

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.18 + phase: 0
(107 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
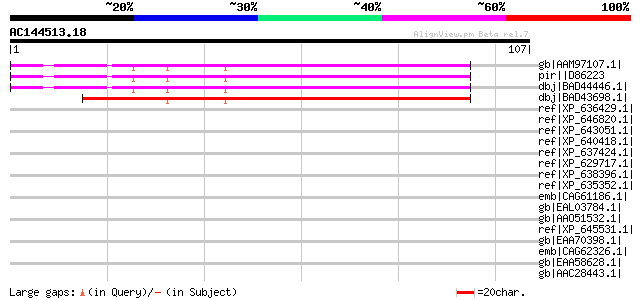
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM97107.1| ATP-dependent Clp protease proteolytic subunit (C... 75 5e-13
pir||D86223 hypothetical protein [imported] - Arabidopsis thalia... 75 5e-13
dbj|BAD44446.1| ClpP protease complex subunit ClpR3 [Arabidopsis... 75 5e-13
dbj|BAD43698.1| ClpP protease complex subunit ClpR3 [Arabidopsis... 74 1e-12
ref|XP_636429.1| hypothetical protein DDB0188216 [Dictyostelium ... 35 0.34
ref|XP_646820.1| hypothetical protein DDB0202057 [Dictyostelium ... 34 0.76
ref|XP_643051.1| hypothetical protein DDB0217837 [Dictyostelium ... 34 0.76
ref|XP_640418.1| hypothetical protein DDB0218351 [Dictyostelium ... 34 0.76
ref|XP_637424.1| hypothetical protein DDB0218889 [Dictyostelium ... 34 0.76
ref|XP_629717.1| hypothetical protein DDB0215500 [Dictyostelium ... 34 0.76
ref|XP_638396.1| hypothetical protein DDB0186183 [Dictyostelium ... 33 1.3
ref|XP_635352.1| hypothetical protein DDB0219987 [Dictyostelium ... 33 1.3
emb|CAG61186.1| unnamed protein product [Candida glabrata CBS138... 33 1.7
gb|EAL03784.1| hypothetical protein CaO19.1481 [Candida albicans... 33 1.7
gb|AAO51532.1| similar to Orgyia pseudotsugata multicapsid polyh... 33 1.7
ref|XP_645531.1| hypothetical protein DDB0216924 [Dictyostelium ... 33 1.7
gb|EAA70398.1| hypothetical protein FG10082.1 [Gibberella zeae P... 33 2.2
emb|CAG62326.1| unnamed protein product [Candida glabrata CBS138... 33 2.2
gb|EAA58628.1| hypothetical protein AN6244.2 [Aspergillus nidula... 33 2.2
gb|AAC28443.1| HeRunt-1 [Heliocidaris erythrogramma] 33 2.2
>gb|AAM97107.1| ATP-dependent Clp protease proteolytic subunit (ClpR3), putative
[Arabidopsis thaliana] gi|18390982|ref|NP_563836.1|
ATP-dependent Clp protease proteolytic subunit, putative
[Arabidopsis thaliana] gi|25083963|gb|AAN72143.1|
ATP-dependent Clp protease proteolytic subunit (ClpR3),
putative [Arabidopsis thaliana]
gi|51971739|dbj|BAD44534.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51971625|dbj|BAD44477.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51971381|dbj|BAD44355.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51971379|dbj|BAD44354.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51971032|dbj|BAD44208.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51969858|dbj|BAD43621.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51969856|dbj|BAD43620.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51969676|dbj|BAD43530.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51968816|dbj|BAD43100.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51968776|dbj|BAD43080.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
gi|51968388|dbj|BAD42886.1| ClpP protease complex
subunit ClpR3 [Arabidopsis thaliana]
Length = 330
Score = 74.7 bits (182), Expect = 5e-13
Identities = 50/110 (45%), Positives = 65/110 (58%), Gaps = 18/110 (16%)
Query: 1 MATCFRVPISMPTSMPASSTRFTP-----------TRLRTLR-TVNAISKSKIPF---NP 45
MA+C + SM + +P SS+ F+P L+T R A + +KIP NP
Sbjct: 1 MASCLQA--SMNSLLPRSSS-FSPHPPLSSNSSGRRNLKTFRYAFRAKASAKIPMPPINP 57
Query: 46 NDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQVSNS 95
DPFLS LAS+A +SPE L N + D P+LDIFDSPQLM++PAQV S
Sbjct: 58 KDPFLSTLASIAANSPEKLLNRPVNADVPPYLDIFDSPQLMSSPAQVERS 107
>pir||D86223 hypothetical protein [imported] - Arabidopsis thaliana
gi|2342674|gb|AAB70396.1| Similar to ATP-dependent Clp
protease (gb|D90915). EST gb|N65461 comes from this
gene. [Arabidopsis thaliana]
Length = 292
Score = 74.7 bits (182), Expect = 5e-13
Identities = 50/110 (45%), Positives = 65/110 (58%), Gaps = 18/110 (16%)
Query: 1 MATCFRVPISMPTSMPASSTRFTP-----------TRLRTLR-TVNAISKSKIPF---NP 45
MA+C + SM + +P SS+ F+P L+T R A + +KIP NP
Sbjct: 1 MASCLQA--SMNSLLPRSSS-FSPHPPLSSNSSGRRNLKTFRYAFRAKASAKIPMPPINP 57
Query: 46 NDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQVSNS 95
DPFLS LAS+A +SPE L N + D P+LDIFDSPQLM++PAQV S
Sbjct: 58 KDPFLSTLASIAANSPEKLLNRPVNADVPPYLDIFDSPQLMSSPAQVERS 107
>dbj|BAD44446.1| ClpP protease complex subunit ClpR3 [Arabidopsis thaliana]
Length = 330
Score = 74.7 bits (182), Expect = 5e-13
Identities = 50/110 (45%), Positives = 65/110 (58%), Gaps = 18/110 (16%)
Query: 1 MATCFRVPISMPTSMPASSTRFTP-----------TRLRTLR-TVNAISKSKIPF---NP 45
MA+C + SM + +P SS+ F+P L+T R A + +KIP NP
Sbjct: 1 MASCLQA--SMNSLLPRSSS-FSPHPPLSSNSSGRRNLKTFRYAFRAKASAKIPMPPINP 57
Query: 46 NDPFLSKLASVAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQVSNS 95
DPFLS LAS+A +SPE L N + D P+LDIFDSPQLM++PAQV S
Sbjct: 58 KDPFLSTLASIAANSPEKLLNRPVNADVPPYLDIFDSPQLMSSPAQVERS 107
>dbj|BAD43698.1| ClpP protease complex subunit ClpR3 [Arabidopsis thaliana]
Length = 315
Score = 73.6 bits (179), Expect = 1e-12
Identities = 43/84 (51%), Positives = 52/84 (61%), Gaps = 4/84 (4%)
Query: 16 PASSTRFTPTRLRTLR-TVNAISKSKIPF---NPNDPFLSKLASVAESSPETLFNPSSSP 71
P SS L+T R A + +KIP NP DPFLS LAS+A +SPE L N +
Sbjct: 9 PLSSNSSGRRNLKTFRYAFRAKASAKIPMPPINPKDPFLSTLASIAANSPEKLLNRPVNA 68
Query: 72 DNLPFLDIFDSPQLMATPAQVSNS 95
D P+LDIFDSPQLM++PAQV S
Sbjct: 69 DVPPYLDIFDSPQLMSSPAQVERS 92
>ref|XP_636429.1| hypothetical protein DDB0188216 [Dictyostelium discoideum]
gi|60464807|gb|EAL62927.1| hypothetical protein
DDB0188216 [Dictyostelium discoideum]
Length = 406
Score = 35.4 bits (80), Expect = 0.34
Identities = 30/105 (28%), Positives = 51/105 (48%), Gaps = 9/105 (8%)
Query: 11 MPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKL-------ASVAESSPET 63
+PTS +++T F P + TL N+I SKI FN P + KL + + E+
Sbjct: 108 LPTSKISNTTYFIPPFITTLIFENSIQDSKIAFNFIPPTIKKLILPIYFNQPLRNTINES 167
Query: 64 LFNPSSSPDNLPFLDIFDSPQLMATPAQVSNSNFSFHF-LQIEEN 107
+F +SS +++ F + F+ L P + S F L++ +N
Sbjct: 168 IFPMNSSLESIDFGESFNK-NLYDLPKSLKQIKLSKDFNLKLNKN 211
>ref|XP_646820.1| hypothetical protein DDB0202057 [Dictyostelium discoideum]
gi|60474825|gb|EAL72762.1| hypothetical protein
DDB0202057 [Dictyostelium discoideum]
Length = 769
Score = 34.3 bits (77), Expect = 0.76
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 7/87 (8%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFL--SKLASVAESSPETLFNP 67
S P+ P ST +P + ++ N+ KS PF SK S++ S+ +P
Sbjct: 653 SKPSKSPLKSTSKSPLKSTSISPSNSPFKSPFKSPSKSPFKLPSKSPSISPSN-----SP 707
Query: 68 SSSPDNLPFLDIFDSPQLMATPAQVSN 94
SP N PF F SP + ++ N
Sbjct: 708 FKSPSNSPFKSPFKSPSKSPSIGKIVN 734
>ref|XP_643051.1| hypothetical protein DDB0217837 [Dictyostelium discoideum]
gi|60471168|gb|EAL69135.1| hypothetical protein
DDB0217837 [Dictyostelium discoideum]
Length = 572
Score = 34.3 bits (77), Expect = 0.76
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 7/87 (8%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFL--SKLASVAESSPETLFNP 67
S P+ P ST +P + ++ N+ KS PF SK S++ S+ +P
Sbjct: 456 SKPSKSPLKSTSKSPLKSTSISPSNSPFKSPFKSPSKSPFKLPSKSPSISPSN-----SP 510
Query: 68 SSSPDNLPFLDIFDSPQLMATPAQVSN 94
SP N PF F SP + ++ N
Sbjct: 511 FKSPSNSPFKSPFKSPSKSPSIGKIVN 537
>ref|XP_640418.1| hypothetical protein DDB0218351 [Dictyostelium discoideum]
gi|60468458|gb|EAL66463.1| hypothetical protein
DDB0218351 [Dictyostelium discoideum]
Length = 572
Score = 34.3 bits (77), Expect = 0.76
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 7/87 (8%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFL--SKLASVAESSPETLFNP 67
S P+ P ST +P + ++ N+ KS PF SK S++ S+ +P
Sbjct: 456 SKPSKSPLKSTSKSPLKSTSISPSNSPFKSPFKSPSKSPFKLPSKSPSISPSN-----SP 510
Query: 68 SSSPDNLPFLDIFDSPQLMATPAQVSN 94
SP N PF F SP + ++ N
Sbjct: 511 FKSPSNSPFKSPFKSPSKSPSIGKIVN 537
>ref|XP_637424.1| hypothetical protein DDB0218889 [Dictyostelium discoideum]
gi|60465849|gb|EAL63923.1| hypothetical protein
DDB0218889 [Dictyostelium discoideum]
Length = 1049
Score = 34.3 bits (77), Expect = 0.76
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 7/87 (8%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFL--SKLASVAESSPETLFNP 67
S P+ P ST +P + ++ N+ KS PF SK S++ S+ +P
Sbjct: 443 SKPSKSPLKSTSKSPLKSTSISPSNSPFKSPFKSPSKSPFKLPSKSPSISPSN-----SP 497
Query: 68 SSSPDNLPFLDIFDSPQLMATPAQVSN 94
SP N PF F SP + ++ N
Sbjct: 498 FKSPSNSPFKSPFKSPSKSPSIGKIVN 524
>ref|XP_629717.1| hypothetical protein DDB0215500 [Dictyostelium discoideum]
gi|60463105|gb|EAL61300.1| hypothetical protein
DDB0215500 [Dictyostelium discoideum]
Length = 589
Score = 34.3 bits (77), Expect = 0.76
Identities = 25/87 (28%), Positives = 38/87 (42%), Gaps = 7/87 (8%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFL--SKLASVAESSPETLFNP 67
S P+ P ST +P + ++ N+ KS PF SK S++ S+ +P
Sbjct: 456 SKPSKSPLKSTSKSPLKSTSISPSNSPFKSPFKSPSKSPFKLPSKSPSISPSN-----SP 510
Query: 68 SSSPDNLPFLDIFDSPQLMATPAQVSN 94
SP N PF F SP + ++ N
Sbjct: 511 FKSPSNSPFKSPFKSPSKSPSIGKIVN 537
>ref|XP_638396.1| hypothetical protein DDB0186183 [Dictyostelium discoideum]
gi|60466996|gb|EAL65038.1| hypothetical protein
DDB0186183 [Dictyostelium discoideum]
Length = 469
Score = 33.5 bits (75), Expect = 1.3
Identities = 26/87 (29%), Positives = 42/87 (47%), Gaps = 10/87 (11%)
Query: 12 PTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSP 71
PTS+ SS+ + T T KS IP P L+ ++ + + + + +PSSSP
Sbjct: 210 PTSILISSS--STLNNTTTSTTTTAPKSPIPI----PSLTPISQIKNLTTKPIVSPSSSP 263
Query: 72 DNLPFLDIFDSPQLMATPAQVSNSNFS 98
L P+++ATP S+S+ S
Sbjct: 264 ----LLSSISPPKVLATPTLSSSSSSS 286
>ref|XP_635352.1| hypothetical protein DDB0219987 [Dictyostelium discoideum]
gi|60463694|gb|EAL61876.1| histone H3 domain-containing
protein [Dictyostelium discoideum]
Length = 619
Score = 33.5 bits (75), Expect = 1.3
Identities = 22/85 (25%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 10 SMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSS 69
S P+S +SS+ TPT+ R R + +K P + ++ L++ + P T SS
Sbjct: 393 SSPSSSSSSSSSLTPTKSR--RETIVTTPTKTPPSTRSKSITPLSTPTRTPPSTRSKTSS 450
Query: 70 SPDNLPFLDIFDSPQLMATPAQVSN 94
+P + P +P + + VS+
Sbjct: 451 TPSSTPSSLSISTPSSTSISSPVSS 475
>emb|CAG61186.1| unnamed protein product [Candida glabrata CBS138]
gi|50291599|ref|XP_448232.1| unnamed protein product
[Candida glabrata]
Length = 958
Score = 33.1 bits (74), Expect = 1.7
Identities = 28/103 (27%), Positives = 48/103 (46%), Gaps = 13/103 (12%)
Query: 7 VPISMPTSMPASSTRFTPTRLRTL--RTVNAISKSKIPFN---------PNDPFLSKLAS 55
+P SMP+SMP+S P+ + + ++ + S IP + P+ S +S
Sbjct: 418 IPSSMPSSMPSSMPSSIPSSMPSSMPSSIPSSIPSSIPSSMPSSMPGSMPSSIPSSMPSS 477
Query: 56 VAESSPETLFNPSSSPDNLPFLDIFDSPQLMATPAQVSNSNFS 98
+ S P ++ PSS P ++P P + P+ VSN + S
Sbjct: 478 IPSSMPSSI--PSSIPSSMPSSMPSSMPSSVIKPSTVSNHSSS 518
>gb|EAL03784.1| hypothetical protein CaO19.1481 [Candida albicans SC5314]
gi|46444362|gb|EAL03637.1| hypothetical protein
CaO19.9056 [Candida albicans SC5314]
Length = 838
Score = 33.1 bits (74), Expect = 1.7
Identities = 24/82 (29%), Positives = 33/82 (39%), Gaps = 2/82 (2%)
Query: 8 PISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNP 67
PIS P + + S + TPT T T KS P S L SP+
Sbjct: 39 PISAPATKESVSAKPTPTP--TAPTTGVAPKSSPLITPLTASQSNLTKTTSISPQLSKAK 96
Query: 68 SSSPDNLPFLDIFDSPQLMATP 89
S+ P P I +P+L++ P
Sbjct: 97 SNQPSQPPCKQIKTNPKLLSNP 118
>gb|AAO51532.1| similar to Orgyia pseudotsugata multicapsid polyhedrosis virus
(OpMNPV). Hypothetical 29.3 kDa protein (ORF92)
[Dictyostelium discoideum]
Length = 762
Score = 33.1 bits (74), Expect = 1.7
Identities = 29/101 (28%), Positives = 41/101 (39%), Gaps = 4/101 (3%)
Query: 6 RVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLF 65
+ P S PTS P S TPT+ T S ++ P + P S +S S T
Sbjct: 601 QTPSSTPTSSPTSLPTSTPTQTPTSSPTPTSSPTQTPTSSPTPTSSPTSSPTSSPTPT-- 658
Query: 66 NPSSSPDNLPFLDIFDSPQLMATPAQVSNSNFSFHFLQIEE 106
P+ + + SP +M TP S S S H + +E
Sbjct: 659 -PTQTQTQTLNPNPTTSPSIMPTPTYTS-SEISSHEIPKKE 697
>ref|XP_645531.1| hypothetical protein DDB0216924 [Dictyostelium discoideum]
gi|60473666|gb|EAL71607.1| hypothetical protein
DDB0216924 [Dictyostelium discoideum]
Length = 814
Score = 33.1 bits (74), Expect = 1.7
Identities = 29/101 (28%), Positives = 41/101 (39%), Gaps = 4/101 (3%)
Query: 6 RVPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLF 65
+ P S PTS P S TPT+ T S ++ P + P S +S S T
Sbjct: 653 QTPSSTPTSSPTSLPTSTPTQTPTSSPTPTSSPTQTPTSSPTPTSSPTSSPTSSPTPT-- 710
Query: 66 NPSSSPDNLPFLDIFDSPQLMATPAQVSNSNFSFHFLQIEE 106
P+ + + SP +M TP S S S H + +E
Sbjct: 711 -PTQTQTQTLNPNPTTSPSIMPTPTYTS-SEISSHEIPKKE 749
>gb|EAA70398.1| hypothetical protein FG10082.1 [Gibberella zeae PH-1]
gi|46137133|ref|XP_390258.1| hypothetical protein
FG10082.1 [Gibberella zeae PH-1]
Length = 595
Score = 32.7 bits (73), Expect = 2.2
Identities = 17/47 (36%), Positives = 23/47 (48%)
Query: 26 RLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPD 72
R+R+L + + S P P +PF A+VA SP T SPD
Sbjct: 470 RMRSLLSARPVLGSDFPVEPPNPFQGIYAAVARRSPHTGRGTDESPD 516
>emb|CAG62326.1| unnamed protein product [Candida glabrata CBS138]
gi|50293877|ref|XP_449350.1| unnamed protein product
[Candida glabrata]
Length = 922
Score = 32.7 bits (73), Expect = 2.2
Identities = 28/92 (30%), Positives = 45/92 (48%), Gaps = 10/92 (10%)
Query: 7 VPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFN 66
+P S+P+SMP+S P+ + TV+ S S P+ S +S+ S P ++
Sbjct: 418 MPSSIPSSMPSSIPSSMPSSVIKPSTVSNHSSS----IPSSIPSSMPSSIPSSIPSSM-- 471
Query: 67 PSSSPDNLPFLDIFDSPQLMATPAQVSNSNFS 98
PSS P ++P P + P+ VSN + S
Sbjct: 472 PSSMPSSIP----SSMPSSVIKPSTVSNHSSS 499
Score = 32.7 bits (73), Expect = 2.2
Identities = 24/99 (24%), Positives = 45/99 (45%), Gaps = 7/99 (7%)
Query: 7 VPISMPTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFN 66
+P S+P+S+P+S P+ + + + I S + + + S +S+ S P ++
Sbjct: 459 MPSSIPSSIPSSMPSSMPSSIPSSMPSSVIKPSTVSNHSSSMPSSIPSSMPSSMPSSVIK 518
Query: 67 P-------SSSPDNLPFLDIFDSPQLMATPAQVSNSNFS 98
P SS P ++P P + P+ VSN + S
Sbjct: 519 PSTVSNHSSSMPSSIPSSIPSSMPSSVIKPSTVSNHSSS 557
>gb|EAA58628.1| hypothetical protein AN6244.2 [Aspergillus nidulans FGSC A4]
gi|67540148|ref|XP_663848.1| hypothetical protein
AN6244_2 [Aspergillus nidulans FGSC A4]
gi|49097842|ref|XP_410381.1| hypothetical protein
AN6244.2 [Aspergillus nidulans FGSC A4]
Length = 391
Score = 32.7 bits (73), Expect = 2.2
Identities = 16/59 (27%), Positives = 30/59 (50%)
Query: 17 ASSTRFTPTRLRTLRTVNAISKSKIPFNPNDPFLSKLASVAESSPETLFNPSSSPDNLP 75
A T P +L ++RT++ S+ P PN ++ + S A + + NPS+ + +P
Sbjct: 291 AHDTDSGPLKLLSIRTIDPPSRLTQPGIPNSENIATVGSTAAAEESLIINPSTGEEEVP 349
>gb|AAC28443.1| HeRunt-1 [Heliocidaris erythrogramma]
Length = 536
Score = 32.7 bits (73), Expect = 2.2
Identities = 18/66 (27%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 12 PTSMPASSTRFTPTRLRTLRTVNAISKSKIPFNPND-PFLSKLASVAESSPETLFNPSSS 70
P++ SS FT + + L + ++ +P PN P + S + S+P TL +P
Sbjct: 327 PSTPAVSSVSFTTSSMSVLSSGGESPRTILPMTPNPFPLSQDIFSSSSSTPVTLTSPPYL 386
Query: 71 PDNLPF 76
P++ P+
Sbjct: 387 PNSPPY 392
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.317 0.129 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 177,778,142
Number of Sequences: 2540612
Number of extensions: 6533853
Number of successful extensions: 21975
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 21885
Number of HSP's gapped (non-prelim): 82
length of query: 107
length of database: 863,360,394
effective HSP length: 83
effective length of query: 24
effective length of database: 652,489,598
effective search space: 15659750352
effective search space used: 15659750352
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC144513.18


