 460
>ref|XP_483324.1| putative SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)] gi|42408812|dbj|BAD10073.1| putative
SPL1-Related2 protein [Oryza sativa (japonica
cultivar-group)]
Length = 1140
Score = 327 bits (839), Expect = 9e-88
Identities = 245/784 (31%), Positives = 383/784 (48%), Gaps = 73/784 (9%)
Query: 256 PTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGS-SRNSGMVSALFSNGSQG 314
P+ DLL L +LA+ N + SN + Q + G + S++ A N +
Sbjct: 388 PSTMDLLAVLSTALATSNPD------SNTSQSQGSSDSSGNNKSKSQSTEPANVVNSHEK 441
Query: 315 SPTVITQHQPVSMNQMQQEMVHTHDVRTSDH---QLISSIKPSI-------------SNS 358
S V + + + EM D T + +L S + + +S
Sbjct: 442 SIRVFSATRKNDALERSPEMYKQPDQETPPYLSLRLFGSTEEDVPCKMDTANKYLSSESS 501
Query: 359 PPAYSETRDSSGQTKMNNFDLNDIYVDSD--DGTEDLERLPVSTNLATSSVD---YPWTQ 413
P + SS F + + D+ D ED+ + VST+ A + + ++
Sbjct: 502 NPLDERSPSSSPPVTHKFFPIRSVDEDARIADYGEDIATVEVSTSRAWRAPPLELFKDSE 561
Query: 414 QDSHQSSPA----QTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQ 469
+ SP Q+ S S S SPS+S+ + Q RT RI+FKLFGKEP+ P LR +
Sbjct: 562 RPIENGSPPNPAYQSCYTSTSCSDHSPSTSNSDGQDRTGRIIFKLFGKEPSTIPGNLRGE 621
Query: 470 ILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTG 529
I++WL SP ++E YIRPGC+VL++YL W+EL +L + L+ SD FW+ G
Sbjct: 622 IVNWLKHSPNEMEGYIRPGCLVLSMYLSMPAIAWDELEENLLQRVNTLVQGSDLDFWRKG 681
Query: 530 WVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRP 589
+R Q+ +G + S +R+ N ++ VSPIAV ++ +KG NL P
Sbjct: 682 RFLVRTDAQLVSYKDGATRLSKS--WRTWNTPELTFVSPIAVVGGRKTSLILKGRNLTIP 739
Query: 590 ATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSS 649
T++ C GKY+ E + + ++ + GR FIE+E++ +S
Sbjct: 740 GTQIHCTSTGKYISKEVLCSAYPGTIYDDSGVETFDLPGEPHLILGRYFIEVENRFRGNS 799
Query: 650 FFPFIVAEEDVCTEIRVLEPLLESSE-----TDPDIEGTGKIKAKSQAMDFIHEMGWLLH 704
FP I+A VC E+R LE LE S+ +D ++K K + + F++E+GWL
Sbjct: 800 -FPVIIANSSVCQELRSLEAELEGSQFVDGSSDDQAHDARRLKPKDEVLHFLNELGWLFQ 858
Query: 705 RSQLKYRMVNL-NSGVDL--FPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGD-H 760
++ +SG+DL F RF +L+ FS + DWC++ K LL +L ++ +
Sbjct: 859 KAAASTSAEKSDSSGLDLMYFSTARFRYLLLFSSERDWCSLTKTLLEILAKRSLASDELS 918
Query: 761 PTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPD 820
+ LSE+ LL+RAV+R S + LL+++V + P+D L Y F P+
Sbjct: 919 QETLEMLSEIHLLNRAVKRKSSHMARLLVQFV------VVCPDDSKL-------YPFLPN 965
Query: 821 AVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYT 880
GP GLTPLH+AA + + D++DALT+DP +G+ W +A D G +PE YA+LR +
Sbjct: 966 VAGPGGLTPLHLAASIEDAVDIVDALTDDPQQIGLSCWHSALDDDGQSPETYAKLRNNNA 1025
Query: 881 YIHLVQKKI--NKTQGAAHVVVEIPSNMTES-NKNPKQNESFTSLEIGKAEVRRSQGNCK 937
Y LV +K+ K +V + +M +S N K + +L+I RS C
Sbjct: 1026 YNELVAQKLVDRKNNQVTIMVGKEEIHMDQSGNVGEKNKSAIQALQI------RSCNQCA 1079
Query: 938 LCDTKISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMF---RPFRWESL 994
+ D + R R ++ RP + SM+AIAAVCVCV + ++ L F R F+WE L
Sbjct: 1080 ILDAGLLRRPMHSRGLLARPYIHSMLAIAAVCVCVCVFMRA----LLRFNSGRSFKWERL 1135
Query: 995 DFGT 998
DFGT
Sbjct: 1136 DFGT 1139
Score = 145 bits (366), Expect = 7e-33
Identities = 107/314 (34%), Positives = 152/314 (48%), Gaps = 57/314 (18%)
Query: 132 GKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
G GG + +CQV+DC ADL+ KDYHRRHKVCE+H K ++ALVGN MQRFCQQC
Sbjct: 172 GGGGNSGGGGGSYPMCQVDDCRADLTNAKDYHRRHKVCEIHGKTTKALVGNQMQRFCQQC 231
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSN 251
SRFH L EFDEGKRSCRRRLAGHN+RRRKT PT D S LL + +N
Sbjct: 232 SRFHPLSEFDEGKRSCRRRLAGHNRRRRKT--------QPT--DVASQLLLPGNQENAAN 281
Query: 252 MHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSAL--FS 309
QD++ + Q G + +++NL++ ++S + +
Sbjct: 282 -----RTQDIVNLITVIARLQGSNVGKLPSIPPIPDKDNLVQ---------IISKINSIN 327
Query: 310 NGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSS 369
NG+ S + ++ ++ + QQ+ S+ + + E + +
Sbjct: 328 NGNSASKSPPSEAVDLNASHSQQQ-------------------DSVQRTTNGF-EKQTNG 367
Query: 370 GQTKMNNFDLNDIYVDSDDGTEDLERLPV-STNLATSSVDYPWTQQDSHQSSPAQTSGNS 428
+ N FD D ++ L V ST LATS+ D S+ +Q+ G+S
Sbjct: 368 LDKQTNGFDKQADGFDKQAVPSTMDLLAVLSTALATSNPD----------SNTSQSQGSS 417
Query: 429 DSASAQSPSSSSGE 442
DS+ S S E
Sbjct: 418 DSSGNNKSKSQSTE 431
>emb|CAB56770.1| SPL1-Related2 protein [Arabidopsis thaliana]
Length = 812
Score = 294 bits (752), Expect = 1e-77
Identities = 235/841 (27%), Positives = 385/841 (44%), Gaps = 117/841 (13%)
Query: 227 PNGSPTNDDQTSSYLLISLLKILSNMHYQPTDQDLLTHL--LRSLASQNDEQGSKNLSNL 284
P GSP D+ LL+IL+ ++ P DL++ L + SLA +N + + N N
Sbjct: 19 PVGSPAVPDREQ------LLQILNKINALPLPMDLVSKLNNIGSLARKNMDHPTVNPQND 72
Query: 285 LREQENLLREGGSSRNSGMVSALFSNGSQGSPTVIT----------QHQPVSMNQMQQEM 334
+ G S +++ L + SP + + ++ + +
Sbjct: 73 MN--------GASPSTMDLLAVLSTTLGSSSPDALAILSQGGFGNKDSEKTKLSSYENGV 124
Query: 335 VHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDL- 393
+ RT + + S SN P S+ DS GQ ++ L +D +
Sbjct: 125 TTNLEKRTFGFSSVGGERSSSSNQSP--SQDSDSRGQDTRSSLSLQLFTSSPEDESRPTV 182
Query: 394 --------------------ERLPVSTNLATSSVDYPWTQQDSHQSSPAQT--------- 424
PV L + +H++S +T
Sbjct: 183 ASSRKYYSSASSNPVEDRSPSSSPVMQELFPLQASPETMRSKNHKNSSPRTGCLPLELFG 242
Query: 425 -----------------SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLR 467
SG + S S SP S + +AQ RT +IVFKL K+P++ P LR
Sbjct: 243 ASNRGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLR 302
Query: 468 AQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWK 527
++I +WLS P+++ESYIRPGC+VL++Y+ + A WE+L L L LL S FW+
Sbjct: 303 SEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWR 362
Query: 528 TGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLM 587
+ Q+A NG+V S +R+ N ++ +VSP+AV A + V+G +L
Sbjct: 363 NARFIVNTGRQLASHKNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLT 420
Query: 588 RPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCS--VPVSNGRGFIEIEDQG 645
+ C G Y+ E Q DEL F P GR FIE+E+ G
Sbjct: 421 NDGISIRCTHMGSYMAMEVTRAVCRQ--TIFDELNVNSFKVQNVHPGFLGRCFIEVEN-G 477
Query: 646 LSSSFFPFIVAEEDVCTEI-RVLEPLLESSE--TDPDIEGTGK-IKAKSQAMDFIHEMGW 701
FP I+A +C E+ R+ E S+ T+ + + + ++ + + F++E+GW
Sbjct: 478 FRGDSFPLIIANASICKELNRLGEEFHPKSQDMTEEQAQSSNRGPTSREEVLCFLNELGW 537
Query: 702 LLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDET-VNKGDH 760
L ++Q L D F L RF +L+ S++ D+CA+++ LL++L++ VN +
Sbjct: 538 LFQKNQTS----ELREQSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELN 592
Query: 761 PTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPD 820
L+E+ LL+RAV+R S ++VELL+ Y+ + ++ + ++F P+
Sbjct: 593 REALDMLAEIQLLNRAVKRKSTKMVELLIHYLVNPSA-----------LSSSRKFVFLPN 641
Query: 821 AVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYT 880
GP G+TPLH+AA GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++
Sbjct: 642 ITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHN 701
Query: 881 YIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCD 940
Y LV +K+ + V + I + + K+ SLE+ K S +C C
Sbjct: 702 YNSLVARKLADKRN-KQVSLNIEHEVVDQTGLSKR----LSLEMNK-----SSSSCASCA 751
Query: 941 T---KISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFG 997
T K R + + + P + SM+A+A VCVCV + + P ++ F W LD+G
Sbjct: 752 TVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYG 810
Query: 998 T 998
+
Sbjct: 811 S 811
>emb|CAB56773.1| Spl1-Related2 protein [Arabidopsis thaliana]
Length = 712
Score = 293 bits (749), Expect = 3e-77
Identities = 194/586 (33%), Positives = 307/586 (52%), Gaps = 42/586 (7%)
Query: 423 QTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIE 482
Q SG + S S SP S + +AQ RT +IVFKL K+P++ P LR++I +WLS P+++E
Sbjct: 158 QQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQLPGTLRSEIYNWLSNIPSEME 217
Query: 483 SYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFI 542
SYIRPGC+VL++Y+ + A WE+L L L LL S FW+ + Q+A
Sbjct: 218 SYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSPSDFWRNARFIVNTGRQLASH 277
Query: 543 FNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYL 602
NG+V S +R+ N ++ +VSP+AV A + V+G +L + C G Y+
Sbjct: 278 KNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVVRGRSLTNDGISIRCTHMGSYM 335
Query: 603 VCEDAHESTDQYSEELDELQCIQFSCS--VPVSNGRGFIEIEDQGLSSSFFPFIVAEEDV 660
E Q DEL F P GR FIE+E+ G FP I+A +
Sbjct: 336 AMEVTRAVCRQ--TIFDELNVNSFKVQNVHPGFLGRCFIEVEN-GFRGDSFPLIIANASI 392
Query: 661 CTEI-RVLEPLLESSE--TDPDIEGTGK-IKAKSQAMDFIHEMGWLLHRSQLKYRMVNLN 716
C E+ R+ E S+ T+ + + + ++ + + F++E+GWL ++Q L
Sbjct: 393 CKELNRLGEEFHPKSQDMTEEQAQSSNRGPTSREEVLCFLNELGWLFQKNQTS----ELR 448
Query: 717 SGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDET-VNKGDHPTLYQALSEMGLLHR 775
D F L RF +L+ S++ D+CA+++ LL++L++ VN + L+E+ LL+R
Sbjct: 449 EQSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREALDMLAEIQLLNR 507
Query: 776 AVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAG 835
AV+R S ++VELL+ Y+ + L + ++F P+ GP G+TPLH+AA
Sbjct: 508 AVKRKSTKMVELLIHYLVN-----------PLTLSSSRKFVFLPNITGPGGITPLHLAAC 556
Query: 836 KDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGA 895
GS+D++D LTNDP +G+ +W RD+TG TP YA +R ++ Y LV +K+ +
Sbjct: 557 TSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRN- 615
Query: 896 AHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDT---KISCRTAVGRS 952
V + I + + K+ SLE+ K S +C C T K R + +
Sbjct: 616 KQVSLNIEHEVVDQTGLSKR----LSLEMNK-----SSSSCASCATVALKYQRRVSGSQR 666
Query: 953 MVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+ P + SM+A+A VCVCV + + P ++ F W LD+G+
Sbjct: 667 LFPTPIIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSWGGLDYGS 711
>emb|CAB56771.1| SPL1-Related3 protein [Arabidopsis thaliana]
Length = 647
Score = 283 bits (725), Expect = 2e-74
Identities = 178/580 (30%), Positives = 298/580 (50%), Gaps = 46/580 (7%)
Query: 425 SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESY 484
SG + S S SP S + AQ RT +I FKLF K+P++ P LR +I WLS P+D+ES+
Sbjct: 107 SGYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESF 166
Query: 485 IRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFN 544
IRPGC++L++Y+ + + WE+L +L + L V D FW + Q+A +
Sbjct: 167 IRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDSEFWSNSRFLVNAGRQLASHKH 224
Query: 545 GQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVC 604
G++ + S +R+ N ++ TVSP+AV A + V+G NL RL CA G Y
Sbjct: 225 GRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASM 282
Query: 605 EDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAEEDVCT 662
E + ++DEL Q + VS GR FIE+E+ GL FP I+A +C
Sbjct: 283 EVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIELEN-GLRGDNFPLIIANATICK 339
Query: 663 EIRVLEPLLESSET-DPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDL 721
E+ LE + + I+ + +++ + + F++E+GWL R + + G
Sbjct: 340 ELNRLEEEFHPKDVIEEQIQNLDRPRSREEVLCFLNELGWLFQR-----KWTSDIHGEPD 394
Query: 722 FPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGD--HPTLYQALSEMGLLHRAVRR 779
F L RF +L+ S++ D+C++++ +L+++++ + K + L+++ LL+RA++R
Sbjct: 395 FSLPRFKFLLVCSVERDYCSLIRTVLDMMVERNLGKDGLLNKESLDMLADIQLLNRAIKR 454
Query: 780 NSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGS 839
+ ++ E L+ Y V +++F P GP +TPLH+AA S
Sbjct: 455 RNTKMAETLIHY---------------SVNPSTRNFIFLPSIAGPGDITPLHLAASTSSS 499
Query: 840 EDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKI-NKTQGAAHV 898
+D++DALTNDP +G+ W D+TG TP YA +R +++Y LV +K+ +K G +
Sbjct: 500 DDMIDALTNDPQEIGLSCWNTLVDATGQTPFSYAAMRDNHSYNTLVARKLADKRNGQISL 559
Query: 899 VVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRPA 958
+E + +K +E++RS C K + + R + P
Sbjct: 560 NIENGIDQIGLSKRL------------SSELKRSCNTCASVALKYQRKVSGSRRLFPTPI 607
Query: 959 MLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGT 998
+ SM+A+A VCVCV + + P ++ F W LD+G+
Sbjct: 608 IHSMLAVATVCVCVCVFMHAFP-MVRQGSHFSWGGLDYGS 646
>gb|AAC34221.1| putative squamosa-promoter binding protein [Arabidopsis thaliana]
Length = 240
Score = 255 bits (652), Expect = 5e-66
Identities = 142/248 (57%), Positives = 171/248 (68%), Gaps = 44/248 (17%)
Query: 6 GAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPL 65
G E FYG +GKRS EW+LNDW+WDGDLF+A++ + G+QFFPL
Sbjct: 8 GGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR--------GRQFFPL 51
Query: 66 GSGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 125
G+ SSN+SSSCS+EG+ +KKRR + ++ D GAL+LNL G
Sbjct: 52 GN-------SSNSSSSCSDEGN--------DKKRRAVAIQ----GDTNGALTLNLNG--- 89
Query: 126 PVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQ 185
E DG A T + AVCQVE+C ADLS+ KDYHRRHKVCEMHSKA+ A VG +Q
Sbjct: 90 ---ESDGLFP--AKKTKSGAVCQVENCEADLSKVKDYHRRHKVCEMHSKATSATVGGILQ 144
Query: 186 RFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISL 245
RFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRKTN E NG+P +DD +S+YLLI+L
Sbjct: 145 RFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNP-SDDHSSNYLLITL 203
Query: 246 LKILSNMH 253
LKILSNMH
Sbjct: 204 LKILSNMH 211
>gb|AAL69483.2| putative SPL1-related protein [Arabidopsis thaliana]
Length = 314
Score = 162 bits (410), Expect = 5e-38
Identities = 101/315 (32%), Positives = 169/315 (53%), Gaps = 31/315 (9%)
Query: 688 AKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLL 747
++ + + F++E+GWL ++Q L D F L RF +L+ S++ D+CA+++ LL
Sbjct: 26 SREEVLCFLNELGWLFQKNQTS----ELREQSD-FSLARFKFLLVCSVERDYCALIRTLL 80
Query: 748 NLLLDET-VNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKA 806
++L++ VN + L+E+ LL+RAV+R S ++VELL+ Y+ +
Sbjct: 81 DMLVERNLVNDELNREALDMLAEIQLLNRAVKRKSTKMVELLIHYLVN-----------P 129
Query: 807 LVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTG 866
L + ++F P+ GP G+TPLH+AA GS+D++D LTNDP +G+ +W RD+TG
Sbjct: 130 LTLSSSRKFVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDATG 189
Query: 867 STPEDYARLRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGK 926
TP YA +R ++ Y LV +K+ + V + I + + K+ SLE+ K
Sbjct: 190 QTPYSYAAIRNNHNYNSLVARKLADKRN-KQVSLNIEHEVVDQTGLSKR----LSLEMNK 244
Query: 927 AEVRRSQGNCKLCDT---KISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVL 983
S +C C T K R + + + P + SM+A+A VCVCV + + P ++
Sbjct: 245 -----SSSSCASCATVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFP-IV 298
Query: 984 YMFRPFRWESLDFGT 998
F W LD+G+
Sbjct: 299 RQGSHFSWGGLDYGS 313
>gb|AAS79566.1| At1g76580 [Arabidopsis thaliana] gi|46367500|emb|CAG25876.1|
hypothetical protein [Arabidopsis thaliana]
Length = 503
Score = 146 bits (368), Expect = 4e-33
Identities = 134/443 (30%), Positives = 201/443 (45%), Gaps = 55/443 (12%)
Query: 256 PTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSAL------FS 309
P+ DLL L SL S E + +E+ R +S + ++L F
Sbjct: 82 PSTMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTSLEKKTLEFP 141
Query: 310 NGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTS-DHQLISSI-----KPSISNSPPAYS 363
+ G T T H P + + + D R+S QL +S +P +++S YS
Sbjct: 142 SFGGGERTSSTNHSPSQYSDSRGQ-----DTRSSLSLQLFTSSPEEESRPKVASSTKYYS 196
Query: 364 -------ETRDSSGQTKMNN-FDLN--------DIYVDSDDGTED----LERLPVSTNLA 403
E R S M F L+ + Y D+ LE S A
Sbjct: 197 SASSNPVEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNRGA 256
Query: 404 TSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFP 463
T++ +Y + HQS G + S S SP S + AQ RT +I FKLF K+P++ P
Sbjct: 257 TANPNYNVLR---HQS------GYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLP 307
Query: 464 LVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDD 523
LR +I WLS P+D+ES+IRPGC++L++Y+ + + WE+L +L + L V D
Sbjct: 308 NTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDS 365
Query: 524 TFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKG 583
FW + Q+A +G++ + S +R+ N ++ TVSP+AV A + V+G
Sbjct: 366 EFWSNSRFLVNAGRQLASHKHGRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRG 423
Query: 584 VNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIEI 641
NL RL CA G Y E + ++DEL Q + VS GR FIE+
Sbjct: 424 RNLTNDGMRLRCAHMGNYASMEVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIEL 481
Query: 642 EDQGLSSSFFPFIVAEEDVCTEI 664
E+ GL FP I+A +C E+
Sbjct: 482 EN-GLRGDNFPLIIANATICKEL 503
>gb|AAY16440.1| squamosa promoter binding-like protein [Betula platyphylla]
Length = 175
Score = 133 bits (334), Expect = 3e-29
Identities = 62/98 (63%), Positives = 70/98 (71%)
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
KK +G G+ + CQ E C ADLS GK YH+RHKVCE H+KA LVG QRFCQQCS
Sbjct: 38 KKKKGVVGSGGKRCCQAEKCTADLSDGKQYHKRHKVCEHHAKAQVVLVGGMRQRFCQQCS 97
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGS 230
RFH L EFDE KRSCRRRLAGHN+RRR+ E+ GS
Sbjct: 98 RFHELSEFDETKRSCRRRLAGHNERRRENTAESHAEGS 135
>gb|AAP31970.1| At1g76580 [Arabidopsis thaliana] gi|22330667|ref|NP_177784.2|
SPL1-Related3 protein (SPL1R3) [Arabidopsis thaliana]
gi|17065188|gb|AAL32748.1| Unknown protein [Arabidopsis
thaliana]
Length = 488
Score = 130 bits (326), Expect = 3e-28
Identities = 125/419 (29%), Positives = 187/419 (43%), Gaps = 54/419 (12%)
Query: 256 PTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSAL------FS 309
P+ DLL L SL S E + +E+ R +S + ++L F
Sbjct: 82 PSTMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTSLEKKTLEFP 141
Query: 310 NGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTS-DHQLISSI-----KPSISNSPPAYS 363
+ G T T H P + + + D R+S QL +S +P +++S YS
Sbjct: 142 SFGGGERTSSTNHSPSQYSDSRGQ-----DTRSSLSLQLFTSSPEEESRPKVASSTKYYS 196
Query: 364 -------ETRDSSGQTKMNN-FDLN--------DIYVDSDDGTED----LERLPVSTNLA 403
E R S M F L+ + Y D+ LE S A
Sbjct: 197 SASSNPVEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNRGA 256
Query: 404 TSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFP 463
T++ +Y + HQS G + S S SP S + AQ RT +I FKLF K+P++ P
Sbjct: 257 TANPNYNVLR---HQS------GYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLP 307
Query: 464 LVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDD 523
LR +I WLS P+D+ES+IRPGC++L++Y+ + + WE+L +L + L V D
Sbjct: 308 NTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDS 365
Query: 524 TFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKG 583
FW + Q+A +G++ + S +R+ N ++ TVSP+AV A + V+G
Sbjct: 366 EFWSNSRFLVNAGRQLASHKHGRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRG 423
Query: 584 VNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIE 640
NL RL CA G Y E + ++DEL Q + VS GR FIE
Sbjct: 424 RNLTNDGMRLRCAHMGNYASMEVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIE 480
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,746,482,893
Number of Sequences: 2540612
Number of extensions: 76448136
Number of successful extensions: 226201
Number of sequences better than 10.0: 411
Number of HSP's better than 10.0 without gapping: 102
Number of HSP's successfully gapped in prelim test: 319
Number of HSP's that attempted gapping in prelim test: 224548
Number of HSP's gapped (non-prelim): 1321
length of query: 999
length of database: 863,360,394
effective HSP length: 138
effective length of query: 861
effective length of database: 512,755,938
effective search space: 441482862618
effective search space used: 441482862618
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Medicago: description of AC144728.1


