

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.9 + phase: 0
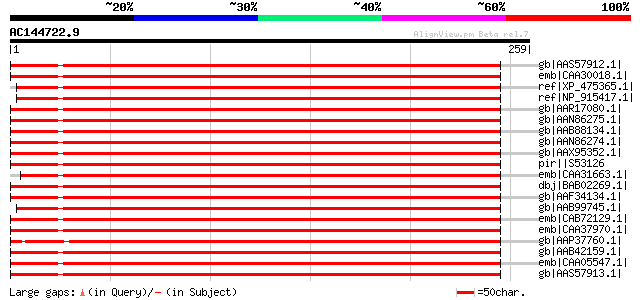
(259 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAS57912.1| 70 kDa heat shock cognate protein 1 [Vigna radiata] 363 3e-99
emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida] gi|123... 362 7e-99
ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar... 361 1e-98
ref|NP_915417.1| putative HSP70 [Oryza sativa (japonica cultivar... 360 2e-98
gb|AAR17080.1| heat shock protein 70-3 [Nicotiana tabacum] 359 4e-98
gb|AAN86275.1| non-cell-autonomous heat shock cognate protein 70... 359 4e-98
gb|AAB88134.1| cytosolic heat shock 70 protein [Spinacia olerace... 359 5e-98
gb|AAN86274.1| non-cell-autonomous heat shock cognate protein 70... 359 5e-98
gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragm... 358 6e-98
pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment) 358 6e-98
emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida] 358 1e-97
dbj|BAB02269.1| 70 kDa heat shock protein [Arabidopsis thaliana]... 358 1e-97
gb|AAF34134.1| high molecular weight heat shock protein [Malus x... 358 1e-97
gb|AAB99745.1| HSP70 [Triticum aestivum] 358 1e-97
emb|CAB72129.1| heat shock protein 70 [Cucumis sativus] 357 1e-97
emb|CAA37970.1| heat shock protein cognate 70 [Lycopersicon escu... 357 2e-97
gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|... 357 2e-97
gb|AAB42159.1| Hsc70 gi|2119719|pir||JC4786 dnaK-type molecular ... 355 5e-97
emb|CAA05547.1| heat shock protein 70 [Arabidopsis thaliana] 355 7e-97
gb|AAS57913.1| 70 kDa heat shock cognate protein 2 [Vigna radiata] 355 9e-97
>gb|AAS57912.1| 70 kDa heat shock cognate protein 1 [Vigna radiata]
Length = 649
Score = 363 bits (932), Expect = 3e-99
Identities = 180/245 (73%), Positives = 207/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVISG DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRFSDASVQSDIKLWPFKVISGAGDKPMIVVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEA+L S VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMKEIAEAFLGSTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>emb|CAA30018.1| heat shock protein 70 [Petunia x hybrida]
gi|123650|sp|P09189|HSP7C_PETHY Heat shock cognate 70
kDa protein
Length = 651
Score = 362 bits (928), Expect = 7e-99
Identities = 179/245 (73%), Positives = 206/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSDP VQ DI WPFKVI G DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIPGPGDKPMIVVTYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>ref|XP_475365.1| putative hsp70 [Oryza sativa (japonica cultivar-group)]
gi|47900318|gb|AAT39165.1| putative hsp70 [Oryza sativa
(japonica cultivar-group)]
Length = 646
Score = 361 bits (926), Expect = 1e-98
Identities = 176/242 (72%), Positives = 206/242 (84%), Gaps = 2/242 (0%)
Query: 4 KHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQ 63
K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AAK+Q
Sbjct: 3 KGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQ 60
Query: 64 AAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAE 123
A+NP NTVFDAKRLIGR+FSDP VQ D+ WPFKV+ G DKPMI V+YKG+EK AE
Sbjct: 61 VAMNPTNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVVPGPGDKPMIVVQYKGEEKQFAAE 120
Query: 124 EISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPT 183
EISS++L M++IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN+PT
Sbjct: 121 EISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 180
Query: 184 AAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDN 243
AAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGEDFDN
Sbjct: 181 AAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDN 240
Query: 244 RM 245
RM
Sbjct: 241 RM 242
>ref|NP_915417.1| putative HSP70 [Oryza sativa (japonica cultivar-group)]
gi|21104622|dbj|BAB93214.1| putative HSP70 [Oryza sativa
(japonica cultivar-group)] gi|15623835|dbj|BAB67894.1|
putative HSP70 [Oryza sativa (japonica cultivar-group)]
Length = 648
Score = 360 bits (924), Expect = 2e-98
Identities = 177/242 (73%), Positives = 206/242 (84%), Gaps = 2/242 (0%)
Query: 4 KHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQ 63
K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AAK+Q
Sbjct: 3 KGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQ 60
Query: 64 AAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAE 123
A+NP NTVFDAKRLIGR+FSDP VQ D+ WPFKVI G DKPMI V+YKG+EK AE
Sbjct: 61 VAMNPINTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPGDKPMIVVQYKGEEKQFSAE 120
Query: 124 EISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPT 183
EISS++L M++IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN+PT
Sbjct: 121 EISSMVLIKMREIAEAYLGSNIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 180
Query: 184 AAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDN 243
AAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGEDFDN
Sbjct: 181 AAAIAYGLDKKATSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDN 240
Query: 244 RM 245
RM
Sbjct: 241 RM 242
>gb|AAR17080.1| heat shock protein 70-3 [Nicotiana tabacum]
Length = 648
Score = 359 bits (922), Expect = 4e-98
Identities = 179/245 (73%), Positives = 206/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVISG DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVISGPGDKPMIVVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEA+L S VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMKEIAEAFLGSTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAN86275.1| non-cell-autonomous heat shock cognate protein 70 [Cucurbita
maxima]
Length = 652
Score = 359 bits (922), Expect = 4e-98
Identities = 177/245 (72%), Positives = 208/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD++RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDNERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI+G DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDSSVQSDIKLWPFKVIAGPGDKPMIVVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEE+SS++L MK+IAEAYL + VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEVSSMVLIKMKEIAEAYLGTTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKSTSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNR+
Sbjct: 239 FDNRL 243
>gb|AAB88134.1| cytosolic heat shock 70 protein [Spinacia oleracea]
gi|425194|gb|AAA62445.1| heat shock protein
gi|11277117|pir||T45522 heat shock protein HSC70-1,
cytosolic [imported] - spinach
Length = 647
Score = 359 bits (921), Expect = 5e-98
Identities = 178/245 (72%), Positives = 207/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ WPFKV+SG +KPMI V YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDASVQADMKHWPFKVVSGPGEKPMIGVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL S VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 AAEEISSMVLTKMKEIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAN86274.1| non-cell-autonomous heat shock cognate protein 70 [Cucurbita
maxima]
Length = 647
Score = 359 bits (921), Expect = 5e-98
Identities = 178/245 (72%), Positives = 207/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI+G DKPMI V YKG++K
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDAPVQSDIKLWPFKVIAGPGDKPMIVVSYKGEDKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL S VKNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNR+
Sbjct: 239 FDNRL 243
>gb|AAX95352.1| dnaK-type molecular chaperone hsp70 - rice (fragment) [Oryza sativa
(japonica cultivar-group)]
Length = 649
Score = 358 bits (920), Expect = 6e-98
Identities = 177/245 (72%), Positives = 207/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI+G DKPMI V+YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVIAGPGDKPMIVVQYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>pir||S53126 dnaK-type molecular chaperone hsp70 - rice (fragment)
Length = 649
Score = 358 bits (920), Expect = 6e-98
Identities = 177/245 (72%), Positives = 207/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI+G DKPMI V+YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVIAGPGDKPMIVVQYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>emb|CAA31663.1| hsp70 (AA 6 - 651) [Petunia x hybrida]
Length = 646
Score = 358 bits (918), Expect = 1e-97
Identities = 176/240 (73%), Positives = 203/240 (84%), Gaps = 2/240 (0%)
Query: 6 DGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAA 65
+G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AAK+Q A
Sbjct: 1 EGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAAKNQVA 58
Query: 66 INPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEI 125
+NP NTVFDAKRLIGR+FSDP VQ DI WPFKVI G DKPMI V YKG+EK AEEI
Sbjct: 59 MNPINTVFDAKRLIGRRFSDPSVQSDIKLWPFKVIPGPGDKPMIVVTYKGEEKQFAAEEI 118
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
SS++L MK+IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN+PTAA
Sbjct: 119 SSMVLTKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAA 178
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGEDFDNRM
Sbjct: 179 AIAYGLDKKASSAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRM 238
>dbj|BAB02269.1| 70 kDa heat shock protein [Arabidopsis thaliana]
gi|16649031|gb|AAL24367.1| 70 kDa heat shock protein
[Arabidopsis thaliana] gi|15809846|gb|AAL06851.1|
AT3g12580/T2E22_110 [Arabidopsis thaliana]
gi|15809832|gb|AAL06844.1| AT3g12580/T2E22_110
[Arabidopsis thaliana] gi|12321973|gb|AAG51030.1| heat
shock protein 70; 34105-36307 [Arabidopsis thaliana]
gi|15230534|ref|NP_187864.1| heat shock protein 70,
putative / HSP70, putative [Arabidopsis thaliana]
Length = 650
Score = 358 bits (918), Expect = 1e-97
Identities = 176/245 (71%), Positives = 208/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR++SDP VQ D WPFKV+SG +KPMI V +KG+EK
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEA+L SPVKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAF34134.1| high molecular weight heat shock protein [Malus x domestica]
Length = 650
Score = 358 bits (918), Expect = 1e-97
Identities = 177/245 (72%), Positives = 207/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR++SDP VQ D+ WPFKVI G DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRYSDPSVQGDMKHWPFKVIPGPGDKPMIVVIYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLVKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAB99745.1| HSP70 [Triticum aestivum]
Length = 648
Score = 358 bits (918), Expect = 1e-97
Identities = 176/242 (72%), Positives = 206/242 (84%), Gaps = 2/242 (0%)
Query: 4 KHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQ 63
K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AAK+Q
Sbjct: 3 KGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKNQ 60
Query: 64 AAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAE 123
A+NP NTVFDAKRLIGR+FSDP VQ D+ WPFKVI G DKPMI V YKG+EK AE
Sbjct: 61 VAMNPTNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPADKPMIVVNYKGEEKQFAAE 120
Query: 124 EISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPT 183
EISS++L M++IAEA+L + VKNAVVTVPAYFNDSQR+AT +AGAIAGLNV+RIIN+PT
Sbjct: 121 EISSMVLIKMREIAEAFLGNSVKNAVVTVPAYFNDSQRQATKDAGAIAGLNVLRIINEPT 180
Query: 184 AAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDN 243
AAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGEDFDN
Sbjct: 181 AAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDN 240
Query: 244 RM 245
RM
Sbjct: 241 RM 242
>emb|CAB72129.1| heat shock protein 70 [Cucumis sativus]
Length = 652
Score = 357 bits (917), Expect = 1e-97
Identities = 176/245 (71%), Positives = 207/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEI ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIFANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ DI WPFKVI+G +DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFSDASVQSDIKLWPFKVIAGPSDKPMIVVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEAYL + VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMKEIAEAYLGTTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVK+TAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKSSSSGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKSTAGDTHLGGED 238
Query: 241 FDNRM 245
FDNR+
Sbjct: 239 FDNRL 243
>emb|CAA37970.1| heat shock protein cognate 70 [Lycopersicon esculentum]
gi|123613|sp|P24629|HSP71_LYCES Heat shock cognate 70
kDa protein 1
Length = 650
Score = 357 bits (916), Expect = 2e-97
Identities = 177/245 (72%), Positives = 206/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ+D+ WPFKVI G DKPMI V YKG+EK
Sbjct: 59 KNQVALNPINTVFDAKRLIGRRFSDASVQEDMKLWPFKVIPGPGDKPMIVVTYKGEEKEF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEA+L S VKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 AAEEISSMVLTKMKEIAEAFLGSTVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSAGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAP37760.1| At1g16030 [Arabidopsis thaliana] gi|15219109|ref|NP_173055.1| heat
shock protein 70, putative / HSP70, putative
[Arabidopsis thaliana] gi|6587810|gb|AAF18501.1|
Identical to gb|AJ002551 heat shock protein 70 from
Arabidopsis thaliana and contains a PF|00012 HSP 70
domain. EST gb|F13893 comes from this gene
gi|25082767|gb|AAN71999.1| heat shock protein hsp70,
putative [Arabidopsis thaliana] gi|25295976|pir||B86295
hypothetical protein T24D18.14 [imported] - Arabidopsis
thaliana
Length = 646
Score = 357 bits (916), Expect = 2e-97
Identities = 177/245 (72%), Positives = 208/245 (84%), Gaps = 3/245 (1%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K + A+GIDLGTTYSCVGVW+ + RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MATKSE-KAIGIDLGTTYSCVGVWMND--RVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 57
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NPQNTVFDAKRLIGRKFSDP VQ DI+ WPFKV+SG +KPMI V YK +EK
Sbjct: 58 KNQVALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQF 117
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
EEISS++L MK++AEA+L VKNAVVTVPAYFNDSQR+AT +AGAI+GLNV+RIIN
Sbjct: 118 SPEEISSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 177
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD KG GE+N+ +FDLGGGTFDVSLLTI+ VFEVKATAG+THLGGED
Sbjct: 178 EPTAAAIAYGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGED 237
Query: 241 FDNRM 245
FDNR+
Sbjct: 238 FDNRL 242
>gb|AAB42159.1| Hsc70 gi|2119719|pir||JC4786 dnaK-type molecular chaperone hsc70-3
- tomato
Length = 651
Score = 355 bits (912), Expect = 5e-97
Identities = 176/245 (71%), Positives = 206/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSDP VQ D+ WPFKVI G DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRFSDPSVQSDMKLWPFKVIPGPADKPMIVVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L MK+IAEA+L + +KNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMKEIAEAFLGTTIKNAVVTVPAYFNDSQRQATKDAGTISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+ + +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKISSTGEKTVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>emb|CAA05547.1| heat shock protein 70 [Arabidopsis thaliana]
Length = 650
Score = 355 bits (911), Expect = 7e-97
Identities = 175/245 (71%), Positives = 206/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR++SDP VQ D WPFKV+SG +KPMI V +KG+EK
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISSI+L M++IAEA+L SPVKNAVV VPAYFNDSQR+ T +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSIVLIKMREIAEAFLGSPVKNAVVIVPAYFNDSQRQGTKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>gb|AAS57913.1| 70 kDa heat shock cognate protein 2 [Vigna radiata]
Length = 648
Score = 355 bits (910), Expect = 9e-97
Identities = 176/245 (71%), Positives = 205/245 (82%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+V FTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVGFTDTERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+F+D VQ DI WPFKV SG DKPMI V YKG++K
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFTDASVQSDIKLWPFKVFSGPGDKPMIQVNYKGEDKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL + VKNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 SAEEISSMVLMKMREIAEAYLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 442,974,302
Number of Sequences: 2540612
Number of extensions: 18317616
Number of successful extensions: 43613
Number of sequences better than 10.0: 2759
Number of HSP's better than 10.0 without gapping: 2396
Number of HSP's successfully gapped in prelim test: 363
Number of HSP's that attempted gapping in prelim test: 34790
Number of HSP's gapped (non-prelim): 3046
length of query: 259
length of database: 863,360,394
effective HSP length: 125
effective length of query: 134
effective length of database: 545,783,894
effective search space: 73135041796
effective search space used: 73135041796
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC144722.9


