

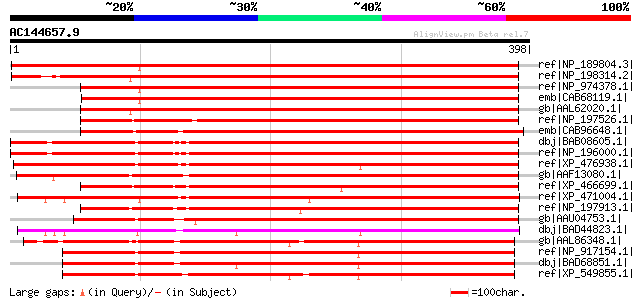
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.9 + phase: 0
(398 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_189804.3| exostosin family protein [Arabidopsis thaliana] 506 e-142
ref|NP_198314.2| exostosin family protein [Arabidopsis thaliana] 494 e-138
ref|NP_974378.1| exostosin family protein [Arabidopsis thaliana] 457 e-127
emb|CAB68119.1| putative protein [Arabidopsis thaliana] gi|11358... 455 e-126
gb|AAL62020.1| AT5g33290/F19N2_10 [Arabidopsis thaliana] gi|1514... 454 e-126
ref|NP_197526.1| exostosin family protein [Arabidopsis thaliana] 436 e-121
emb|CAB96648.1| putative protein [Arabidopsis thaliana] gi|15238... 435 e-120
dbj|BAB08605.1| unnamed protein product [Arabidopsis thaliana] 425 e-117
ref|NP_196000.1| exostosin family protein / pentatricopeptide (P... 425 e-117
ref|XP_476938.1| putative pectin-glucuronyltransferase [Oryza sa... 394 e-108
gb|AAF13080.1| hypothetical protein [Arabidopsis thaliana] gi|15... 390 e-107
ref|XP_466699.1| exostosin-like protein [Oryza sativa (japonica ... 373 e-102
ref|XP_471004.1| OSJNBa0027H06.12 [Oryza sativa (japonica cultiv... 364 3e-99
ref|NP_197913.1| exostosin family protein [Arabidopsis thaliana] 357 5e-97
gb|AAU04753.1| EXO [Cucumis melo] 354 3e-96
dbj|BAD44823.1| putative pectin-glucuronyltransferase [Oryza sat... 340 5e-92
gb|AAL86348.1| unknown protein [Arabidopsis thaliana] gi|2698390... 298 3e-79
ref|NP_917154.1| limonene cyclase-like protein [Oryza sativa (ja... 283 7e-75
dbj|BAD68851.1| pectin-glucuronyltransferase-like [Oryza sativa ... 278 2e-73
ref|XP_549855.1| pectin-glucuronyltransferase -like [Oryza sativ... 275 1e-72
>ref|NP_189804.3| exostosin family protein [Arabidopsis thaliana]
Length = 425
Score = 506 bits (1303), Expect = e-142
Identities = 242/393 (61%), Positives = 304/393 (76%), Gaps = 4/393 (1%)
Query: 2 TSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQ-SFVPKGSIYLNPHAFHQSHKEMVKRFK 60
++LEK EE+L + RA I+RA+R K T+N + +++P G IY N AFHQSH EM+K FK
Sbjct: 31 SNLEKREEELRKARAAIRRAVRFKNCTSNEEVITYIPTGQIYRNSFAFHQSHIEMMKTFK 90
Query: 61 VWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSP---FKATHPELAHVFFLPFSVS 117
VW YKEGEQPLVHDGPVN+ Y IEGQFIDE+ P F+A+ PE AH FFLPFSV+
Sbjct: 91 VWSYKEGEQPLVHDGPVNDIYGIEGQFIDELSYVMGGPSGRFRASRPEEAHAFFLPFSVA 150
Query: 118 KVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVS 177
++ YVY+P S +D+N RL + DY+ +VA+K+P+WN S GADHF++SCHDW P V
Sbjct: 151 NIVHYVYQPITSPADFNRARLHRIFNDYVDVVAHKHPFWNQSNGADHFMVSCHDWAPDVP 210
Query: 178 YANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFA 237
+ P+ FK+F+R LCNANTSEGF N D SIP++N+P KL PP Q+P NRTIL FFA
Sbjct: 211 DSKPEFFKNFMRGLCNANTSEGFRRNIDFSIPEINIPKRKLKPPFMGQNPENRTILAFFA 270
Query: 238 GGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEA 297
G AHG IR+ L WK KD++VQV+++L KGQ+Y +L+G SKFCLCPSG+EVASPR VEA
Sbjct: 271 GRAHGYIREVLFSHWKGKDKDVQVYDHLTKGQNYHELIGHSKFCLCPSGYEVASPREVEA 330
Query: 298 IYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRR 357
IY+GCVPV+I DNYSLPF+DVL+WS+FS+EI VD+IP+IK ILQ I KY +Y NV +
Sbjct: 331 IYSGCVPVVISDNYSLPFNDVLDWSKFSVEIPVDKIPDIKKILQEIPHDKYLRMYRNVMK 390
Query: 358 VRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
VR+HF +NRPA+PFD+IHMILHSVWLRRLN RL
Sbjct: 391 VRRHFVVNRPAQPFDVIHMILHSVWLRRLNIRL 423
>ref|NP_198314.2| exostosin family protein [Arabidopsis thaliana]
Length = 500
Score = 494 bits (1271), Expect = e-138
Identities = 241/392 (61%), Positives = 295/392 (74%), Gaps = 13/392 (3%)
Query: 2 TSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKV 61
+ L+KIE DLA+ RA I++A + Q++V S+Y NP AFHQSH EM+ RFKV
Sbjct: 117 SGLDKIESDLAKARAAIKKAAST--------QNYV--SSLYKNPAAFHQSHTEMMNRFKV 166
Query: 62 WVYKEGEQPLVHDGPVNNKYSIEGQFIDEM---DTSNKSPFKATHPELAHVFFLPFSVSK 118
W Y EGE PL HDGPVN+ Y IEGQF+DEM ++S F+A PE AHVFF+PFSV+K
Sbjct: 167 WTYTEGEVPLFHDGPVNDIYGIEGQFMDEMCVDGPKSRSRFRADRPENAHVFFIPFSVAK 226
Query: 119 VIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSY 178
VI +VYKP S ++ RL L+EDY+ +VA K+PYWN SQG DHF++SCHDW P V
Sbjct: 227 VIHFVYKPITSVEGFSRARLHRLIEDYVDVVATKHPYWNRSQGGDHFMVSCHDWAPDVID 286
Query: 179 ANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAG 238
NPKLF+ FIR LCNANTSEGF PN DVSIP++ LP GKLGP + P R+IL FFAG
Sbjct: 287 GNPKLFEKFIRGLCNANTSEGFRPNVDVSIPEIYLPKGKLGPSFLGKSPRVRSILAFFAG 346
Query: 239 GAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAI 298
+HG+IRK L + WK+ D EVQV++ LP G+DYTK MG+SKFCLCPSG EVASPR VEAI
Sbjct: 347 RSHGEIRKILFQHWKEMDNEVQVYDRLPPGKDYTKTMGMSKFCLCPSGWEVASPREVEAI 406
Query: 299 YAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRV 358
YAGCVPVII DNYSLPFSDVLNW FS++I V RI EIKTILQ+++ +Y +Y V V
Sbjct: 407 YAGCVPVIISDNYSLPFSDVLNWDSFSIQIPVSRIKEIKTILQSVSLVRYLKMYKRVLEV 466
Query: 359 RKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
++HF +NRPAKP+D++HM+LHS+WLRRLN RL
Sbjct: 467 KQHFVLNRPAKPYDVMHMMLHSIWLRRLNLRL 498
>ref|NP_974378.1| exostosin family protein [Arabidopsis thaliana]
Length = 341
Score = 457 bits (1176), Expect = e-127
Identities = 215/339 (63%), Positives = 266/339 (78%), Gaps = 3/339 (0%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSP---FKATHPELAHVFF 111
M+K FKVW YKEGEQPLVHDGPVN+ Y IEGQFIDE+ P F+A+ PE AH FF
Sbjct: 1 MMKTFKVWSYKEGEQPLVHDGPVNDIYGIEGQFIDELSYVMGGPSGRFRASRPEEAHAFF 60
Query: 112 LPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHD 171
LPFSV+ ++ YVY+P S +D+N RL + DY+ +VA+K+P+WN S GADHF++SCHD
Sbjct: 61 LPFSVANIVHYVYQPITSPADFNRARLHRIFNDYVDVVAHKHPFWNQSNGADHFMVSCHD 120
Query: 172 WGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRT 231
W P V + P+ FK+F+R LCNANTSEGF N D SIP++N+P KL PP Q+P NRT
Sbjct: 121 WAPDVPDSKPEFFKNFMRGLCNANTSEGFRRNIDFSIPEINIPKRKLKPPFMGQNPENRT 180
Query: 232 ILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVAS 291
IL FFAG AHG IR+ L WK KD++VQV+++L KGQ+Y +L+G SKFCLCPSG+EVAS
Sbjct: 181 ILAFFAGRAHGYIREVLFSHWKGKDKDVQVYDHLTKGQNYHELIGHSKFCLCPSGYEVAS 240
Query: 292 PRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVL 351
PR VEAIY+GCVPV+I DNYSLPF+DVL+WS+FS+EI VD+IP+IK ILQ I KY +
Sbjct: 241 PREVEAIYSGCVPVVISDNYSLPFNDVLDWSKFSVEIPVDKIPDIKKILQEIPHDKYLRM 300
Query: 352 YSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
Y NV +VR+HF +NRPA+PFD+IHMILHSVWLRRLN RL
Sbjct: 301 YRNVMKVRRHFVVNRPAQPFDVIHMILHSVWLRRLNIRL 339
>emb|CAB68119.1| putative protein [Arabidopsis thaliana] gi|11358246|pir||T46112
hypothetical protein T27B3.50 - Arabidopsis thaliana
Length = 340
Score = 455 bits (1171), Expect = e-126
Identities = 214/338 (63%), Positives = 265/338 (78%), Gaps = 3/338 (0%)
Query: 56 VKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSP---FKATHPELAHVFFL 112
+K FKVW YKEGEQPLVHDGPVN+ Y IEGQFIDE+ P F+A+ PE AH FFL
Sbjct: 1 MKTFKVWSYKEGEQPLVHDGPVNDIYGIEGQFIDELSYVMGGPSGRFRASRPEEAHAFFL 60
Query: 113 PFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDW 172
PFSV+ ++ YVY+P S +D+N RL + DY+ +VA+K+P+WN S GADHF++SCHDW
Sbjct: 61 PFSVANIVHYVYQPITSPADFNRARLHRIFNDYVDVVAHKHPFWNQSNGADHFMVSCHDW 120
Query: 173 GPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTI 232
P V + P+ FK+F+R LCNANTSEGF N D SIP++N+P KL PP Q+P NRTI
Sbjct: 121 APDVPDSKPEFFKNFMRGLCNANTSEGFRRNIDFSIPEINIPKRKLKPPFMGQNPENRTI 180
Query: 233 LTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASP 292
L FFAG AHG IR+ L WK KD++VQV+++L KGQ+Y +L+G SKFCLCPSG+EVASP
Sbjct: 181 LAFFAGRAHGYIREVLFSHWKGKDKDVQVYDHLTKGQNYHELIGHSKFCLCPSGYEVASP 240
Query: 293 RVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLY 352
R VEAIY+GCVPV+I DNYSLPF+DVL+WS+FS+EI VD+IP+IK ILQ I KY +Y
Sbjct: 241 REVEAIYSGCVPVVISDNYSLPFNDVLDWSKFSVEIPVDKIPDIKKILQEIPHDKYLRMY 300
Query: 353 SNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
NV +VR+HF +NRPA+PFD+IHMILHSVWLRRLN RL
Sbjct: 301 RNVMKVRRHFVVNRPAQPFDVIHMILHSVWLRRLNIRL 338
>gb|AAL62020.1| AT5g33290/F19N2_10 [Arabidopsis thaliana]
gi|15146187|gb|AAK83577.1| AT5g33290/F19N2_10
[Arabidopsis thaliana]
Length = 341
Score = 454 bits (1169), Expect = e-126
Identities = 216/339 (63%), Positives = 261/339 (76%), Gaps = 3/339 (0%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEM---DTSNKSPFKATHPELAHVFF 111
M+ RFKVW Y EGE PL HDGPVN+ Y IEGQF+DEM ++S F+A PE AHVFF
Sbjct: 1 MMNRFKVWTYTEGEVPLFHDGPVNDIYGIEGQFMDEMCVDGPKSRSRFRADRPENAHVFF 60
Query: 112 LPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHD 171
+PFSV+KVI +VYKP S ++ RL L+EDY+ +VA K+PYWN SQG DHF++SCHD
Sbjct: 61 IPFSVAKVIHFVYKPITSVEGFSRARLHRLIEDYVDVVATKHPYWNRSQGGDHFMVSCHD 120
Query: 172 WGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRT 231
W P NPKLF+ FIR LCNANTSEGF PN DVSIP++ LP GKLGP + P R+
Sbjct: 121 WAPDEIDGNPKLFEKFIRGLCNANTSEGFRPNVDVSIPEIYLPKGKLGPSFLGKSPRVRS 180
Query: 232 ILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVAS 291
IL FFAG +HG+IRK L + WK+ D EVQV++ LP G+DYTK MG+SKFCLCPSG EVAS
Sbjct: 181 ILAFFAGRSHGEIRKILFQHWKEMDNEVQVYDRLPPGKDYTKTMGMSKFCLCPSGWEVAS 240
Query: 292 PRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVL 351
PR VEAIYAGCVPVII DNYSLPFSDVLNW FS++I V RI EIKTILQ+++ +Y +
Sbjct: 241 PREVEAIYAGCVPVIISDNYSLPFSDVLNWDSFSIQIPVSRIKEIKTILQSVSLVRYLKM 300
Query: 352 YSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
Y V V++HF +NRPAKP+D++HM+LHS+WLRRLN RL
Sbjct: 301 YKRVLEVKQHFVLNRPAKPYDVMHMMLHSIWLRRLNLRL 339
>ref|NP_197526.1| exostosin family protein [Arabidopsis thaliana]
Length = 334
Score = 436 bits (1120), Expect = e-121
Identities = 207/337 (61%), Positives = 264/337 (77%), Gaps = 5/337 (1%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M K+FKVWVY+EGE PLVH GP+NN YSIEGQF+DE++T SPF A +PE AH F LP
Sbjct: 1 MEKKFKVWVYREGETPLVHMGPMNNIYSIEGQFMDEIETG-MSPFAANNPEEAHAFLLPV 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
SV+ ++ Y+Y+P + S H++ L DY+ +VA+KYPYWN S GADHF +SCHDW P
Sbjct: 60 SVANIVHYLYRPLVTYSREQLHKVFL---DYVDVVAHKYPYWNRSLGADHFYVSCHDWAP 116
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHP-NNRTIL 233
VS +NP+L K+ IR LCNANTSEGF P RDVSIP++N+P G LGPP + ++R IL
Sbjct: 117 DVSGSNPELMKNLIRVLCNANTSEGFMPQRDVSIPEINIPGGHLGPPRLSRSSGHDRPIL 176
Query: 234 TFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPR 293
FFAGG+HG IR+ LL+ WKDKDEEVQVHEYL K +DY KLM ++FCLCPSG+EVASPR
Sbjct: 177 AFFAGGSHGYIRRILLQHWKDKDEEVQVHEYLAKNKDYFKLMATARFCLCPSGYEVASPR 236
Query: 294 VVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYS 353
VV AI GCVPVII D+Y+LPFSDVL+W++F++ + +IPEIKTIL++I+ +YRVL
Sbjct: 237 VVAAINLGCVPVIISDHYALPFSDVLDWTKFTIHVPSKKIPEIKTILKSISWRRYRVLQR 296
Query: 354 NVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
V +V++HF +NRP++PFD++ M+LHSVWLRRLN RL
Sbjct: 297 RVLQVQRHFVINRPSQPFDMLRMLLHSVWLRRLNLRL 333
>emb|CAB96648.1| putative protein [Arabidopsis thaliana]
gi|15238967|ref|NP_196674.1| exostosin family protein
[Arabidopsis thaliana]
Length = 336
Score = 435 bits (1119), Expect = e-120
Identities = 200/340 (58%), Positives = 262/340 (76%), Gaps = 4/340 (1%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M KRFK+W Y+EGE PL H GP+NN Y+IEGQF+DE++ N S FKA PE A VF++P
Sbjct: 1 MEKRFKIWTYREGEAPLFHKGPLNNIYAIEGQFMDEIENGN-SRFKAASPEEATVFYIPV 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
+ +IR+VY+P S Y RLQ +V+DYI +++N+YPYWN S+GADHF LSCHDW P
Sbjct: 60 GIVNIIRFVYRPYTS---YARDRLQNIVKDYISLISNRYPYWNRSRGADHFFLSCHDWAP 116
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILT 234
VS +P+L+KHFIRALCNAN+SEGF P RDVS+P++N+P +LG +T + P NR +L
Sbjct: 117 DVSAVDPELYKHFIRALCNANSSEGFTPMRDVSLPEINIPHSQLGFVHTGEPPQNRKLLA 176
Query: 235 FFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRV 294
FFAGG+HG +RK L + WK+KD++V V+E LPK +YTK+M +KFCLCPSG EVASPR+
Sbjct: 177 FFAGGSHGDVRKILFQHWKEKDKDVLVYENLPKTMNYTKMMDKAKFCLCPSGWEVASPRI 236
Query: 295 VEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSN 354
VE++Y+GCVPVII D Y LPFSDVLNW FS+ I + ++P+IK IL+ ITE +Y +
Sbjct: 237 VESLYSGCVPVIIADYYVLPFSDVLNWKTFSVHIPISKMPDIKKILEAITEEEYLNMQRR 296
Query: 355 VRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRLHLKQ 394
V VRKHF +NRP+KP+D++HMI+HS+WLRRLN R+ L Q
Sbjct: 297 VLEVRKHFVINRPSKPYDMLHMIMHSIWLRRLNVRIPLSQ 336
>dbj|BAB08605.1| unnamed protein product [Arabidopsis thaliana]
Length = 408
Score = 425 bits (1092), Expect = e-117
Identities = 207/390 (53%), Positives = 282/390 (72%), Gaps = 9/390 (2%)
Query: 1 MTSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFK 60
+++LEKIE L + RA I+ A +VP G +Y N FH+S+ EM K+FK
Sbjct: 25 LSNLEKIEFKLQKARASIKAASMDDPVDD---PDYVPLGPMYWNAKVFHRSYLEMEKQFK 81
Query: 61 VWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVI 120
++VYKEGE PL HDGP + YS+EG FI E++T + F+ +P+ AHVF+LPFSV K++
Sbjct: 82 IYVYKEGEPPLFHDGPCKSIYSMEGSFIYEIETDTR--FRTNNPDKAHVFYLPFSVVKMV 139
Query: 121 RYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYAN 180
RYVY+ R SR D++P ++ V+DYI +V +KYPYWN S GADHF+LSCHDWGP S+++
Sbjct: 140 RYVYE-RNSR-DFSP--IRNTVKDYINLVGDKYPYWNRSIGADHFILSCHDWGPEASFSH 195
Query: 181 PKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGA 240
P L + IRALCNANTSE F P +DVSIP++NL G L P++R IL FFAGG
Sbjct: 196 PHLGHNSIRALCNANTSERFKPRKDVSIPEINLRTGSLTGLVGGPSPSSRPILAFFAGGV 255
Query: 241 HGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
HG +R LL+ W++KD +++VH+YLP+G Y+ +M SKFC+CPSG+EVASPR+VEA+Y+
Sbjct: 256 HGPVRPVLLQHWENKDNDIRVHKYLPRGTSYSDMMRNSKFCICPSGYEVASPRIVEALYS 315
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
GCVPV+I Y PFSDVLNW FS+ ++V+ IP +KTIL +I+ +Y +Y V +VR+
Sbjct: 316 GCVPVLINSGYVPPFSDVLNWRSFSVIVSVEDIPNLKTILTSISPRQYLRMYRRVLKVRR 375
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
HFE+N PAK FD+ HMILHS+W+RRLN ++
Sbjct: 376 HFEVNSPAKRFDVFHMILHSIWVRRLNVKI 405
>ref|NP_196000.1| exostosin family protein / pentatricopeptide (PPR) repeat-containing
protein [Arabidopsis thaliana]
Length = 1388
Score = 425 bits (1092), Expect = e-117
Identities = 207/390 (53%), Positives = 282/390 (72%), Gaps = 9/390 (2%)
Query: 1 MTSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFK 60
+++LEKIE L + RA I+ A +VP G +Y N FH+S+ EM K+FK
Sbjct: 1005 LSNLEKIEFKLQKARASIKAASMDDPVDD---PDYVPLGPMYWNAKVFHRSYLEMEKQFK 1061
Query: 61 VWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVI 120
++VYKEGE PL HDGP + YS+EG FI E++T + F+ +P+ AHVF+LPFSV K++
Sbjct: 1062 IYVYKEGEPPLFHDGPCKSIYSMEGSFIYEIETDTR--FRTNNPDKAHVFYLPFSVVKMV 1119
Query: 121 RYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYAN 180
RYVY+ R SR D++P ++ V+DYI +V +KYPYWN S GADHF+LSCHDWGP S+++
Sbjct: 1120 RYVYE-RNSR-DFSP--IRNTVKDYINLVGDKYPYWNRSIGADHFILSCHDWGPEASFSH 1175
Query: 181 PKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGA 240
P L + IRALCNANTSE F P +DVSIP++NL G L P++R IL FFAGG
Sbjct: 1176 PHLGHNSIRALCNANTSERFKPRKDVSIPEINLRTGSLTGLVGGPSPSSRPILAFFAGGV 1235
Query: 241 HGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
HG +R LL+ W++KD +++VH+YLP+G Y+ +M SKFC+CPSG+EVASPR+VEA+Y+
Sbjct: 1236 HGPVRPVLLQHWENKDNDIRVHKYLPRGTSYSDMMRNSKFCICPSGYEVASPRIVEALYS 1295
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
GCVPV+I Y PFSDVLNW FS+ ++V+ IP +KTIL +I+ +Y +Y V +VR+
Sbjct: 1296 GCVPVLINSGYVPPFSDVLNWRSFSVIVSVEDIPNLKTILTSISPRQYLRMYRRVLKVRR 1355
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
HFE+N PAK FD+ HMILHS+W+RRLN ++
Sbjct: 1356 HFEVNSPAKRFDVFHMILHSIWVRRLNVKI 1385
>ref|XP_476938.1| putative pectin-glucuronyltransferase [Oryza sativa (japonica
cultivar-group)] gi|34394611|dbj|BAC83913.1| putative
pectin-glucuronyltransferase [Oryza sativa (japonica
cultivar-group)] gi|50508944|dbj|BAD31848.1| putative
pectin-glucuronyltransferase [Oryza sativa (japonica
cultivar-group)]
Length = 606
Score = 394 bits (1012), Expect = e-108
Identities = 203/392 (51%), Positives = 273/392 (68%), Gaps = 11/392 (2%)
Query: 4 LEKIEEDLAQTRALIQRAIRSKKSTTNMK-QSFVPKGSIYLNPHAFHQSHKEMVKRFKVW 62
LE +E LA+ RA I+ AI++K + + + +VP G +Y N +AFH+S+ EM K FKV+
Sbjct: 216 LELLELGLAKARATIREAIQNKDNKPPLTDKDYVPVGPVYRNAYAFHRSYLEMEKVFKVF 275
Query: 63 VYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRY 122
VY+EGE P+ HDGP + YS EG+FI M+ N+ + P+ AHVFFLPFSV K+++
Sbjct: 276 VYEEGEPPVFHDGPCRSIYSTEGRFIYAMEMENR--MRTRDPDQAHVFFLPFSVVKMVKM 333
Query: 123 VYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPK 182
+Y+P D +P L+ + DYI +V+ KYP+WN S GADHF+LSCHDWGP VS AN
Sbjct: 334 IYEPNSH--DMDP--LRRTISDYINVVSTKYPHWNRSLGADHFMLSCHDWGPYVSSANGH 389
Query: 183 LFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGAHG 242
LF + IR LCNANTSEGF P+RDVS+P++NL + ++R IL FFAGG HG
Sbjct: 390 LFSNSIRVLCNANTSEGFDPSRDVSLPEINLRSDVVDRQVGGPSASHRPILAFFAGGDHG 449
Query: 243 KIRKKLLKSW-KDKDEEVQVHEYLPK--GQDYTKLMGLSKFCLCPSGHEVASPRVVEAIY 299
+R LL+ W K +D ++QV EYLP+ G YT +M S+FCLCPSG+EVASPRVVEAIY
Sbjct: 450 PVRPLLLQHWGKGQDADIQVSEYLPRRHGMSYTDMMRRSRFCLCPSGYEVASPRVVEAIY 509
Query: 300 AGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVR 359
CVPV+I D+Y+LPF+DVLNW+ FS+ +AV IP +K IL ++ +Y + VR VR
Sbjct: 510 LECVPVVIGDDYTLPFADVLNWAAFSVRVAVGDIPRLKEILAAVSPRQYIRMQRRVRAVR 569
Query: 360 KHFEMNRPA-KPFDLIHMILHSVWLRRLNFRL 390
+HF ++ A + FD+ HMILHS+WLRRLN R+
Sbjct: 570 RHFMVSDGAPRRFDVFHMILHSIWLRRLNVRV 601
>gb|AAF13080.1| hypothetical protein [Arabidopsis thaliana]
gi|15231488|ref|NP_187419.1| exostosin family protein
[Arabidopsis thaliana]
Length = 470
Score = 390 bits (1003), Expect = e-107
Identities = 188/388 (48%), Positives = 266/388 (68%), Gaps = 8/388 (2%)
Query: 6 KIEEDLAQTRALIQRAIRSKKSTTNMK---QSFVPKGSIYLNPHAFHQSHKEMVKRFKVW 62
K+E +LA R LI+ A + STT+ + +VP G IY NP+AFH+S+ M K FK++
Sbjct: 87 KVEAELATARVLIREAQLNYSSTTSSPLGDEDYVPHGDIYRNPYAFHRSYLLMEKMFKIY 146
Query: 63 VYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRY 122
VY+EG+ P+ H G + YS+EG F++ M+ ++ ++ P+ AHV+FLPFSV ++ +
Sbjct: 147 VYEEGDPPIFHYGLCKDIYSMEGLFLNFME-NDVLKYRTRDPDKAHVYFLPFSVVMILHH 205
Query: 123 VYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPK 182
++ P L+ ++ DY++I++ KYPYWN S G DHF+LSCHDWG R ++ K
Sbjct: 206 LFDPVVRDKAV----LERVIADYVQIISKKYPYWNTSDGFDHFMLSCHDWGHRATWYVKK 261
Query: 183 LFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGAHG 242
LF + IR LCNAN SE F P +D P++NL G + P +RT L FFAG +HG
Sbjct: 262 LFFNSIRVLCNANISEYFNPEKDAPFPEINLLTGDINNLTGGLDPISRTTLAFFAGKSHG 321
Query: 243 KIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGC 302
KIR LL WK+KD+++ V+E LP G DYT++M S+FC+CPSGHEVASPRV EAIY+GC
Sbjct: 322 KIRPVLLNHWKEKDKDILVYENLPDGLDYTEMMRKSRFCICPSGHEVASPRVPEAIYSGC 381
Query: 303 VPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRKHF 362
VPV+I +NY LPFSDVLNW +FS+ ++V IPE+K IL +I E +Y LY V++V++H
Sbjct: 382 VPVLISENYVLPFSDVLNWEKFSVSVSVKEIPELKRILMDIPEERYMRLYEGVKKVKRHI 441
Query: 363 EMNRPAKPFDLIHMILHSVWLRRLNFRL 390
+N P K +D+ +MI+HS+WLRRLN +L
Sbjct: 442 LVNDPPKRYDVFNMIIHSIWLRRLNVKL 469
>ref|XP_466699.1| exostosin-like protein [Oryza sativa (japonica cultivar-group)]
gi|47497631|dbj|BAD19700.1| exostosin-like protein
[Oryza sativa (japonica cultivar-group)]
Length = 345
Score = 373 bits (958), Expect = e-102
Identities = 179/347 (51%), Positives = 241/347 (68%), Gaps = 15/347 (4%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M +RFK+W Y+EGE P+ H GP + YSIEGQF+ EMD +S F A P+ AH F LP
Sbjct: 1 MERRFKIWTYREGEPPVAHIGPGTDIYSIEGQFMYEMDDP-RSRFAARRPDDAHAFLLPI 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
SV ++ YVY+ + D P L+ LV DY+++VA +YPYWN S+GADH ++SCHDW P
Sbjct: 60 SVCNLVHYVYR-LNATGDLAP--LRGLVADYVRVVAERYPYWNRSRGADHVIVSCHDWAP 116
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILT 234
V+ A+ +L+ + IR LCNANTSEGF P +D ++P++NL G L P P NRT L
Sbjct: 117 MVTSAHRQLYGNAIRVLCNANTSEGFRPRKDATLPEVNLADGVLRRPTAGLPPENRTTLA 176
Query: 235 FFAGGAHGKIRKKLLKSWK-----------DKDEEVQVHEYLPKGQDYTKLMGLSKFCLC 283
FFAGG HG IR+ LL+ W D D +++VHEYLP G+DY M ++FCLC
Sbjct: 177 FFAGGRHGHIRESLLRHWLIGNKGGAAADGDGDGDMRVHEYLPAGEDYHAQMAAARFCLC 236
Query: 284 PSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNI 343
PSG EVASPRVVE+++AGCVPVII + Y PF DVL+W + S+ + RIPE++ IL+ +
Sbjct: 237 PSGFEVASPRVVESVFAGCVPVIISEGYPPPFGDVLDWGKMSVAVPAARIPELRAILRRV 296
Query: 344 TETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
+E +YRVL + V + ++HF ++RPA+ FD+IHM+LHS+WLRRLN RL
Sbjct: 297 SERRYRVLRARVLQAQRHFVLHRPARRFDMIHMVLHSIWLRRLNVRL 343
>ref|XP_471004.1| OSJNBa0027H06.12 [Oryza sativa (japonica cultivar-group)]
gi|38345576|emb|CAE01775.2| OSJNBa0027H06.12 [Oryza
sativa (japonica cultivar-group)]
Length = 441
Score = 364 bits (934), Expect = 3e-99
Identities = 189/406 (46%), Positives = 261/406 (63%), Gaps = 25/406 (6%)
Query: 7 IEEDLAQTRALIQRAIRSKK-------STTNMKQSFVPKGS------IYLNPHAFHQSHK 53
+EE+L RA I+RA R + + N + + + +Y NP AF++S+
Sbjct: 39 VEEELDAARAAIRRAARQHRRGGGGDVGSANWLRFYGGEADYDLLSRVYRNPAAFYRSYV 98
Query: 54 EMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSP----FKATHPELAHV 109
EM +RFKV+VY+EGE P++H+GP N Y+IEG FI++++ + S + P AH
Sbjct: 99 EMERRFKVYVYEEGEPPILHEGPCKNIYTIEGSFIEQLELMSPSDAGGGVRTWDPTRAHA 158
Query: 110 FFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSC 169
FFLPFSVS+++++VY+P D P L+ +V DY+++VA ++P+WN S GADHF+LSC
Sbjct: 159 FFLPFSVSQMVKFVYRPPSQ--DRAP--LRAIVADYVRVVAARHPFWNRSAGADHFMLSC 214
Query: 170 HDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPN- 228
HDWGP S P+L+ + IRALCNANTSEGF P +DVS+P++NL G + P
Sbjct: 215 HDWGPYASRGQPELYTNAIRALCNANTSEGFRPGKDVSVPEINLYDGDMPRELLAPAPGL 274
Query: 229 -NRTILTFFAGGAHGKIRKKLLKSWKDKDEEV-QVHEY-LPKGQDYTKLMGLSKFCLCPS 285
+R +L FFAGG HG +R LL+ WK +D V+EY LP DY M ++FCLCPS
Sbjct: 275 ESRPLLAFFAGGRHGHVRDLLLRHWKGRDAATFPVYEYDLPAAGDYYSFMRRARFCLCPS 334
Query: 286 GHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITE 345
GHEVASPRVVEAI A CVPV+I D Y+LPF+DVL W FS+ +AV IP ++ L+ I
Sbjct: 335 GHEVASPRVVEAIQAECVPVVIADGYALPFADVLRWEAFSVAVAVGDIPRLRERLERIPA 394
Query: 346 TKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRLH 391
+ L VR V++H + +P + D+ +MILHSVWLR LN RLH
Sbjct: 395 AEVERLRRGVRLVKRHLMLQQPPRRLDMFNMILHSVWLRGLNLRLH 440
>ref|NP_197913.1| exostosin family protein [Arabidopsis thaliana]
Length = 334
Score = 357 bits (915), Expect = 5e-97
Identities = 173/339 (51%), Positives = 235/339 (69%), Gaps = 9/339 (2%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M KRFKV+VY+EGE PLVHDGP + Y++EG+FI EM+ ++ F+ P A+V+FLPF
Sbjct: 1 MEKRFKVYVYEEGEPPLVHDGPCKSVYAVEGRFITEME-KRRTKFRTYDPNQAYVYFLPF 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
SV+ ++RY+Y+ SD P L+ V DYI++V+ +P+WN + GADHF+L+CHDWGP
Sbjct: 60 SVTWLVRYLYE---GNSDAKP--LKTFVSDYIRLVSTNHPFWNRTNGADHFMLTCHDWGP 114
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPP---NTDQHPNNRT 231
S AN LF IR +CNAN+SEGF P +DV++P++ L G++ + + R
Sbjct: 115 LTSQANRDLFNTSIRVMCNANSSEGFNPTKDVTLPEIKLYGGEVDHKLRLSKTLSASPRP 174
Query: 232 ILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVAS 291
L FFAGG HG +R LLK WK +D ++ V+EYLPK +Y M SKFC CPSG+EVAS
Sbjct: 175 YLGFFAGGVHGPVRPILLKHWKQRDLDMPVYEYLPKHLNYYDFMRSSKFCFCPSGYEVAS 234
Query: 292 PRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVL 351
PRV+EAIY+ C+PVI+ N+ LPF+DVL W FS+ + V IP +K IL +I+ KY L
Sbjct: 235 PRVIEAIYSECIPVILSVNFVLPFTDVLRWETFSVLVDVSEIPRLKEILMSISNEKYEWL 294
Query: 352 YSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
SN+R VR+HFE+N P + FD H+ LHS+WLRRLN +L
Sbjct: 295 KSNLRYVRRHFELNDPPQRFDAFHLTLHSIWLRRLNLKL 333
>gb|AAU04753.1| EXO [Cucumis melo]
Length = 343
Score = 354 bits (908), Expect = 3e-96
Identities = 170/344 (49%), Positives = 234/344 (67%), Gaps = 12/344 (3%)
Query: 50 QSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHV 109
+S+ EM + K++VYKEGE P+ H GP + YS EG+FI EM+ N + P+ A +
Sbjct: 7 RSYLEMERLLKIYVYKEGEPPMFHGGPCKSIYSTEGRFIHEMEKGNL--YTTNDPDQALL 64
Query: 110 FFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLL---VEDYIKIVANKYPYWNISQGADHFL 166
+FLPFSV +++Y+Y P N H + + + DYI +++ K+P+W+ S GADHF+
Sbjct: 65 YFLPFSVVNLVQYLYVP-------NSHEVNAIGRAITDYINVISKKHPFWDRSLGADHFM 117
Query: 167 LSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQH 226
LSCHDWGPR + P LF + IR LCNAN SEGF P++D S P+++L G++
Sbjct: 118 LSCHDWGPRTTSYVPLLFNNSIRVLCNANVSEGFLPSKDASFPEIHLRTGEIDGLIGGLS 177
Query: 227 PNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSG 286
P+ R++L FFAG HG IR LL+ WK+KDE+V V+E LP G Y ++ S+FCLCPSG
Sbjct: 178 PSRRSVLAFFAGRLHGHIRYLLLQEWKEKDEDVLVYEELPSGISYNSMLKKSRFCLCPSG 237
Query: 287 HEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITET 346
+EVASPRVVEAIYA CVPV+I ++Y PFSDVLNW FS++I V IP IK IL+ I++T
Sbjct: 238 YEVASPRVVEAIYAECVPVLISESYVPPFSDVLNWKSFSVQIQVKDIPNIKKILKGISQT 297
Query: 347 KYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
+Y + V++V++HF +N K FD HMILHS+WLRRLN +
Sbjct: 298 QYLRMQRRVKQVQRHFALNGTPKRFDAFHMILHSIWLRRLNIHI 341
>dbj|BAD44823.1| putative pectin-glucuronyltransferase [Oryza sativa (japonica
cultivar-group)] gi|52075645|dbj|BAD44815.1| putative
pectin-glucuronyltransferase [Oryza sativa (japonica
cultivar-group)]
Length = 514
Score = 340 bits (872), Expect = 5e-92
Identities = 189/451 (41%), Positives = 264/451 (57%), Gaps = 70/451 (15%)
Query: 7 IEEDLAQTRALIQRAIRSKK----------STTNMKQ----SFVPKGS------IYLNPH 46
+E +L RA I+RA R ++ S++N+ SF +Y NP
Sbjct: 66 VERELDAARAAIRRAARRRRHGDLAGGEGRSSSNVSSAKWLSFFGDADHARLERVYRNPA 125
Query: 47 AFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNK--SPFKATHP 104
AF++S+ EM +RFKV+VY+EGE P+ H+GP N Y++EG+FI+E++ + P
Sbjct: 126 AFYRSYVEMERRFKVYVYEEGEPPIAHEGPCKNIYAVEGRFIEELELMAPPLGGVRTWDP 185
Query: 105 ELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADH 164
AH FLP SVS++++ Y+P Y+ L+ +V DY+ +VA+++ +WN S GADH
Sbjct: 186 ARAHALFLPLSVSQMVQLAYRPLS----YDLSPLRAIVADYVAVVASRHRFWNRSAGADH 241
Query: 165 FLLSCHDW------------------------GPRVSYANPKLFKHFIRALCNANTSEGF 200
F+LSCHDW GP S +P+L+ + IRALCNANTSEGF
Sbjct: 242 FMLSCHDWAIHTPSVQRDSISGFPTFRVQRLIGPHASRGHPELYANAIRALCNANTSEGF 301
Query: 201 WPNRDVSIPQLNLPVGKLGPPN-TDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEV 259
P++DVSIP++NL G + P + P R L FFAGG HG +R LL+ WK +D V
Sbjct: 302 RPDKDVSIPEINLYDGDMPPELLSPAPPPPRPFLAFFAGGRHGHVRDLLLRHWKGRDPAV 361
Query: 260 -QVHEY-LPK-----------------GQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
V+EY LP G Y M S+FCLCPSGHEVASPRVVEAI+A
Sbjct: 362 FPVYEYDLPSIPVSVSGDGDTDAGGEGGNPYYWYMRRSRFCLCPSGHEVASPRVVEAIHA 421
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
GCVPV++ D Y+ PF+DVL W FS+ +AV +P ++ +L+ I + L VR V++
Sbjct: 422 GCVPVVVADGYAPPFADVLRWEAFSVAVAVADVPRLRELLERIPAPEVERLRDGVRLVKR 481
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLNFRLH 391
HF +++P + D+ HMILHSVWLRRLN RL+
Sbjct: 482 HFMLHQPPERLDMFHMILHSVWLRRLNLRLN 512
>gb|AAL86348.1| unknown protein [Arabidopsis thaliana] gi|26983908|gb|AAN86206.1|
unknown protein [Arabidopsis thaliana]
gi|30683790|ref|NP_567512.2| exostosin family protein
[Arabidopsis thaliana]
Length = 542
Score = 298 bits (762), Expect = 3e-79
Identities = 156/387 (40%), Positives = 240/387 (61%), Gaps = 26/387 (6%)
Query: 11 LAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKVWVYKEGEQP 70
L + IQRA N F P ++ N F +S++ M KV++Y +G++P
Sbjct: 159 LTYAKLEIQRA----PEVINDTDLFAP---LFRNLSVFKRSYELMELILKVYIYPDGDKP 211
Query: 71 LVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRYVYKPRKSR 130
+ H+ +N Y+ EG F+ M+ SNK F +PE AH+F++P+SV ++ + ++ P
Sbjct: 212 IFHEPHLNGIYASEGWFMKLME-SNKQ-FVTKNPERAHLFYMPYSVKQLQKSIFVP---- 265
Query: 131 SDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLFKHFIRA 190
+N L + + DY+ +++ KYP+WN + G+DHFL++CHDWGP +P+L ++ I+A
Sbjct: 266 GSHNIKPLSIFLRDYVNMLSIKYPFWNRTHGSDHFLVACHDWGPYTVNEHPELKRNAIKA 325
Query: 191 LCNANTSEG-FWPNRDVSIPQLNL-----PVGKLGPPNTDQHPNNRTILTFFAGGAHGKI 244
LCNA+ S+G F P +DVS+P+ ++ P+ +G N + R IL FFAG HG++
Sbjct: 326 LCNADLSDGIFVPGKDVSLPETSIRNAGRPLRNIGNGN---RVSQRPILAFFAGNLHGRV 382
Query: 245 RKKLLKSWKDKDEEVQVHEYLP----KGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
R KLLK W++KDE+++++ LP + Y + M SK+CLCP G+EV SPR+VEAIY
Sbjct: 383 RPKLLKHWRNKDEDMKIYGPLPHNVARKMTYVQHMKSSKYCLCPMGYEVNSPRIVEAIYY 442
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
CVPV+I DN+ LPFSDVL+WS FS+ + IP +K IL I +Y + SNV+ V++
Sbjct: 443 ECVPVVIADNFMLPFSDVLDWSAFSVVVPEKEIPRLKEILLEIPMRRYLKMQSNVKMVQR 502
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLN 387
HF + + +D+ HMILHS+W LN
Sbjct: 503 HFLWSPKPRKYDVFHMILHSIWFNLLN 529
>ref|NP_917154.1| limonene cyclase-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 499
Score = 283 bits (724), Expect = 7e-75
Identities = 146/354 (41%), Positives = 225/354 (63%), Gaps = 13/354 (3%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
++ N F +S++ M + KV++Y++G +P+ H P++ Y+ EG F+ + S + F
Sbjct: 146 LFKNVSQFKRSYELMERILKVYIYQDGRRPIFHTPPLSGIYASEGWFMKLLKESRR--FA 203
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
T P AH+F+LP+S ++ +Y P +N L + D++K +A KYP+WN ++
Sbjct: 204 VTDPAKAHLFYLPYSSQQLRISLYVP----DSHNLRPLAAYLRDFVKGLAAKYPFWNRTR 259
Query: 161 GADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEG-FWPNRDVSIPQLNLPVGKLG 219
GADHFL++CHDWG + A+ L ++ ++ALCNA++SEG F P RDVS+P+ + +
Sbjct: 260 GADHFLVACHDWGSYTTTAHGDLRRNTVKALCNADSSEGIFTPGRDVSLPETTIRTPRRP 319
Query: 220 PPNTDQHP-NNRTILTFFAGGAHGKIRKKLLKSWKD-KDEEVQVHEYLP----KGQDYTK 273
P + R IL FFAG HG++R LLK W D +D++++V+ LP + Y +
Sbjct: 320 LRYVGGLPVSRRGILAFFAGNVHGRVRPVLLKHWGDGRDDDMRVYGPLPARVSRRMSYIQ 379
Query: 274 LMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRI 333
M S+FCLCP G+EV SPR+VEA+Y CVPVII DN+ LP SDVL+WS F++ +A +
Sbjct: 380 HMKNSRFCLCPMGYEVNSPRIVEALYYECVPVIIADNFVLPLSDVLDWSAFAVVVAEKDV 439
Query: 334 PEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLN 387
P++K ILQ IT KY ++ V+R+++HF + +DL HMILHS+WL R+N
Sbjct: 440 PDLKKILQGITLRKYVAMHGCVKRLQRHFLWHARPLRYDLFHMILHSIWLSRVN 493
>dbj|BAD68851.1| pectin-glucuronyltransferase-like [Oryza sativa (japonica
cultivar-group)]
Length = 501
Score = 278 bits (711), Expect = 2e-73
Identities = 146/356 (41%), Positives = 225/356 (63%), Gaps = 15/356 (4%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
++ N F +S++ M + KV++Y++G +P+ H P++ Y+ EG F+ + S + F
Sbjct: 146 LFKNVSQFKRSYELMERILKVYIYQDGRRPIFHTPPLSGIYASEGWFMKLLKESRR--FA 203
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
T P AH+F+LP+S ++ +Y P +N L + D++K +A KYP+WN ++
Sbjct: 204 VTDPAKAHLFYLPYSSQQLRISLYVP----DSHNLRPLAAYLRDFVKGLAAKYPFWNRTR 259
Query: 161 GADHFLLSCHDW--GPRVSYANPKLFKHFIRALCNANTSEG-FWPNRDVSIPQLNLPVGK 217
GADHFL++CHDW G + A+ L ++ ++ALCNA++SEG F P RDVS+P+ + +
Sbjct: 260 GADHFLVACHDWLQGSYTTTAHGDLRRNTVKALCNADSSEGIFTPGRDVSLPETTIRTPR 319
Query: 218 LGPPNTDQHP-NNRTILTFFAGGAHGKIRKKLLKSWKD-KDEEVQVHEYLP----KGQDY 271
P + R IL FFAG HG++R LLK W D +D++++V+ LP + Y
Sbjct: 320 RPLRYVGGLPVSRRGILAFFAGNVHGRVRPVLLKHWGDGRDDDMRVYGPLPARVSRRMSY 379
Query: 272 TKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVD 331
+ M S+FCLCP G+EV SPR+VEA+Y CVPVII DN+ LP SDVL+WS F++ +A
Sbjct: 380 IQHMKNSRFCLCPMGYEVNSPRIVEALYYECVPVIIADNFVLPLSDVLDWSAFAVVVAEK 439
Query: 332 RIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLN 387
+P++K ILQ IT KY ++ V+R+++HF + +DL HMILHS+WL R+N
Sbjct: 440 DVPDLKKILQGITLRKYVAMHGCVKRLQRHFLWHARPLRYDLFHMILHSIWLSRVN 495
>ref|XP_549855.1| pectin-glucuronyltransferase -like [Oryza sativa (japonica
cultivar-group)] gi|52076236|dbj|BAD44890.1|
pectin-glucuronyltransferase -like [Oryza sativa
(japonica cultivar-group)] gi|52076197|dbj|BAD44851.1|
pectin-glucuronyltransferase -like [Oryza sativa
(japonica cultivar-group)]
Length = 550
Score = 275 bits (704), Expect = 1e-72
Identities = 144/357 (40%), Positives = 219/357 (61%), Gaps = 20/357 (5%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
++ N F +S++ M + KV+VY +G +P+ H + Y+ EG F+ M+ + F
Sbjct: 199 LFRNVSVFRRSYELMERLLKVFVYHDGAKPIFHSPELKGIYASEGWFMKLMEGNQH--FV 256
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
P AH+F+LP+S ++ +Y P + + L + V+ YI ++ K+PYWN ++
Sbjct: 257 VRDPNRAHLFYLPYSSRQLEHNLYVPGSNTIE----PLSIFVKKYIDFISTKFPYWNRTK 312
Query: 161 GADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEG-FWPNRDVSIPQLNL-----P 214
GADHF ++CHDWGP + + +L K+ I+ALCNA+ SEG F RDVS+P+ L P
Sbjct: 313 GADHFFVACHDWGPYTTKLHDELRKNTIKALCNADLSEGVFIHGRDVSLPETFLRSPRRP 372
Query: 215 VGKLGPPNTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLP----KGQD 270
+ +G + R+IL FFAG HG++R LL+ W KD ++++++ LP + +
Sbjct: 373 LRGIGGKPAAE----RSILAFFAGQMHGRVRPVLLQYWGGKDADMRIYDRLPHRITRRMN 428
Query: 271 YTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAV 330
Y + M SK+C+CP G+EV SPR+VEAIY CVPVII DN+ LPF D LNWS FS+ I
Sbjct: 429 YIQHMKSSKYCICPMGYEVNSPRIVEAIYYECVPVIIADNFVLPFDDALNWSAFSVVIPE 488
Query: 331 DRIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLN 387
+P++K IL I + +Y + SNV+RV+KHF + +D+ HMILHS+W R+N
Sbjct: 489 KDVPKLKQILLAIPDDQYMAMQSNVQRVQKHFIWHPNPIKYDIFHMILHSIWYSRVN 545
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.321 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 724,700,951
Number of Sequences: 2540612
Number of extensions: 32196316
Number of successful extensions: 66308
Number of sequences better than 10.0: 209
Number of HSP's better than 10.0 without gapping: 179
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 65924
Number of HSP's gapped (non-prelim): 247
length of query: 398
length of database: 863,360,394
effective HSP length: 130
effective length of query: 268
effective length of database: 533,080,834
effective search space: 142865663512
effective search space used: 142865663512
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144657.9


