

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144645.6 - phase: 0 /pseudo
(570 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
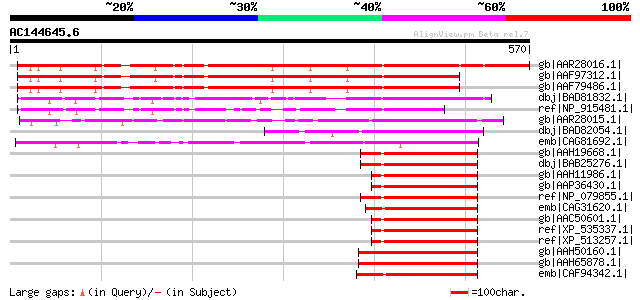
Score E
Sequences producing significant alignments: (bits) Value
gb|AAR28016.1| TAF12b [Arabidopsis thaliana] gi|20259253|gb|AAM1... 551 e-155
gb|AAF97312.1| Unknown protein [Arabidopsis thaliana] 493 e-138
gb|AAF79486.1| F1L3.13 [Arabidopsis thaliana] 484 e-135
dbj|BAD81832.1| transcription initiation factor IID (TFIID) subu... 301 3e-80
ref|NP_915481.1| P0446B05.23 [Oryza sativa (japonica cultivar-gr... 213 2e-53
gb|AAR28015.1| TAF12 [Arabidopsis thaliana] gi|6143870|gb|AAF044... 194 8e-48
dbj|BAD82054.1| transcription initiation factor IID (TFIID) subu... 175 4e-42
emb|CAG81692.1| unnamed protein product [Yarrowia lipolytica CLI... 144 8e-33
gb|AAH19668.1| TAF12 RNA polymerase II, TATA box binding protein... 121 5e-26
dbj|BAB25276.1| unnamed protein product [Mus musculus] 121 5e-26
gb|AAH11986.1| TAF12 protein [Homo sapiens] gi|30582901|gb|AAP35... 119 3e-25
gb|AAP36430.1| Homo sapiens TAF12 RNA polymerase II, TATA box bi... 119 3e-25
ref|NP_079855.1| TAF12 RNA polymerase II, TATA box binding prote... 119 3e-25
emb|CAG31620.1| hypothetical protein [Gallus gallus] 119 3e-25
gb|AAC50601.1| TAF15 119 3e-25
ref|XP_535337.1| PREDICTED: similar to Transcription initiation ... 118 6e-25
ref|XP_513257.1| PREDICTED: hypothetical protein XP_513257 [Pan ... 118 6e-25
gb|AAH50160.1| Unknown (protein for MGC:56398) [Danio rerio] 117 1e-24
gb|AAH65878.1| TAF12 RNA polymerase II, TATA box binding protein... 117 1e-24
emb|CAF94342.1| unnamed protein product [Tetraodon nigroviridis] 116 2e-24
>gb|AAR28016.1| TAF12b [Arabidopsis thaliana] gi|20259253|gb|AAM14362.1| unknown
protein [Arabidopsis thaliana]
gi|15810159|gb|AAL07223.1| unknown protein [Arabidopsis
thaliana] gi|30685323|ref|NP_849680.1| transcription
initiation factor IID (TFIID) subunit A family protein
[Arabidopsis thaliana] gi|18394483|ref|NP_564023.1|
transcription initiation factor IID (TFIID) subunit A
family protein [Arabidopsis thaliana]
Length = 683
Score = 551 bits (1420), Expect = e-155
Identities = 349/628 (55%), Positives = 413/628 (65%), Gaps = 85/628 (13%)
Query: 9 PDSQIQNPSSSNP----SIPSPSPIS------------------------QQQQSPSIHM 40
P+ NP+SSNP IPSPSP+S QQQQ
Sbjct: 74 PNISSPNPTSSNPPIGAQIPSPSPLSHPSSSLDQQTQTQQLVQQTQQLPQQQQQIMQQIS 133
Query: 41 SNSSPSLSQDQQQL----HTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQV---- 92
S+ P LS QQQ+ H + P S++Q+ Q+LQRSPS+SRL+QIQ QQQQQ
Sbjct: 134 SSPIPQLSPQQQQILQQQHMTSQQIPMSSYQIAQSLQRSPSLSRLSQIQQQQQQQHQGQY 193
Query: 93 --VARQQAALYGGQMNFGGSAAV--SAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPM 148
V RQQA LYG MNFGGS +V S Q QQ+ N+SR+ L+GQSGH PM
Sbjct: 194 GNVLRQQAGLYG-TMNFGGSGSVQQSQQNQQMVN---------PNMSRAGLVGQSGHLPM 243
Query: 149 LSGA-GAAQFN-----LLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLAS 202
L+GA GAAQ N L SPRQK GMVQ SQF +S GQ LQGMQ A+GMMGS NL S
Sbjct: 244 LNGAAGAAQMNIQPQLLAASPRQKSGMVQGSQFHPGSS-GQQLQGMQ-AMGMMGSLNLTS 301
Query: 203 QMRTNGGLYTQQQQIRLTPAQMRQQLSQQ-ALNSQQVQGIPRSSSLAFMNSQLSGLSQNG 261
QMR N LY QQ R+ P QMRQQLSQQ AL S QVQ + R+SSLAFMN QLSGL+QNG
Sbjct: 302 QMRGNPALYAQQ---RINPGQMRQQLSQQNALTSPQVQNLQRTSSLAFMNPQLSGLAQNG 358
Query: 262 QPGMMHNSLTQSQWLKQMPAMSGPAS---------PLRLQQHQRQQQQLASSGQLQQNSM 312
Q GMM NSL+Q QWLKQM ++ P S L LQQ Q+QQQQL SS QL Q+SM
Sbjct: 359 QAGMMQNSLSQQQWLKQMSGITSPNSFRLQPSQRQALLLQQQQQQQQQL-SSPQLHQSSM 417
Query: 313 TLNQQQLSQFMQQQKS---MGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLH 369
+LNQQQ+SQ +QQQ+ +GQ Q++Q Q QQ QQ LQ Q Q Q+Q QQQ +
Sbjct: 418 SLNQQQISQIIQQQQQQSQLGQSQMNQSHSQQQLQQMQQQLQQQPQQQMQQQQQQQQQMQ 477
Query: 370 ------SPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDL 423
SPR+ GQKS+SLTGSQP+AT SG TTPGGSSSQGTEA TNQ+LGKRKIQDL
Sbjct: 478 INQQQPSPRMLSHAGQKSVSLTGSQPEATQSGTTTPGGSSSQGTEA-TNQLLGKRKIQDL 536
Query: 424 VAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDL 483
V+QVD KLDP+V DLLLE ADDFIDSVT+ C LAKHRKSS LE KD+LLHLEKN L
Sbjct: 537 VSQVDVHAKLDPDVEDLLLEVADDFIDSVTSFACSLAKHRKSSVLEPKDILLHLEKNLHL 596
Query: 484 TIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSKDISRQGHPNPAGS 543
TIPG+SSE+K+ Q++ + +LHK+RL VR L+ESS PE+ +NSK+ RQ NP G
Sbjct: 597 TIPGFSSEDKR-QTKTVPTDLHKKRLAMVRALLESSK-PETNASNSKETMRQAMVNPNGP 654
Query: 544 HHLMRP-LSSDQLVSHSTSSQMLQQMTR 570
+HL+RP SS+QLVS ++ +LQ MTR
Sbjct: 655 NHLLRPSQSSEQLVSQTSGPHILQHMTR 682
>gb|AAF97312.1| Unknown protein [Arabidopsis thaliana]
Length = 674
Score = 493 bits (1269), Expect = e-138
Identities = 312/551 (56%), Positives = 361/551 (64%), Gaps = 82/551 (14%)
Query: 9 PDSQIQNPSSSNP----SIPSPSPIS------------------------QQQQSPSIHM 40
P+ NP+SSNP IPSPSP+S QQQQ
Sbjct: 74 PNISSPNPTSSNPPIGAQIPSPSPLSHPSSSLDQQTQTQQLVQQTQQLPQQQQQIMQQIS 133
Query: 41 SNSSPSLSQDQQQL----HTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQV---- 92
S+ P LS QQQ+ H + P S++Q+ Q+LQRSPS+SRL+QIQ QQQQQ
Sbjct: 134 SSPIPQLSPQQQQILQQQHMTSQQIPMSSYQIAQSLQRSPSLSRLSQIQQQQQQQHQGQY 193
Query: 93 --VARQQAALYGGQMNFGGSAAV--SAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPM 148
V RQQA LYG MNFGGS +V S Q QQ+ N+SR+ L+GQSGH PM
Sbjct: 194 GNVLRQQAGLYG-TMNFGGSGSVQQSQQNQQMVN---------PNMSRAGLVGQSGHLPM 243
Query: 149 LSGA-GAAQFN-----LLTSPRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLAS 202
L+GA GAAQ N L SPRQK GMVQ SQF +S GQ LQGMQ A+GMMGS NL S
Sbjct: 244 LNGAAGAAQMNIQPQLLAASPRQKSGMVQGSQFHPGSS-GQQLQGMQ-AMGMMGSLNLTS 301
Query: 203 QMRTNGGLYTQQQQIRLTPAQMRQQLSQQ-ALNSQQVQGIPRSSSLAFMNSQLSGLSQNG 261
QMR N LY QQ R+ P QMRQQLSQQ AL S QVQ + R+SSLAFMN QLSGL+QNG
Sbjct: 302 QMRGNPALYAQQ---RINPGQMRQQLSQQNALTSPQVQNLQRTSSLAFMNPQLSGLAQNG 358
Query: 262 QPGMMHNSLTQSQWLKQMPAMSGPAS---------PLRLQQHQRQQQQLASSGQLQQNSM 312
Q GMM NSL+Q QWLKQM ++ P S L LQQ Q+QQQQL SS QL Q+SM
Sbjct: 359 QAGMMQNSLSQQQWLKQMSGITSPNSFRLQPSQRQALLLQQQQQQQQQL-SSPQLHQSSM 417
Query: 313 TLNQQQLSQFMQQQKS---MGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLH 369
+LNQQQ+SQ +QQQ+ +GQ Q++Q Q QQ QQ LQ Q Q Q+Q QQQ +
Sbjct: 418 SLNQQQISQIIQQQQQQSQLGQSQMNQSHSQQQLQQMQQQLQQQPQQQMQQQQQQQQQMQ 477
Query: 370 ------SPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDL 423
SPR+ GQKS+SLTGSQP+AT SG TTPGGSSSQGTEA TNQ+LGKRKIQDL
Sbjct: 478 INQQQPSPRMLSHAGQKSVSLTGSQPEATQSGTTTPGGSSSQGTEA-TNQLLGKRKIQDL 536
Query: 424 VAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDL 483
V+QVD KLDP+V DLLLE ADDFIDSVT+ C LAKHRKSS LE KD+LLHLEKN L
Sbjct: 537 VSQVDVHAKLDPDVEDLLLEVADDFIDSVTSFACSLAKHRKSSVLEPKDILLHLEKNLHL 596
Query: 484 TIPGYSSEEKK 494
TIPG+SSE+K+
Sbjct: 597 TIPGFSSEDKR 607
>gb|AAF79486.1| F1L3.13 [Arabidopsis thaliana]
Length = 734
Score = 484 bits (1245), Expect = e-135
Identities = 307/548 (56%), Positives = 357/548 (65%), Gaps = 79/548 (14%)
Query: 9 PDSQIQNPSSSNP----SIPSPSPIS------------------------QQQQSPSIHM 40
P+ NP+SSNP IPSPSP+S QQQQ
Sbjct: 74 PNISSPNPTSSNPPIGAQIPSPSPLSHPSSSLDQQTQTQQLVQQTQQLPQQQQQIMQQIS 133
Query: 41 SNSSPSLSQDQQQL----HTINTINPNSNFQLQQTLQRSPSMSRLNQIQPQQQQQV---- 92
S+ P LS QQQ+ H + P S++Q+ Q+LQRSPS+SRL+QIQ QQQQQ
Sbjct: 134 SSPIPQLSPQQQQILQQQHMTSQQIPMSSYQIAQSLQRSPSLSRLSQIQQQQQQQHQGQY 193
Query: 93 --VARQQAALYGGQMNFGGSAAV--SAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPM 148
V RQQA LYG MNFGGS +V S Q QQ+ N+SR+ L+GQSGH PM
Sbjct: 194 GNVLRQQAGLYG-TMNFGGSGSVQQSQQNQQMVN---------PNMSRAGLVGQSGHLPM 243
Query: 149 LSGA-GAAQFNLLTS--PRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMR 205
L+GA GAAQ N+ K GMVQ SQF +S GQ LQGMQ A+GMMGS NL SQMR
Sbjct: 244 LNGAAGAAQMNIQPQLLAASKSGMVQGSQFHPGSS-GQQLQGMQ-AMGMMGSLNLTSQMR 301
Query: 206 TNGGLYTQQQQIRLTPAQMRQQLSQQ-ALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPG 264
N LY QQ R+ P QMRQQLSQQ AL S QVQ + R+SSLAFMN QLSGL+QNGQ G
Sbjct: 302 GNPALYAQQ---RINPGQMRQQLSQQNALTSPQVQNLQRTSSLAFMNPQLSGLAQNGQAG 358
Query: 265 MMHNSLTQSQWLKQMPAMSGPAS---------PLRLQQHQRQQQQLASSGQLQQNSMTLN 315
MM NSL+Q QWLKQM ++ P S L LQQ Q+QQQQL SS QL Q+SM+LN
Sbjct: 359 MMQNSLSQQQWLKQMSGITSPNSFRLQPSQRQALLLQQQQQQQQQL-SSPQLHQSSMSLN 417
Query: 316 QQQLSQFMQQQKS---MGQPQLHQQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLH--- 369
QQQ+SQ +QQQ+ +GQ Q++Q Q QQ QQ LQ Q Q Q+Q QQQ +
Sbjct: 418 QQQISQIIQQQQQQSQLGQSQMNQSHSQQQLQQMQQQLQQQPQQQMQQQQQQQQQMQINQ 477
Query: 370 ---SPRVAGPTGQKSISLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQ 426
SPR+ GQKS+SLTGSQP+AT SG TTPGGSSSQGTEA TNQ+LGKRKIQDLV+Q
Sbjct: 478 QQPSPRMLSHAGQKSVSLTGSQPEATQSGTTTPGGSSSQGTEA-TNQLLGKRKIQDLVSQ 536
Query: 427 VDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIP 486
VD KLDP+V DLLLE ADDFIDSVT+ C LAKHRKSS LE KD+LLHLEKN LTIP
Sbjct: 537 VDVHAKLDPDVEDLLLEVADDFIDSVTSFACSLAKHRKSSVLEPKDILLHLEKNLHLTIP 596
Query: 487 GYSSEEKK 494
G+SSE+K+
Sbjct: 597 GFSSEDKR 604
>dbj|BAD81832.1| transcription initiation factor IID (TFIID) subunit A-like protein
[Oryza sativa (japonica cultivar-group)]
gi|56784033|dbj|BAD82661.1| transcription initiation
factor IID (TFIID) subunit A-like protein [Oryza sativa
(japonica cultivar-group)]
Length = 542
Score = 301 bits (772), Expect = 3e-80
Identities = 232/554 (41%), Positives = 298/554 (52%), Gaps = 87/554 (15%)
Query: 9 PDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSN----SSPSLSQDQQQLHTINTINPNS 64
PD Q+ + S P P+P+P+ Q PS +S+ SSP L ++ P
Sbjct: 15 PD-QLAAAAVSTPQNPNPNPLLSPQIPPSPTVSDLSAISSPQLDPSAAGGGAMD--YPPR 71
Query: 65 NFQLQQ--------------TLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGS 110
Q+Q + RS S SRL + Q A A +YG Q+NF G
Sbjct: 72 PPQMQAPSPGQAAAGAGGFGQIHRSGSGSRLAAVGQLPQYAAAA---ARMYGSQVNFSGG 128
Query: 111 AAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAA--------QFNLLTS 162
QQQQ +AA R+A++ Q G ML G G A Q ++
Sbjct: 129 GGQVGQQQQQQQQLAA---------RAAMLSQ-GQIGMLQGQGNAASAAHYGLQSQMMAQ 178
Query: 163 PRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPA 222
PRQK GMVQ +QF++AN+A Q+LQGM Q++G+MG MR NG + QQ+ A
Sbjct: 179 PRQK-GMVQGAQFNTANAA-QALQGM-QSMGVMGG------MRGNGTIPYNQQRFAHAQA 229
Query: 223 QMR-QQLSQQALNSQQV--QGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQ----W 275
Q+R QQ SQQ S QV QG+ R++S+A +N QL G S NG M SL Q Q W
Sbjct: 230 QLRPQQTSQQGTLSPQVVGQGLTRTASIAALNPQLPGSSTNGPMAQM--SLPQKQQQAAW 287
Query: 276 LKQMPAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLH 335
LKQM + G SP+ QQ Q QQ+ L QLQQ S LNQ Q++Q QQ + L
Sbjct: 288 LKQMQSSLG--SPVSPQQFQHQQRMLLIH-QLQQQS-GLNQHQIAQTQQQHPHLNTQLLQ 343
Query: 336 QQQPSPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASG 395
QQ Q QQQQQ SPR++ QKS++LTGSQP SG
Sbjct: 344 QQHILQQLQQQQQ---------------------SPRISASGSQKSMNLTGSQPGTPLSG 382
Query: 396 ATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTH 455
T GGS+SQG E TNQ+LGKRKIQDLV+QVDP GK+DPEV DLLLE ADDFIDSVT
Sbjct: 383 GTMTGGSASQGAE-VTNQLLGKRKIQDLVSQVDPLGKVDPEVEDLLLEIADDFIDSVTAF 441
Query: 456 GCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRML 515
C LAKHRKSS LE+KD+LLHLEKNW L++PG+ E+K Q P+ + ++ +
Sbjct: 442 ACTLAKHRKSSVLEAKDVLLHLEKNWHLSVPGFLREDKNPQRHPVKVSVDPQQPECDAAG 501
Query: 516 MESSSVPESIVNNS 529
+ S+ + ++NNS
Sbjct: 502 IRSTG-NKLVINNS 514
>ref|NP_915481.1| P0446B05.23 [Oryza sativa (japonica cultivar-group)]
Length = 448
Score = 213 bits (541), Expect = 2e-53
Identities = 187/498 (37%), Positives = 248/498 (49%), Gaps = 94/498 (18%)
Query: 9 PDSQIQNPSSSNPSIPSPSPISQQQQSPSIHMSN----SSPSLSQDQQQLHTINTINPNS 64
PD Q+ + S P P+P+P+ Q PS +S+ SSP L ++ P
Sbjct: 15 PD-QLAAAAVSTPQNPNPNPLLSPQIPPSPTVSDLSAISSPQLDPSAAGGGAMDY--PPR 71
Query: 65 NFQLQQT--------------LQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGS 110
Q+Q + RS S SRL + Q A A +YG Q+NF G
Sbjct: 72 PPQMQAPSPGQAAAGAGGFGQIHRSGSGSRLAAVGQLPQYAAAA---ARMYGSQVNFSGG 128
Query: 111 AAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAA--------QFNLLTS 162
QQQQ +AA R+A++ Q G ML G G A Q ++
Sbjct: 129 GGQVGQQQQQQQQLAA---------RAAMLSQ-GQIGMLQGQGNAASAAHYGLQSQMMAQ 178
Query: 163 PRQKGGMVQSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPA 222
PRQKG MVQ +QF++AN+A Q+LQGMQ ++G+MG MR NG + QQ+ A
Sbjct: 179 PRQKG-MVQGAQFNTANAA-QALQGMQ-SMGVMGG------MRGNGTIPYNQQRFAHAQA 229
Query: 223 QMR-QQLSQQALNSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWL--KQM 279
Q+R QQ SQQ S QV L F S L +N + ++ T W+ K
Sbjct: 230 QLRPQQTSQQGTLSPQV--------LDFQGCG-SRLDKNSIDCCIKSTAT---WIVDKWA 277
Query: 280 PAMSGPASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQP 339
+ P+S + A+ ++ N++ ++ F + + ++
Sbjct: 278 DGTNVPSS------------ETAAGSMVKTNAIIAG---ITCFSTTISASTKDAINTSTS 322
Query: 340 SPQQQQQQQLLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTP 399
+ QQQ L Q QQQ Q SPR++ QKS++LTGSQP SG T
Sbjct: 323 TALLQQQHILQQLQQQQQ------------SPRISASGSQKSMNLTGSQPGTPLSGGTMT 370
Query: 400 GGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCIL 459
GGS+SQG E TNQ+LGKRKIQDLV+QVDP GK+DPEV DLLLE ADDFIDSVT C L
Sbjct: 371 GGSASQGAEV-TNQLLGKRKIQDLVSQVDPLGKVDPEVEDLLLEIADDFIDSVTAFACTL 429
Query: 460 AKHRKSSTLESKDLLLHL 477
AKHRKSS LE+KD+LLHL
Sbjct: 430 AKHRKSSVLEAKDVLLHL 447
Score = 37.7 bits (86), Expect = 1.0
Identities = 59/237 (24%), Positives = 86/237 (35%), Gaps = 12/237 (5%)
Query: 213 QQQQIRLTPAQMRQQLSQQALNSQQVQGIPRSSSLAFMNS-QLSGLSQNGQPGMMHNSLT 271
Q Q+ Q + L S Q+ P S L+ ++S QL + G G M
Sbjct: 14 QPDQLAAAAVSTPQNPNPNPLLSPQIPPSPTVSDLSAISSPQLDPSAAGG--GAMDYPPR 71
Query: 272 QSQWLKQMPAMSGPASPLRLQQHQRQQ-QQLASSGQLQQNSMTLNQQQLSQ--FMQQQKS 328
Q P + + Q H+ +LA+ GQL Q + + SQ F
Sbjct: 72 PPQMQAPSPGQAAAGAGGFGQIHRSGSGSRLAAVGQLPQYAAAAARMYGSQVNFSGGGGQ 131
Query: 329 MGQPQLHQQQPSPQQQQ--QQQLLQPQQQSQLQASVH---QQQHLHSPRVAGPT-GQKSI 382
+GQ Q QQQ + + Q Q+ Q Q ++ H Q Q + PR G G +
Sbjct: 132 VGQQQQQQQQLAARAAMLSQGQIGMLQGQGNAASAAHYGLQSQMMAQPRQKGMVQGAQFN 191
Query: 383 SLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVID 439
+ +Q GG GT Q + Q Q QG L P+V+D
Sbjct: 192 TANAAQALQGMQSMGVMGGMRGNGTIPYNQQRFAHAQAQLRPQQTSQQGTLSPQVLD 248
>gb|AAR28015.1| TAF12 [Arabidopsis thaliana] gi|6143870|gb|AAF04417.1| unknown
protein [Arabidopsis thaliana]
gi|62320727|dbj|BAD95394.1| hypothetical protein
[Arabidopsis thaliana] gi|13492646|gb|AAK28289.1|
putative TBP-associated 58 kDa subunit protein
[Arabidopsis thaliana] gi|18398741|ref|NP_566367.1|
transcription initiation factor IID (TFIID) subunit A
family protein [Arabidopsis thaliana]
Length = 539
Score = 194 bits (492), Expect = 8e-48
Identities = 184/552 (33%), Positives = 257/552 (46%), Gaps = 78/552 (14%)
Query: 11 SQIQNPSSSNPS---IPSPSPISQQQQSPSIHMSNSSPSLSQD------QQQLHTINTIN 61
+QIQ S+NPS + S P S QSPS++ + + P ++ QQQ H
Sbjct: 29 TQIQPTPSTNPSPSSVVSSIPSSPAPQSPSLNPNPNPPQYTRPVTSPATQQQQH------ 82
Query: 62 PNSNFQLQQTLQRSPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGSAAVSAQQQQLS 121
L Q L R P + +P QQ +A ++ + A S+ +S
Sbjct: 83 ------LSQPLVRPPPQA---YSRPWQQHSSYTHFSSAS-SPLLSSSSAPASSSSSLPIS 132
Query: 122 G---GVAAMGGSASNLSRSALIGQSGHFPM-LSGAGAAQFNLLTSPRQKGGMVQSSQFSS 177
G G A+G AS + S S H P G+ Q+ L G V S+ +S
Sbjct: 133 GQQRGGMAIGVPASPIP-SPSPTPSQHSPSAFPGSFGQQYGGLGR-----GTVGMSEATS 186
Query: 178 ANSAGQ--SLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQ--QIRLTPAQMRQQLSQQAL 233
S+ Q +QG Q IGMMG+ SQ+R +G QQ+ Q L PA S A
Sbjct: 187 NTSSPQVRMMQGTQ-GIGMMGTLGSGSQIRPSGMTQHQQRPTQSSLRPASSTSTQSPVAQ 245
Query: 234 NSQQVQGIPRSSSLAFMNSQLSGLSQNGQPGMMHNSLTQSQWLKQMPAMSGPASPLRLQQ 293
N Q + R S ++ N Q +G SQ + WL P P P
Sbjct: 246 NFQG-HSLMRPSPISSPNVQSTGASQQSLQAI------NQPWLSSTPQGKPPLPP----- 293
Query: 294 HQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQQQQLLQPQ 353
S + Q NS ++ QQ QQ S P PQQQQ QQ QPQ
Sbjct: 294 ---------PSYRPQVNSPSM--QQRPHIPQQHISTSAAT-----PQPQQQQSQQQHQPQ 337
Query: 354 QQ-SQLQASVHQQQHLHSP-RVAGPTGQKSIS-LTGSQPDATASGATTPGGSSSQGTEAA 410
+Q QL++ H H P RV G QK S + SQP G + S TE +
Sbjct: 338 EQLQQLRSPQQPLAHPHQPTRVQGLVNQKVTSPVMPSQPPVAQPG--NHAKTVSAETEPS 395
Query: 411 TNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAKHRKSSTLES 470
+++LGKR I +L+ Q+DP KLDPEV D+L + A+DF++S+TT GC LAKHRKS LE+
Sbjct: 396 DDRILGKRSIHELLQQIDPSEKLDPEVEDILSDIAEDFVESITTFGCSLAKHRKSDILEA 455
Query: 471 KDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRMLMESSSVPESIVNNSK 530
KD+LLH+E+NW++ PG+SS+E K +PL+ ++HK RL A++ SV + N++
Sbjct: 456 KDILLHVERNWNIRPPGFSSDEFKTFRKPLTTDIHKERLAAIK-----KSVTATEAANAR 510
Query: 531 DISRQGHPNPAG 542
+ G N G
Sbjct: 511 NQFGHGTANARG 522
>dbj|BAD82054.1| transcription initiation factor IID (TFIID) subunit A-like protein
[Oryza sativa (japonica cultivar-group)]
Length = 295
Score = 175 bits (443), Expect = 4e-42
Identities = 105/248 (42%), Positives = 143/248 (57%), Gaps = 14/248 (5%)
Query: 280 PAMSGPASPLRLQQHQR--QQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQ 337
PA+S P + L QQ Q+ Q Q L Q QQ L QQQ QQQ+ Q Q QQ
Sbjct: 29 PAISHPQNALLTQQQQKLPQHQHLQQQQQQQQQQQKLQQQQ-----QQQQQKLQQQQQQQ 83
Query: 338 QPSPQQQQQQQLLQPQ---QQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATAS 394
Q QQQQQQQ QPQ QQSQ ++ QQ + + A + L P +
Sbjct: 84 QQKLQQQQQQQQNQPQHSSQQSQQTTTLRNQQQISQQQTARTPVSMAQKL--DSPAVLKA 141
Query: 395 GATTPGGSSSQGTEA--ATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSV 452
G +S +A + N++L KR I +LVAQ+DP KLDPEV D+L++ A+DF++SV
Sbjct: 142 TNVQSGDMASVDVDAGGSGNRLLSKRSIHELVAQIDPSEKLDPEVEDVLIDIAEDFVESV 201
Query: 453 TTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAV 512
T C LAKHRKSS LE+KD+LLH E++W++T+PG+S +E K +P N++H+ RL +
Sbjct: 202 ATFACSLAKHRKSSILEAKDVLLHAERSWNITLPGFSGDEIKLYKKPHVNDIHRERLTLI 261
Query: 513 RMLMESSS 520
+ M S S
Sbjct: 262 KKSMASES 269
>emb|CAG81692.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50547847|ref|XP_501393.1| hypothetical protein
[Yarrowia lipolytica]
Length = 652
Score = 144 bits (363), Expect = 8e-33
Identities = 153/533 (28%), Positives = 229/533 (42%), Gaps = 65/533 (12%)
Query: 7 GSPD-SQIQNPSSS--NPSIPSPSPISQQQ--QSPSIHMSNSSPSLSQ---DQQQLHTIN 58
G+P+ NP+++ NPS P+ +P QQ Q+ + + P+ ++ +QQQ +
Sbjct: 154 GTPNPGNTPNPANATPNPSNPTNNPRMNQQLLQAQTFLLRRQPPNFNEMTYEQQQTFRVK 213
Query: 59 TINPNSNFQ--LQQTLQR------SPSMSRLNQIQPQQQQQVVARQQAALYGGQMNFGGS 110
+ + Q QQ++++ P+ + + Q QQQ + ++ L ++ G
Sbjct: 214 NLESSRRTQHEWQQSMEQMQQVIGDPNTTDDRRQQLQQQLDQIRLKKEGLRAIEIVLGQQ 273
Query: 111 AAVSAQQQQLSGGVAAMGGSASNLSRSALIGQSGHFPMLSGAGAAQFNLLTSPRQKGGMV 170
A +A+ G V+ G GQ P + A Q L R G M
Sbjct: 274 HAAAARAH---GDVSVTGAQ----------GQGQQTPNQAQAQVPQNQGL---RPMGQMQ 317
Query: 171 QSSQFSSANSAGQSLQGMQQAIGMMGSPNLASQMRTNGGLYTQQQQIRLTPAQMRQ-QLS 229
+Q QGM Q G N M G+ Q Q + QMRQ Q +
Sbjct: 318 GQNQMQG--------QGMNQ-----GQMNQGQGMAQGQGMGQGQAQGQRPMMQMRQGQQA 364
Query: 230 QQALNSQQVQG--IPRSSSLAFMNSQLSGLSQN-GQP-GMMHNSLTQSQWLKQMPAMSGP 285
Q QQ QG +P+ + Q G+ Q G P G M Q Q Q P
Sbjct: 365 QGQQGGQQAQGPGMPQGQGI-----QGQGMPQGQGMPQGQMRPGQPQGQPQGQPQGQMRP 419
Query: 286 ASPLRLQQHQRQQQQLASSGQLQQNSMTLNQQQLSQFMQQQKSMGQPQLHQQQPSPQQQQ 345
P Q Q Q GQ Q + Q Q +Q Q GQ Q +P+P QQQ
Sbjct: 420 GQPQGQGMPQGQGQGQMRPGQPQPQAPQAAQAQAAQGQQ-----GQGQGPSSRPNPAQQQ 474
Query: 346 QQQ-LLQPQQQSQLQASVHQQQHLHSPRVAGPTGQKSISLTGSQPDATASGATTPGGSSS 404
Q ++ P + + + QQ +V + +LTG A P + +
Sbjct: 475 QPSAMVSPTRAAVNPFPIPQQLQARQGQVPPMAARPRATLTGGN-GAPLPSLNHPPHTRA 533
Query: 405 QGTEAATNQVLGKRKIQDLVAQV---DPQGKLDPEVIDLLLEFADDFIDSVTTHGCILAK 461
+ ++VL KRK+ +LV V D + +D +V +LLL+ AD+F+ SVT C LAK
Sbjct: 534 PPVDMMGDRVLSKRKLSELVRSVAGEDAEATVDGDVEELLLDLADEFVSSVTAFSCRLAK 593
Query: 462 HRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVRM 514
HRKS TLESKDL LHLE+NW++ IPGYS +E + R + H ++L + M
Sbjct: 594 HRKSDTLESKDLQLHLERNWNIRIPGYSGDEVRSVRRLAPTQGHVQKLAGITM 646
>gb|AAH19668.1| TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated
factor [Mus musculus] gi|47117262|sp|Q8VE65|TAF12_MOUSE
Transcription initiation factor TFIID subunit 12
(Transcription initiation factor TFIID 20 kDa subunits)
(TAFII-20) (TAFII20)
Length = 161
Score = 121 bits (304), Expect = 5e-26
Identities = 64/130 (49%), Positives = 87/130 (66%), Gaps = 4/130 (3%)
Query: 386 GSQPDATASG--ATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLE 443
GS ++T G A TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+
Sbjct: 29 GSMANSTTVGKIAGTPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQ 86
Query: 444 FADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNE 503
ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E
Sbjct: 87 IADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTE 146
Query: 504 LHKRRLDAVR 513
HK+R+ +R
Sbjct: 147 AHKQRMALIR 156
>dbj|BAB25276.1| unnamed protein product [Mus musculus]
Length = 161
Score = 121 bits (304), Expect = 5e-26
Identities = 64/130 (49%), Positives = 87/130 (66%), Gaps = 4/130 (3%)
Query: 386 GSQPDATASG--ATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLE 443
GS ++T G A TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+
Sbjct: 29 GSMANSTTVGKIAGTPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQ 86
Query: 444 FADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNE 503
ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E
Sbjct: 87 IADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTE 146
Query: 504 LHKRRLDAVR 513
HK+R+ +R
Sbjct: 147 AHKQRMALIR 156
>gb|AAH11986.1| TAF12 protein [Homo sapiens] gi|30582901|gb|AAP35678.1| TAF12 RNA
polymerase II, TATA box binding protein (TBP)-associated
factor, 20kDa [Homo sapiens] gi|56206092|emb|CAI22291.1|
TAF12 RNA polymerase II, TATA box binding protein
(TBP)-associated factor, 20kDa [Homo sapiens]
gi|61360319|gb|AAX41843.1| TAF12 RNA polymerase II TATA
box binding protein-associated factor [synthetic
construct] gi|61360311|gb|AAX41842.1| TAF12 RNA
polymerase II TATA box binding protein-associated factor
[synthetic construct] gi|27501932|gb|AAO13491.1| TAF12
RNA polymerase II, TATA box binding protein
(TBP)-associated factor, 20kDa [Homo sapiens]
gi|5032153|ref|NP_005635.1| TAF12 RNA polymerase II,
TATA box binding protein (TBP)-associated factor, 20 kD
[Homo sapiens] gi|3024708|sp|Q16514|TAF12_HUMAN
Transcription initiation factor TFIID subunit 12
(Transcription initiation factor TFIID 20/15 kDa
subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15)
gi|1373377|gb|AAC50600.1| TAF20
gi|791055|emb|CAA58826.1| PolII transcription factor
TFTIID [Homo sapiens] gi|1345404|dbj|BAA09112.1| TFIID
subunit p22 [Homo sapiens]
Length = 161
Score = 119 bits (297), Expect = 3e-25
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 43 TPGAGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 100
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 101 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 156
>gb|AAP36430.1| Homo sapiens TAF12 RNA polymerase II, TATA box binding protein
(TBP)-associated factor, 20kDa [synthetic construct]
gi|61370130|gb|AAX43442.1| TAF12 RNA polymerase II TATA
box binding protein-associated factor [synthetic
construct] gi|61370125|gb|AAX43441.1| TAF12 RNA
polymerase II TATA box binding protein-associated factor
[synthetic construct]
Length = 162
Score = 119 bits (297), Expect = 3e-25
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 43 TPGAGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 100
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 101 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 156
>ref|NP_079855.1| TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated
factor [Mus musculus] gi|12850306|dbj|BAB28669.1|
unnamed protein product [Mus musculus]
Length = 161
Score = 119 bits (297), Expect = 3e-25
Identities = 63/130 (48%), Positives = 86/130 (65%), Gaps = 4/130 (3%)
Query: 386 GSQPDATASG--ATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLE 443
GS ++T G A TPG E NQVL K+K+QDLV +VDP +L +V ++LL+
Sbjct: 29 GSMANSTTVGKIAGTPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLGEDVEEMLLQ 86
Query: 444 FADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNE 503
ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E
Sbjct: 87 IADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNIWIPGFGSEEIRPYKKACTTE 146
Query: 504 LHKRRLDAVR 513
HK+R+ +R
Sbjct: 147 AHKQRMALIR 156
>emb|CAG31620.1| hypothetical protein [Gallus gallus]
Length = 161
Score = 119 bits (297), Expect = 3e-25
Identities = 60/123 (48%), Positives = 83/123 (66%), Gaps = 2/123 (1%)
Query: 391 ATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFID 450
A A TP G E+ NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+
Sbjct: 36 AVAKMPGTPSGGGRLSPES--NQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIE 93
Query: 451 SVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLD 510
SV T C LA+HRKS+TLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+
Sbjct: 94 SVVTAACQLARHRKSNTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMA 153
Query: 511 AVR 513
+R
Sbjct: 154 LIR 156
>gb|AAC50601.1| TAF15
Length = 131
Score = 119 bits (297), Expect = 3e-25
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 13 TPGAGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 70
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 71 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 126
>ref|XP_535337.1| PREDICTED: similar to Transcription initiation factor TFIID subunit
12 (Transcription initiation factor TFIID 20/15 kDa
subunits) (TAFII-20/TAFII-15) (TAFII20/TAFII15) [Canis
familiaris]
Length = 213
Score = 118 bits (295), Expect = 6e-25
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 95 TPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 152
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 153 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 208
>ref|XP_513257.1| PREDICTED: hypothetical protein XP_513257 [Pan troglodytes]
Length = 131
Score = 118 bits (295), Expect = 6e-25
Identities = 59/116 (50%), Positives = 80/116 (68%), Gaps = 2/116 (1%)
Query: 398 TPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDLLLEFADDFIDSVTTHGC 457
TPG E NQVL K+K+QDLV +VDP +LD +V ++LL+ ADDFI+SV T C
Sbjct: 13 TPGTGGRLSPE--NNQVLTKKKLQDLVREVDPNEQLDEDVEEMLLQIADDFIESVVTAAC 70
Query: 458 ILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSNELHKRRLDAVR 513
LA+HRKSSTLE KD+ LHLE+ W++ IPG+ SEE + + + E HK+R+ +R
Sbjct: 71 QLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSEEIRPYKKACTTEAHKQRMALIR 126
>gb|AAH50160.1| Unknown (protein for MGC:56398) [Danio rerio]
Length = 183
Score = 117 bits (292), Expect = 1e-24
Identities = 58/131 (44%), Positives = 86/131 (65%), Gaps = 1/131 (0%)
Query: 384 LTGSQPDATASGATTPGGSSSQGTEAATN-QVLGKRKIQDLVAQVDPQGKLDPEVIDLLL 442
L+ S ++T + PG G + QVL K+K+QDLV ++DP +LD +V ++LL
Sbjct: 27 LSSSMANSTVAPGKLPGTPGPAGRLSPEGPQVLSKKKLQDLVREIDPNEQLDEDVEEMLL 86
Query: 443 EFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSN 502
+ ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ S+E + + +
Sbjct: 87 QIADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSDEIRPYKKACTT 146
Query: 503 ELHKRRLDAVR 513
E HK+R+ +R
Sbjct: 147 EAHKQRMALIR 157
>gb|AAH65878.1| TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated
factor [Danio rerio] gi|34785133|gb|AAH56696.1| TAF12
RNA polymerase II, TATA box binding protein
(TBP)-associated factor [Danio rerio]
gi|38198645|ref|NP_938182.1| TAF12 RNA polymerase II,
TATA box binding protein (TBP)-associated factor [Danio
rerio]
Length = 162
Score = 117 bits (292), Expect = 1e-24
Identities = 58/131 (44%), Positives = 86/131 (65%), Gaps = 1/131 (0%)
Query: 384 LTGSQPDATASGATTPGGSSSQGTEAATN-QVLGKRKIQDLVAQVDPQGKLDPEVIDLLL 442
L+ S ++T + PG G + QVL K+K+QDLV ++DP +LD +V ++LL
Sbjct: 27 LSSSMANSTVAPGKLPGTPGPAGRLSPEGPQVLSKKKLQDLVREIDPNEQLDEDVEEMLL 86
Query: 443 EFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPLSN 502
+ ADDFI+SV T C LA+HRKSSTLE KD+ LHLE+ W++ IPG+ S+E + + +
Sbjct: 87 QIADDFIESVVTAACQLARHRKSSTLEVKDVQLHLERQWNMWIPGFGSDEIRPYKKACTT 146
Query: 503 ELHKRRLDAVR 513
E HK+R+ +R
Sbjct: 147 EAHKQRMALIR 157
>emb|CAF94342.1| unnamed protein product [Tetraodon nigroviridis]
Length = 160
Score = 116 bits (291), Expect = 2e-24
Identities = 57/133 (42%), Positives = 87/133 (64%), Gaps = 2/133 (1%)
Query: 381 SISLTGSQPDATASGATTPGGSSSQGTEAATNQVLGKRKIQDLVAQVDPQGKLDPEVIDL 440
S+++ + T TPG + E + QVL K+K+QDLV ++DP +LD +V ++
Sbjct: 25 SVAMNMANSTTTVMKVMTPGPAGRSSPEGS--QVLTKKKLQDLVREIDPNEQLDEDVEEM 82
Query: 441 LLEFADDFIDSVTTHGCILAKHRKSSTLESKDLLLHLEKNWDLTIPGYSSEEKKYQSRPL 500
LL+ ADDFI+SV T C LA+HRKS+TLE KD+ LHLE+ W++ IPG+ S+E + +
Sbjct: 83 LLQIADDFIESVVTAACQLARHRKSNTLEVKDVQLHLERQWNMWIPGFGSDEIRPFKKAC 142
Query: 501 SNELHKRRLDAVR 513
+ E HK+R+ +R
Sbjct: 143 TTEAHKQRMALIR 155
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.309 0.121 0.328
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 910,086,094
Number of Sequences: 2540612
Number of extensions: 38828748
Number of successful extensions: 705616
Number of sequences better than 10.0: 8659
Number of HSP's better than 10.0 without gapping: 4172
Number of HSP's successfully gapped in prelim test: 4767
Number of HSP's that attempted gapping in prelim test: 251056
Number of HSP's gapped (non-prelim): 89128
length of query: 570
length of database: 863,360,394
effective HSP length: 133
effective length of query: 437
effective length of database: 525,458,998
effective search space: 229625582126
effective search space used: 229625582126
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC144645.6


