

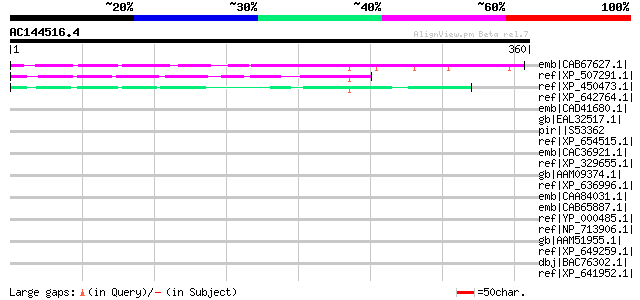
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144516.4 + phase: 0
(360 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB67627.1| putative protein [Arabidopsis thaliana] gi|21689... 137 4e-31
ref|XP_507291.1| PREDICTED OSJNBa0016N23.125 gene product [Oryza... 87 8e-16
ref|XP_450473.1| hypothetical protein [Oryza sativa (japonica cu... 61 6e-08
ref|XP_642764.1| hypothetical protein DDB0169144 [Dictyostelium ... 47 0.001
emb|CAD41680.1| OSJNBa0019K04.27 [Oryza sativa (japonica cultiva... 45 0.003
gb|EAL32517.1| GA10981-PA [Drosophila pseudoobscura] 45 0.003
pir||S53362 mucin 5AC (clone JER47) - human (fragment) 44 0.006
ref|XP_654515.1| hypothetical protein 51.t00023 [Entamoeba histo... 44 0.010
emb|CAC36921.1| SPAPB1E7.04c [Schizosaccharomyces pombe] gi|1911... 43 0.013
ref|XP_329655.1| predicted protein [Neurospora crassa] gi|289196... 43 0.013
gb|AAM09374.1| similar to Leptospira interrogans serovar lai str... 43 0.017
ref|XP_636996.1| hypothetical protein DDB0187577 [Dictyostelium ... 42 0.022
emb|CAA84031.1| mucin [Homo sapiens] 42 0.029
emb|CAB65887.1| EG:33C11.1 [Drosophila melanogaster] gi|20428645... 42 0.029
ref|YP_000485.1| putative lipoprotein [Leptospira interrogans se... 42 0.029
ref|NP_713906.1| hypothetical protein LA3726 [Leptospira interro... 42 0.029
gb|AAM51955.1| GH24653p [Drosophila melanogaster] 42 0.029
ref|XP_649259.1| hypothetical protein 326.t00008 [Entamoeba hist... 42 0.029
dbj|BAC76302.1| ORF515 [Cyanidioschyzon merolae] gi|30468253|ref... 42 0.029
ref|XP_641952.1| hypothetical protein DDB0206180 [Dictyostelium ... 42 0.029
>emb|CAB67627.1| putative protein [Arabidopsis thaliana] gi|21689689|gb|AAM67466.1|
unknown protein [Arabidopsis thaliana]
gi|20259405|gb|AAM14023.1| unknown protein [Arabidopsis
thaliana] gi|15230911|ref|NP_191358.1| expressed protein
[Arabidopsis thaliana] gi|11357955|pir||T46021
hypothetical protein T10K17.200 - Arabidopsis thaliana
Length = 367
Score = 137 bits (346), Expect = 4e-31
Identities = 119/396 (30%), Positives = 179/396 (45%), Gaps = 68/396 (17%)
Query: 1 MKLSLKFQNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPT 60
MK S+KF+ Q + AKVP++I P S I A + + S +LST F +
Sbjct: 1 MKASMKFREE-------QKPLFRAKVPLSILGLPFQSGIVAGES---KELSLNLSTFFES 50
Query: 61 GPTFKLSYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFS 120
GP+ K++Y P + + PFSL +K+G G FGSP S ++ SA F+L + PSF
Sbjct: 51 GPSLKVAYRPNDSWN--PFSLIVKTGSGSFGSPISSSMLMSAEFNLLGQGN-----PSFM 103
Query: 121 LHFKPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNN 180
LHFKPQFG FS+ K+ S E+ + NGS S + ++E
Sbjct: 104 LHFKPQFGDFSIKKSHSSSG-----------FERNLIKSMNGSVSE-DDSSIEVVDTPAV 151
Query: 181 NNVGVGV-GVGVGPDGKNNEKCGLSPCVGVMARTVMPVTQGLLLNFRWGVSFPGK----- 234
N G G V V P + GL V V ART +PV +LNFRWGV P +
Sbjct: 152 NGCGGGFRKVTVLPSTSAGDIAGLLSGVEVAARTSLPVRGRAVLNFRWGVRVPTEIRRDF 211
Query: 235 ------SVLKMPYLAVNKIGLERVE--EMKKDEWNND-DNCEGNLQVLKEMKID--LENV 283
S+ + P+L +NKIG+E V+ + K + D G Q + +E +
Sbjct: 212 DPTAAISLRRFPFLVMNKIGIEHVDGADAKVTKSTGDPGKVSGPAQFTTSGDVAEVIEEL 271
Query: 284 EKENKEMKRVLDEMKMGVSK-----------GKVSKQKHAGESVQSWGSNKNGRKENEKK 332
ENK++KR +++++ +S G SK + + + + +N N +
Sbjct: 272 RTENKQLKRAVEDLREVISNVRPYSPATIDYGSHSKYRESERNNNNNNNNNNNNNGRSRA 331
Query: 333 QEIKPQNIVVSDF-----------ESELEKAIKAAA 357
+ SD+ EL+KA+K AA
Sbjct: 332 DRWSSERTTTSDYGGKKSKEEGNVAEELKKALKGAA 367
>ref|XP_507291.1| PREDICTED OSJNBa0016N23.125 gene product [Oryza sativa (japonica
cultivar-group)] gi|50947535|ref|XP_483295.1| unknown
protein [Oryza sativa (japonica cultivar-group)]
gi|28411873|dbj|BAC57403.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 378
Score = 87.0 bits (214), Expect = 8e-16
Identities = 78/275 (28%), Positives = 121/275 (43%), Gaps = 62/275 (22%)
Query: 1 MKLSLKFQNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPT 60
MK S+KF++++ ++ A+VPI + PL S ++A + + LST F
Sbjct: 1 MKASIKFRDDDR-------PLLRARVPIRVLGLPLHSGLSAGGD--PRELRLDLSTAFSF 51
Query: 61 GPTFKLSYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFS 120
GP +LSY P +LPFS+S+++G+G GSP +P +A F +L S P P+F
Sbjct: 52 GPAIRLSYRPN--DPALPFSVSVRAGVGPLGSPARAPFSLAAEF---NLLSGNPGSPAFF 106
Query: 121 LHFKPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNN 180
L KP+ G FSL T+ S S P + G V + +G + +N + F
Sbjct: 107 LLLKPRLGDFSLSHTLRS--------SPHPGNKVGEVSDGDGH---GREVNYKAFSFAAA 155
Query: 181 NNVGVGVGVGVGPDGKNNEKCGLSPCVGVMARTVMPVTQGLLLNFRWGVSFPGK------ 234
G GVG L + + R+V+P+ L F WG+ P +
Sbjct: 156 GKSGGGVG-------------ALLSGMRLTTRSVLPLWGRASLRFNWGLRAPPELQAALA 202
Query: 235 ------------------SVLKMPYLAVNKIGLER 251
V KMP L ++KI +E+
Sbjct: 203 ADDAMVGASRSRKGGARVPVSKMPLLVIDKISIEQ 237
>ref|XP_450473.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|49388831|dbj|BAD26021.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 274
Score = 60.8 bits (146), Expect = 6e-08
Identities = 73/333 (21%), Positives = 126/333 (36%), Gaps = 89/333 (26%)
Query: 1 MKLSLKFQNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPT 60
MK S+K + + ++ AK+P+ +F+ P ++++TA + ++ SL+T P
Sbjct: 1 MKASVKLREDGA------APLLRAKLPVALFSVPAVASLTAGD---PANLRLSLATAAPA 51
Query: 61 GPTFKLSYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFS 120
P+ +LSY P +S P SL++ G G GSP+ S SA ++ +T SFS
Sbjct: 52 LPSIRLSYAPNRATS--PLSLAVVLGSGPGGSPSSSGAAASA--ITMAVEVNTAGAVSFS 107
Query: 121 LHFKPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNN 180
L KP G F++ K
Sbjct: 108 LALKPSLGDFAVRKRF-------------------------------------------- 123
Query: 181 NNVGVGVGVGVGPDGKNNEKCGLSPCVGVMARTVMPVTQG-LLLNFRWGVSFPGK----- 234
++ G G G G S V R+ +PV G ++ RWGV P +
Sbjct: 124 DSAAAGGGGGSGSSA--------SAASEVTMRSAIPVRGGAAAVSVRWGVRIPAEVTAGG 175
Query: 235 -------SVLKMPYLAVNKIGLERVEEMKKDEWNNDDNCEGNLQVLKEMKIDLENVEKEN 287
++ ++P+L + K+ +ER + +E +EN
Sbjct: 176 EEGAAALALRRLPFLVLGKVTVERRPPPPPASTAEETT-----------TTTVEKTRREN 224
Query: 288 KEMKRVLDEMKMGVSKGKVSKQKHAGESVQSWG 320
+ + R LDE++ ++ K A +S G
Sbjct: 225 ERLTRELDELRAAATEKTERKMTSAAAGRRSSG 257
>ref|XP_642764.1| hypothetical protein DDB0169144 [Dictyostelium discoideum]
gi|28828651|gb|AAO51254.1| similar to Plasmodium
falciparum. Hypothetical protein [Dictyostelium
discoideum] gi|60470841|gb|EAL68813.1| hypothetical
protein DDB0169144 [Dictyostelium discoideum]
Length = 1123
Score = 46.6 bits (109), Expect = 0.001
Identities = 51/202 (25%), Positives = 85/202 (41%), Gaps = 24/202 (11%)
Query: 4 SLKFQNNNENQQTPQTQIMTAKVPIT----IFNNPLLSTITATNNYSTSDF--------- 50
++ F N +N TP + ++ ++ ++P LS I +N+ STS+
Sbjct: 336 NIHFLTNQQNSDTPLSNSSSSTPTLSPAQSAISSPTLSNIINSNSNSTSNLNDLIKDSSQ 395
Query: 51 -------SFSLSTNFPTGPTFKLSYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSAN 103
S S S++ + + S + +++SSS S S S S + S S++
Sbjct: 396 PSTSTSTSTSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 455
Query: 104 FSLSSLSSSTPLLPSFSLHFKPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFV---PET 160
S SS SSS P L S + P+ H SL +++ N N + +K P
Sbjct: 456 SSSSSSSSSQPTLNQHSKYI-PKSLHSSLSSIPINNKENNNNNNNRDNRDKDICTTSPNL 514
Query: 161 NGSCSGWQNLNLEPFGHNNNNN 182
N S + +L + NNNNN
Sbjct: 515 NDSINNTLDLFNKKINSNNNNN 536
>emb|CAD41680.1| OSJNBa0019K04.27 [Oryza sativa (japonica cultivar-group)]
gi|50928131|ref|XP_473593.1| OSJNBa0019K04.27 [Oryza
sativa (japonica cultivar-group)]
Length = 180
Score = 45.4 bits (106), Expect = 0.003
Identities = 29/66 (43%), Positives = 37/66 (55%), Gaps = 7/66 (10%)
Query: 60 TGPTFKLSYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSF 119
+GP +L Y P S PF+LS+ + LG GSP +P S F+L L S+ LP F
Sbjct: 21 SGPALRLLYRPN--GSQHPFALSVCTSLGPLGSPTRAPFAISTEFNL--LCSN---LPVF 73
Query: 120 SLHFKP 125
SL FKP
Sbjct: 74 SLLFKP 79
>gb|EAL32517.1| GA10981-PA [Drosophila pseudoobscura]
Length = 522
Score = 45.1 bits (105), Expect = 0.003
Identities = 41/131 (31%), Positives = 62/131 (47%), Gaps = 11/131 (8%)
Query: 69 TPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQFG 128
TPT++SSS S S ++G +P S + F N + + S T L + F+P G
Sbjct: 343 TPTSSSSSSSNSSSNQTGSAA-STPTSSAIEFVKNELVDAQSPYTTALSAERFLFEPSDG 401
Query: 129 HFSL-HKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLN-----LEPFGHN-NNN 181
+ H + +S+ N L+++ + G +G Q+ N L F H NN
Sbjct: 402 FPDIKHAVSVLPTSDINSLNMASVVHAN---SGGGGAAGHQHNNQHICLLVDFHHGLQNN 458
Query: 182 NVGVGVGVGVG 192
+GVGVGVGVG
Sbjct: 459 GLGVGVGVGVG 469
>pir||S53362 mucin 5AC (clone JER47) - human (fragment)
Length = 477
Score = 44.3 bits (103), Expect = 0.006
Identities = 32/143 (22%), Positives = 66/143 (45%), Gaps = 1/143 (0%)
Query: 8 QNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKLS 67
Q+ + Q++ T ++T T + + T+T + T+ + + +T+ + P +S
Sbjct: 148 QSTSSWQKSRTTTLVTTSTTSTPQTSTTSAPTTSTTSAPTTSTTSAPTTSTTSTPQTSIS 207
Query: 68 YTPTTTSSSLPFSLSL-KSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQ 126
PTT+++S P S ++ ++ + +P S F + S+ ++ST P+ S PQ
Sbjct: 208 SAPTTSTTSAPTSSTISRATTSIISAPTTSTTSFPTTSTTSATTTSTTSAPTTSTTSTPQ 267
Query: 127 FGHFSLHKTVLSDSSNTNPLSVS 149
S + + S T P V+
Sbjct: 268 TSKTSAATSSTTSGSGTTPSPVT 290
>ref|XP_654515.1| hypothetical protein 51.t00023 [Entamoeba histolytica HM-1:IMSS]
gi|56471568|gb|EAL49127.1| hypothetical protein
51.t00023 [Entamoeba histolytica HM-1:IMSS]
Length = 568
Score = 43.5 bits (101), Expect = 0.010
Identities = 65/301 (21%), Positives = 115/301 (37%), Gaps = 39/301 (12%)
Query: 7 FQNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKL 66
F N + TP T T T NN + N++TS+ + + T TG
Sbjct: 215 FGNFTPSNTTPTTGNNTTGTTTTTGNNTTGTGSNPFGNFTTSNSTTTTGTTTTTGNN-TT 273
Query: 67 SYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQ 126
+ T TTT ++ + +G FG+ F+ N + +S +S+TP S
Sbjct: 274 TGTTTTTGTTTTTGTTTTTGSNPFGN-------FNTN-NTTSNASTTPTTGS------NP 319
Query: 127 FGHFSLHKTVLSDSSNT----NPLS--VSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNN 180
FG+F+ T + + T NP + P T G+ + PFG+ +
Sbjct: 320 FGNFTTSNTTPTTGTTTTTGSNPFGNFTNTTTSSATTPNTTGTNTS------NPFGNFTS 373
Query: 181 NNVGVGVGVGVG-PDGKNNEKCGLSPCVGVMARTVMPVTQGLLLNFRWGVSFPGKSVLKM 239
+ G G P G NN S G+ P T + + + ++
Sbjct: 374 STTNTTTTNGSGNPFGTNN--LLNSSTTGIQTNGNQPTTNDIQSSI--------QKEAEL 423
Query: 240 PYLAVNKIGLER-VEEMKKDEWNNDDNCEGNLQVLKEMKIDLENVEKENKEMKRVLDEMK 298
+ +GL+ +EE KD ++ + + +K + EKE +E+K+ + +M
Sbjct: 424 LFSRAKTLGLDDCIEEWNKDLQSDAKRFQEISEKIKAQNESIMKAEKEIEELKKFVQQMS 483
Query: 299 M 299
+
Sbjct: 484 V 484
>emb|CAC36921.1| SPAPB1E7.04c [Schizosaccharomyces pombe]
gi|19115042|ref|NP_594130.1| hypothetical protein
SPAPB1E7.04c [Schizosaccharomyces pombe 972h-]
Length = 1236
Score = 43.1 bits (100), Expect = 0.013
Identities = 42/154 (27%), Positives = 76/154 (49%), Gaps = 6/154 (3%)
Query: 9 NNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKLSY 68
+++ + +PQ+ + T+ ++ ++ LLS +A + S+S S S+ ++ T S
Sbjct: 485 SSSSSASSPQSTLSTSSEVVSEVSSTLLSGSSAIPSTSSSTPSSSIISSPMTSVLSSSSS 544
Query: 69 TPTTTSSSLPFSL-SLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQF 127
PT++SS S+ ++ SG+ S + P FS+ S+ S S+S+P S S+
Sbjct: 545 IPTSSSSDFSSSITTISSGI----SSSSIPSTFSSVSSILSSSTSSPSSTSLSISSSSTS 600
Query: 128 GHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETN 161
FS T S SS ++ +S S I P T+
Sbjct: 601 STFSSAST-SSPSSISSSISSSSTILSSPTPSTS 633
>ref|XP_329655.1| predicted protein [Neurospora crassa] gi|28919655|gb|EAA29070.1|
predicted protein [Neurospora crassa]
Length = 589
Score = 43.1 bits (100), Expect = 0.013
Identities = 49/180 (27%), Positives = 73/180 (40%), Gaps = 34/180 (18%)
Query: 16 TPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSL-STNFPTGPTFKLSYTPTTTS 74
TPQ + P + ++PL ST + +S S + PT P+F +S PTT
Sbjct: 98 TPQA-LNAGASPFSPASSPLTSTSSTGATIGSSSVEVDTPSASIPTTPSFSISPNPTT-- 154
Query: 75 SSLPFSLSLKSGLGLFGSPN-HSPLVFSA-NFSLSSLSSSTPLLPSFSLHFKPQFGHFSL 132
L+ SP+ SP FS+ NF + TP+ P+ S + P FGH +
Sbjct: 155 -------------NLWASPSAGSPASFSSENFD----TIDTPITPAHSSNL-PFFGHLAS 196
Query: 133 HKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGH----------NNNNN 182
S S+N N + + G + +L+L +GH NNNNN
Sbjct: 197 ASGPASSSNNNNDNLTNTGLPSGNYGMIYSPIAPISSLSLPFYGHSTSGSGSASSNNNNN 256
>gb|AAM09374.1| similar to Leptospira interrogans serovar lai str. 56601.
Leucine-rich repeat containing protein [Dictyostelium
discoideum]
Length = 725
Score = 42.7 bits (99), Expect = 0.017
Identities = 56/182 (30%), Positives = 73/182 (39%), Gaps = 31/182 (17%)
Query: 9 NNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKLSY 68
NNN N T T T T NN L T++Y +S S S P+G + S+
Sbjct: 401 NNNNNTTTTTTTTTTTTTTSTTNNNNTL----FTDSYLSSASSSSFLNKSPSGLSLVGSF 456
Query: 69 TPT-----TTSSSLPFSLS--LKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSL 121
T + SS++ S S + G GL N L+ S +FS S S T SFS
Sbjct: 457 NGTIKKSISNSSNMNGSGSNGVGHGSGLTPKLNFKSLITSRDFS-SDSDSDTDSTSSFS- 514
Query: 122 HFKPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFV-PETNGSCSGWQNLNLEPFGHNNN 180
T S + NT S+SP+ +N S S N N G+NNN
Sbjct: 515 -------------TYSSSTINTKRASISPRTSLTLSNSASNDSVSAVANGN----GNNNN 557
Query: 181 NN 182
NN
Sbjct: 558 NN 559
>ref|XP_636996.1| hypothetical protein DDB0187577 [Dictyostelium discoideum]
gi|60465411|gb|EAL63496.1| hypothetical protein
DDB0187577 [Dictyostelium discoideum]
Length = 742
Score = 42.4 bits (98), Expect = 0.022
Identities = 50/153 (32%), Positives = 68/153 (43%), Gaps = 9/153 (5%)
Query: 35 LLSTITATNNYSTSDFSFSLSTNFPTGPTFKLSYTPT--TTSSSLPFSLSLKSGLGLF-- 90
L S+IT++ TS S S ST+ T P F S++ TTS+ + LS K+ L L
Sbjct: 205 LSSSITSS---ITSSCSSSTSTSLTTSPIFNKSHSNRNLTTSTIIANRLSTKAALLLSRD 261
Query: 91 GSPNHSPLVFSA-NFSLSSLSSSTPLLPSFSLHFKPQFGHFSLHKTVLSDSSNTNPLSVS 149
+HS + NF SS S TP S P+ S T S SS+T S+
Sbjct: 262 DDEDHSKTILDLKNFYSSSSSLFTPSGSPRSTTISPR-SITSPTTTTTSLSSSTPSFSII 320
Query: 150 PQIEKGFVPETNGSCSGWQNLNLEPFGHNNNNN 182
Q+E T + + N N +NNNNN
Sbjct: 321 TQLENNTNITTQDNNNNNNNNNNNNNNNNNNNN 353
Score = 33.9 bits (76), Expect = 7.8
Identities = 29/109 (26%), Positives = 50/109 (45%), Gaps = 3/109 (2%)
Query: 9 NNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSF--SLSTNFPTGPTFKL 66
NNN N +T+ I F +L + S+F+F SLS++ P+ +
Sbjct: 123 NNNNNNNNNNNNNITSDFNIHSFFK-ILKQFKSVERVCLSNFNFITSLSSSSPSSSSSSS 181
Query: 67 SYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPL 115
S + +++SSS S L S L S S + S + S S+ +++P+
Sbjct: 182 SSSSSSSSSSSSSSSLLLSSLSSLSSSITSSITSSCSSSTSTSLTTSPI 230
>emb|CAA84031.1| mucin [Homo sapiens]
Length = 477
Score = 42.0 bits (97), Expect = 0.029
Identities = 31/143 (21%), Positives = 65/143 (44%), Gaps = 1/143 (0%)
Query: 8 QNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKLS 67
Q+ + Q++ T ++T T + + T+T + T+ + + +T+ + P +S
Sbjct: 148 QSTSSWQKSRTTTLVTTSTTSTPQTSTTSAPTTSTTSAPTTSTTSAPTTSTTSTPQTSIS 207
Query: 68 YTPTTTSSSLPFSLSLKSGL-GLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQ 126
PT++++S P S ++ + + +P S F + S+ ++ST P+ S PQ
Sbjct: 208 SAPTSSTTSAPTSSTISARTTSIISAPTTSTTSFPTTSTTSATTTSTTSAPTSSTTSTPQ 267
Query: 127 FGHFSLHKTVLSDSSNTNPLSVS 149
S + + S T P V+
Sbjct: 268 TSKTSAATSSTTSGSGTTPSPVT 290
>emb|CAB65887.1| EG:33C11.1 [Drosophila melanogaster] gi|20428645|ref|NP_620473.1|
CG11405-PA [Drosophila melanogaster]
gi|7290135|gb|AAF45599.1| CG11405-PA [Drosophila
melanogaster]
Length = 613
Score = 42.0 bits (97), Expect = 0.029
Identities = 56/207 (27%), Positives = 79/207 (38%), Gaps = 47/207 (22%)
Query: 9 NNNENQQ--TPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKL 66
NN +QQ P +++ VP S + NN + ++ S S+N +
Sbjct: 375 NNVIDQQHANPSPSLLSDYVPNCDGLTGSASNHPSHNNNNNNNNSSGASSNTSNNNSNIS 434
Query: 67 SYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQ 126
S++ TSS+ P + S S + F N + S S T L + F+P
Sbjct: 435 SHSSNATSSTTPTATS-------------SAIEFVKNELVDSQSPYTTALSAERFLFEPS 481
Query: 127 FGHFSL-HKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNNNN--- 182
G + H VL N L+++ V TNGS N + HNNNNN
Sbjct: 482 DGFPDIKHACVLPSPCGINGLNMAS------VVNTNGSTGNNNNSH-----HNNNNNNNN 530
Query: 183 -----------------VGVGVGVGVG 192
VGVGVGVGVG
Sbjct: 531 HLMDFHQGLQHGNIGTGVGVGVGVGVG 557
>ref|YP_000485.1| putative lipoprotein [Leptospira interrogans serovar Copenhageni
str. Fiocruz L1-130] gi|45599633|gb|AAS69122.1| putative
lipoprotein [Leptospira interrogans serovar Copenhageni
str. Fiocruz L1-130]
Length = 510
Score = 42.0 bits (97), Expect = 0.029
Identities = 46/189 (24%), Positives = 75/189 (39%), Gaps = 17/189 (8%)
Query: 21 IMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFK-----LSYTPTTTSS 75
I+ P+ + N L TIT N S SFS+S N P G F ++ TPT T
Sbjct: 90 ILNRSYPVLVSNMFLTPTITQDPNTSLEFKSFSISPNLPAGIFFDSFTGWITGTPTVTVP 149
Query: 76 SLPFSLSLKSGL------GLFGSPNHSPLV-FSANF----SLSSLSSSTPLLP-SFSLHF 123
S +++SL + L+ N + + FS N+ + S L +L + +
Sbjct: 150 SSEYTISLNYSIRNNKDRKLYQDQNTTAKISFSTNYDPTLTYSFLQPGNNILSLGVGITY 209
Query: 124 KPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNNNNV 183
P F S S T P ++ G + T + +G N + N +++V
Sbjct: 210 LPTVNGFGSGTITYSISPATLPAGLNFNTSNGTIFGTPTTVTGSTNYTITATNVNASDSV 269
Query: 184 GVGVGVGVG 192
+ V +G
Sbjct: 270 SFSLQVAIG 278
>ref|NP_713906.1| hypothetical protein LA3726 [Leptospira interrogans serovar Lai
str. 56601] gi|24197719|gb|AAN50924.1| conserved
hypothetical protein [Leptospira interrogans serovar lai
str. 56601]
Length = 510
Score = 42.0 bits (97), Expect = 0.029
Identities = 46/189 (24%), Positives = 75/189 (39%), Gaps = 17/189 (8%)
Query: 21 IMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFK-----LSYTPTTTSS 75
I+ P+ + N L TIT N S SFS+S N P G F ++ TPT T
Sbjct: 90 ILNRSYPVLVSNMFLTPTITQDPNTSLEFKSFSISPNLPAGIFFDSFTGWITGTPTVTVP 149
Query: 76 SLPFSLSLKSGL------GLFGSPNHSPLV-FSANF----SLSSLSSSTPLLP-SFSLHF 123
S +++SL + L+ N + + FS N+ + S L +L + +
Sbjct: 150 SSEYTISLNYSIRNNKDRKLYQDQNTTAKISFSTNYDPTLTYSFLQPGNNILSLGVGITY 209
Query: 124 KPQFGHFSLHKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNNNNV 183
P F S S T P ++ G + T + +G N + N +++V
Sbjct: 210 LPTVNGFGSGTITYSISPATLPAGLNFNTSNGTIFGTPTTVTGSTNYTITATNVNASDSV 269
Query: 184 GVGVGVGVG 192
+ V +G
Sbjct: 270 SFSLQVAIG 278
>gb|AAM51955.1| GH24653p [Drosophila melanogaster]
Length = 295
Score = 42.0 bits (97), Expect = 0.029
Identities = 56/207 (27%), Positives = 79/207 (38%), Gaps = 47/207 (22%)
Query: 9 NNNENQQ--TPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKL 66
NN +QQ P +++ VP S + NN + ++ S S+N +
Sbjct: 57 NNVIDQQHANPSPSLLSDYVPNCDGLTGSASNHPSHNNNNNNNNSSGASSNTSNNNSNIS 116
Query: 67 SYTPTTTSSSLPFSLSLKSGLGLFGSPNHSPLVFSANFSLSSLSSSTPLLPSFSLHFKPQ 126
S++ TSS+ P + S S + F N + S S T L + F+P
Sbjct: 117 SHSSNATSSTTPTATS-------------SAIEFVKNELVDSQSPYTTALSAERFLFEPS 163
Query: 127 FGHFSL-HKTVLSDSSNTNPLSVSPQIEKGFVPETNGSCSGWQNLNLEPFGHNNNNN--- 182
G + H VL N L+++ V TNGS N + HNNNNN
Sbjct: 164 DGFPDIKHACVLPSPCGINGLNMAS------VVNTNGSTGNNNNSH-----HNNNNNNNN 212
Query: 183 -----------------VGVGVGVGVG 192
VGVGVGVGVG
Sbjct: 213 HLMDFHQGLQHGNIGTGVGVGVGVGVG 239
>ref|XP_649259.1| hypothetical protein 326.t00008 [Entamoeba histolytica HM-1:IMSS]
gi|56465658|gb|EAL43878.1| hypothetical protein
326.t00008 [Entamoeba histolytica HM-1:IMSS]
Length = 554
Score = 42.0 bits (97), Expect = 0.029
Identities = 32/107 (29%), Positives = 57/107 (52%), Gaps = 10/107 (9%)
Query: 253 EEMKKDEWNN--DDNCEGNLQVLKEMKIDLENVEKENKEMKRVLDEMKM--GVSKGKVSK 308
EE++ ++ NN D E ++ L E+K + V EN+E+K++L+E+K+ G + ++ +
Sbjct: 290 EELENEKKNNQKDRINEEQIKELNEIKEMNQKVITENEELKKILEELKIKEGALQKEIGE 349
Query: 309 QKHAGESVQSWGSNKNGRKENEK----KQEIKPQNIVVSDFESELEK 351
K G+ +Q + RK NE+ QE+K Q V E +K
Sbjct: 350 NKEKGQKLQD--EKEEFRKANEEMKITTQELKQQLSQVKTTNEEEQK 394
>dbj|BAC76302.1| ORF515 [Cyanidioschyzon merolae] gi|30468253|ref|NP_849140.1|
ORF515 [Cyanidioschyzon merolae strain 10D]
Length = 515
Score = 42.0 bits (97), Expect = 0.029
Identities = 31/108 (28%), Positives = 60/108 (54%), Gaps = 10/108 (9%)
Query: 253 EEMKKDEWNNDDNCE--GNLQVLKEMKIDLE--------NVEKENKEMKRVLDEMKMGVS 302
E++ ++E N + N E LQ L+E + +LE N E+ N+E+ + L E ++ +
Sbjct: 242 EQVDQEESNEESNYELIKLLQSLEESEQELEKASKQEQVNQEESNQELLQSLGESELQLE 301
Query: 303 KGKVSKQKHAGESVQSWGSNKNGRKENEKKQEIKPQNIVVSDFESELE 350
K +V++++ E +QS G ++ ++ + KQE Q ++ S E EL+
Sbjct: 302 KEQVNQEESNQELLQSLGESELQLEKEQVKQEESNQELLQSLGEPELQ 349
Score = 41.2 bits (95), Expect = 0.049
Identities = 28/99 (28%), Positives = 56/99 (56%), Gaps = 4/99 (4%)
Query: 256 KKDEWNNDDNCEGNLQVLKEMKIDLE----NVEKENKEMKRVLDEMKMGVSKGKVSKQKH 311
K+++ N +++ + LQ L E ++ LE N E+ N+E+ + L E ++ + K +V +++
Sbjct: 276 KQEQVNQEESNQELLQSLGESELQLEKEQVNQEESNQELLQSLGESELQLEKEQVKQEES 335
Query: 312 AGESVQSWGSNKNGRKENEKKQEIKPQNIVVSDFESELE 350
E +QS G + ++ + KQE Q ++ S E EL+
Sbjct: 336 NQELLQSLGEPELQLEKEQVKQEEFNQELLQSLGEPELQ 374
>ref|XP_641952.1| hypothetical protein DDB0206180 [Dictyostelium discoideum]
gi|60470006|gb|EAL67987.1| hypothetical protein
DDB0206180 [Dictyostelium discoideum]
Length = 1350
Score = 42.0 bits (97), Expect = 0.029
Identities = 38/116 (32%), Positives = 51/116 (43%), Gaps = 11/116 (9%)
Query: 8 QNNNENQQTPQTQIMTAKVPITIFNNPLLSTITATNNYSTSDFSFSLSTNFPTGPTFKLS 67
Q N + +P T T++ IT N +T T TNN ++ + SLSTN +
Sbjct: 1235 QQNPGSTSSPHTLNTTSQHQITTTTNTTTTTTTTTNNSPNNNNNSSLSTNSTPNQIQGSN 1294
Query: 68 YTPTTTSSSLPFSLSLKSGLGLFGSP--NHSPLVFSANFSLSSLSSSTPLLPSFSL 121
TPT L S L L SP N SPL + + + S + S P PSF L
Sbjct: 1295 TTPT---------LEKLSSLNLALSPIQNTSPLDSAPSSNESPCAQSLPNTPSFYL 1341
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.310 0.128 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 652,478,963
Number of Sequences: 2540612
Number of extensions: 30877118
Number of successful extensions: 115537
Number of sequences better than 10.0: 655
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 622
Number of HSP's that attempted gapping in prelim test: 112366
Number of HSP's gapped (non-prelim): 2646
length of query: 360
length of database: 863,360,394
effective HSP length: 129
effective length of query: 231
effective length of database: 535,621,446
effective search space: 123728554026
effective search space used: 123728554026
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144516.4


