

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144513.13 + phase: 0
(544 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
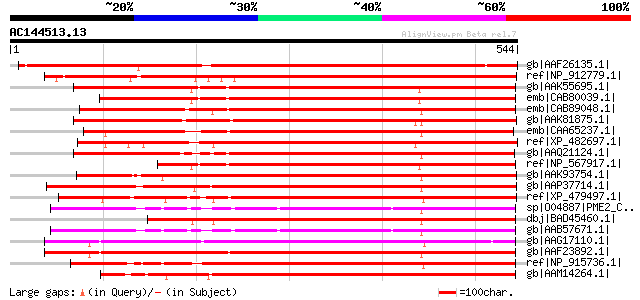
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF26135.1| putative pectinesterase [Arabidopsis thaliana] gi... 679 0.0
ref|NP_912779.1| unnamed protein product [Oryza sativa (japonica... 512 e-143
gb|AAK55695.1| AT4g33220/F4I10_150 [Arabidopsis thaliana] 484 e-135
emb|CAB80039.1| pectinesterase-like protein [Arabidopsis thalian... 482 e-134
emb|CAB89048.1| pectinesterase-like protein [Arabidopsis thalian... 477 e-133
gb|AAK81875.1| pectin methylesterase PME1 [Vitis vinifera] 469 e-130
emb|CAA65237.1| pectinesterase [Prunus persica] gi|6093744|sp|Q4... 465 e-129
ref|XP_482697.1| putative pectinesterase [Oryza sativa (japonica... 454 e-126
gb|AAQ21124.1| pectinesterase [Fragaria x ananassa] 449 e-125
ref|NP_567917.1| pectinesterase family protein [Arabidopsis thal... 432 e-119
gb|AAK93754.1| putative pectinesterase [Arabidopsis thaliana] gi... 431 e-119
gb|AAP37714.1| At3g49220 [Arabidopsis thaliana] gi|6723392|emb|C... 423 e-117
ref|XP_479497.1| putative pectinesterase [Oryza sativa (japonica... 416 e-115
sp|O04887|PME2_CITSI Pectinesterase-2 precursor (Pectin methyles... 416 e-115
dbj|BAD45460.1| putative pectinesterase [Oryza sativa (japonica ... 414 e-114
gb|AAB57671.1| pectinesterase [Citrus sinensis] gi|7447381|pir||... 413 e-114
gb|AAG17110.1| putative pectin methylesterase 3 [Linum usitatiss... 400 e-110
gb|AAF23892.1| pectin methyl esterase [Solanum tuberosum] 400 e-110
ref|NP_915736.1| putative pectin esterase [Oryza sativa (japonic... 399 e-110
gb|AAM14264.1| putative pectinesterase [Arabidopsis thaliana] gi... 399 e-110
>gb|AAF26135.1| putative pectinesterase [Arabidopsis thaliana]
gi|15230020|ref|NP_187213.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 543
Score = 679 bits (1752), Expect = 0.0
Identities = 341/546 (62%), Positives = 418/546 (76%), Gaps = 21/546 (3%)
Query: 10 ILILLPSFDQVLSFPDEIPSDIQTQDMQALIAQACMDIENQNSCLTNIHNELTRTGPP-S 68
+L++L S S+ I + + ++L+A+AC I+ C++NI + +G +
Sbjct: 8 LLVMLMSV-HTSSYETTILKPYKEDNFRSLVAKACQFIDAHELCVSNIWTHVKESGHGLN 66
Query: 69 PTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
P SV+ AA++ ++A A+ + + S+ +REQ+AIEDCKEL+ FSV+ELAWS+ EM
Sbjct: 67 PHSVLRAAVKEAHDKAKLAMERIPTVMMLSIRSREQVAIEDCKELVGFSVTELAWSMLEM 126
Query: 129 RRIRAGDR---------TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQ 179
++ G A GNL+ WLSAA+SNQDTC+EGFEGT+R+ E I GS+ Q
Sbjct: 127 NKLHGGGGIDLDDGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTERKYEELIKGSLRQ 186
Query: 180 VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELL-KSKPHGKI 238
VTQL+SNVL +YTQLN LPF+ RN ++ + PEW+TE D+ L+ + P
Sbjct: 187 VTQLVSNVLDMYTQLNALPFKASRNESV--------IASPEWLTETDESLMMRHDPSVMH 238
Query: 239 ADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGI 298
+ VVA+DG G+YRTINEA+N AP+HS +R+VIYVKKG+YKENID+KKK TNIM+VGDGI
Sbjct: 239 PNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGI 298
Query: 299 GQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAF 358
GQTI+T +RNFMQG TTFRTAT AVSG+GFIAKD+TFRNTAGP N QAVALRVDSDQSAF
Sbjct: 299 GQTIITGDRNFMQGLTTFRTATVAVSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAF 358
Query: 359 FRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
+RCS+EG QDTLYAHSLRQFYR+CEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT
Sbjct: 359 YRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTIT 418
Query: 419 AQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
AQGRKSP+Q+TGF IQ+SYVLA+QPTYLGRPWK YSRTVY+NTYMS +VQPRGWLEW GN
Sbjct: 419 AQGRKSPNQNTGFVIQNSYVLATQPTYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGN 478
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
FALDTLWYGEY N GPG +GRVKWPGYH++ D A FTV F++G WLP TGV F
Sbjct: 479 FALDTLWYGEYNNIGPGWRSSGRVKWPGYHIM-DKRTALSFTVGSFIDGRRWLPATGVTF 537
Query: 539 TAGLSN 544
TAGL+N
Sbjct: 538 TAGLAN 543
>ref|NP_912779.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|5922617|dbj|BAA84618.1| putative pectinesterase
[Oryza sativa (japonica cultivar-group)]
gi|6016850|dbj|BAA85193.1| unnamed protein product
[Oryza sativa (japonica cultivar-group)]
Length = 611
Score = 512 bits (1318), Expect = e-143
Identities = 270/570 (47%), Positives = 375/570 (65%), Gaps = 69/570 (12%)
Query: 38 ALIAQACMDIE----------NQNSCLTNIHNELTRTGPPS-PTSVINAALRTTINEAIG 86
AL+ QAC ++ + C++ + + R G S P V+ AA+R T+ EA+G
Sbjct: 45 ALLQQACFNVTAFGGGGGGGGGEGGCVSRL--DTARGGAGSGPVPVLRAAVRDTLGEAVG 102
Query: 87 AINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM------------------ 128
A+ + +++ S + RE++A+ DC EL+ +SV EL W+L M
Sbjct: 103 AVAAVAGLASLSNHAREEMAVRDCVELVGYSVDELGWALDAMADPDGGVAAAEEEDETEP 162
Query: 129 ---RRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLIS 185
RR R G R E ++ AWLSAA+ NQ TC++GF GTD RL + +VTQ+TQL+S
Sbjct: 163 ETRRRRRRGARA---EDDIHAWLSAAMGNQGTCLDGFHGTDSRLLRRVESAVTQLTQLVS 219
Query: 186 NVLSLYTQLNRLP--------FRPPRNTTLHETS---------TDESLEFPEWMTEA--- 225
N+L+++ +L + P N + T+ T S + P W+T+
Sbjct: 220 NLLAMHKKLRDITPQHQHQHHHHPGNNNNKNGTADGAAAGGDDTGPSSDLPPWVTDVVND 279
Query: 226 -DQELLKSKPHGKIA---------DAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKK 275
++E+ ++ G+ + D VVA DGSG++RT++EAV APSHS RR+VIYVK+
Sbjct: 280 VEEEVTATRGRGRSSSSGRKAMRVDVVVAQDGSGRWRTVSEAVARAPSHSRRRYVIYVKR 339
Query: 276 GLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTF 335
G+Y+EN++++KK TNI++VG+G+G+T++T +R+ GWTTFR+ATFAVSG GFIA+DMT
Sbjct: 340 GVYEENVEVRKKKTNIVIVGEGMGETVITGSRSMAAGWTTFRSATFAVSGAGFIARDMTI 399
Query: 336 RNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNG 395
RNTAGP HQAVALRVDSD+SAFFR ++EG+QDTLYAHSLRQFYR+C + GT+DFIFGNG
Sbjct: 400 RNTAGPAAHQAVALRVDSDRSAFFRIAVEGHQDTLYAHSLRQFYRDCRVSGTVDFIFGNG 459
Query: 396 AAVLQNCKIYTRVPLPLQKV-TITAQGRKSPHQSTGFTIQDSYVLASQPTYLGRPWKEYS 454
AV+Q I T P Q ++TAQGR+ P+Q+TGF + V A PTYLGRPWK +S
Sbjct: 460 IAVIQRTTISTLPPAAGQNAGSVTAQGRRDPNQNTGFALHACIVEAKYPTYLGRPWKPFS 519
Query: 455 RTVYINTYMSSMVQPRGWLEWLGNFA-LDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDA 513
R V + +Y+ + VQPRGWLEW G+ L TL+YGEYRNYGPG+++ GRV+WPGYHVI DA
Sbjct: 520 RVVVMESYLGAGVQPRGWLEWDGDGGELATLFYGEYRNYGPGANIGGRVRWPGYHVIMDA 579
Query: 514 SAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
+ A FTV+RF++G +WLP TGV FTA L+
Sbjct: 580 AVAVRFTVRRFIDGLAWLPSTGVTFTADLN 609
>gb|AAK55695.1| AT4g33220/F4I10_150 [Arabidopsis thaliana]
Length = 525
Score = 484 bits (1246), Expect = e-135
Identities = 246/496 (49%), Positives = 331/496 (66%), Gaps = 25/496 (5%)
Query: 69 PTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
P S +++ T + + +++ + FS + R Q A+ DC +LLDFS EL WS
Sbjct: 31 PASEFVSSINTIVVVIRQVSSILSQFADFSGDRRLQNAVSDCLDLLDFSSEELTWSASAS 90
Query: 129 RRIRA-GDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNV 187
+ G+ T + WLSAALSNQ TC+EGF+GT ++S ++GS+ Q+ ++ +
Sbjct: 91 ENPKGKGNGTGDVGSDTRTWLSAALSNQATCMEGFDGTSGLVKSLVAGSLDQLYSMLREL 150
Query: 188 LSLYTQ------------LNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPH 235
L L + + P PP L +T DESL+FP+W+ D++LL+S +
Sbjct: 151 LPLVQPEQKPKAVSKPGPIAKGPKAPP-GRKLRDTDEDESLQFPDWVRPDDRKLLES--N 207
Query: 236 GKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVG 295
G+ D VALDG+G + I +A+ AP +S+ R VIY+KKGLY EN+++KKK NI+M+G
Sbjct: 208 GRTYDVSVALDGTGNFTKIMDAIKKAPDYSSTRFVIYIKKGLYLENVEIKKKKWNIVMLG 267
Query: 296 DGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQ 355
DGI T+++ NR+F+ GWTTFR+ATFAVSG+GF+A+D+TF+NTAGP HQAVALR DSD
Sbjct: 268 DGIDVTVISGNRSFIDGWTTFRSATFAVSGRGFLARDITFQNTAGPEKHQAVALRSDSDL 327
Query: 356 SAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKV 415
S FFRC++ G QDTLY H++RQFYREC I GT+DFIFG+G V QNC+I + LP QK
Sbjct: 328 SVFFRCAMRGYQDTLYTHTMRQFYRECTITGTVDFIFGDGTVVFQNCQILAKRGLPNQKN 387
Query: 416 TITAQGRKSPHQSTGFTIQDSYV---------LASQPTYLGRPWKEYSRTVYINTYMSSM 466
TITAQGRK +Q +GF+IQ S + L + TYLGRPWK YSRTV+I MS +
Sbjct: 388 TITAQGRKDVNQPSGFSIQFSNISADADLVPYLNTTRTYLGRPWKLYSRTVFIRNNMSDV 447
Query: 467 VQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLN 526
V+P GWLEW +FALDTL+YGE+ NYGPGS L+ RVKWPGYHV ++ A FTV +F+
Sbjct: 448 VRPEGWLEWNADFALDTLFYGEFMNYGPGSGLSSRVKWPGYHVFNNSDQANNFTVSQFIK 507
Query: 527 GGSWLPRTGVKFTAGL 542
G WLP TGV F+ GL
Sbjct: 508 GNLWLPSTGVTFSDGL 523
>emb|CAB80039.1| pectinesterase-like protein [Arabidopsis thaliana]
gi|4455336|emb|CAB36796.1| pectinesterase-like protein
[Arabidopsis thaliana] gi|7447375|pir||T05202
pectinesterase homolog F4I10.150 - Arabidopsis thaliana
Length = 477
Score = 482 bits (1240), Expect = e-134
Identities = 243/468 (51%), Positives = 319/468 (67%), Gaps = 25/468 (5%)
Query: 97 FSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRA-GDRTAQYEGNLEAWLSAALSNQ 155
FS + R Q A+ DC +LLDFS EL WS + G+ T + WLSAALSNQ
Sbjct: 11 FSGDRRLQNAVSDCLDLLDFSSEELTWSASASENPKGKGNGTGDVGSDTRTWLSAALSNQ 70
Query: 156 DTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQ------------LNRLPFRPPR 203
TC+EGF+GT ++S ++GS+ Q+ ++ +L L + + P PP
Sbjct: 71 ATCMEGFDGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPP- 129
Query: 204 NTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPS 263
L +T DESL+FP+W+ D++LL+S +G+ D VALDG+G + I +A+ AP
Sbjct: 130 GRKLRDTDEDESLQFPDWVRPDDRKLLES--NGRTYDVSVALDGTGNFTKIMDAIKKAPD 187
Query: 264 HSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAV 323
+S+ R VIY+KKGLY EN+++KKK NI+M+GDGI T+++ NR+F+ GWTTFR+ATFAV
Sbjct: 188 YSSTRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAV 247
Query: 324 SGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECE 383
SG+GF+A+D+TF+NTAGP HQAVALR DSD S FFRC++ G QDTLY H++RQFYREC
Sbjct: 248 SGRGFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECT 307
Query: 384 IYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYV----- 438
I GT+DFIFG+G V QNC+I + LP QK TITAQGRK +Q +GF+IQ S +
Sbjct: 308 ITGTVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADAD 367
Query: 439 ----LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGP 494
L + TYLGRPWK YSRTV+I MS +V+P GWLEW +FALDTL+YGE+ NYGP
Sbjct: 368 LVPYLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFALDTLFYGEFMNYGP 427
Query: 495 GSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
GS L+ RVKWPGYHV ++ A FTV +F+ G WLP TGV F+ GL
Sbjct: 428 GSGLSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 475
>emb|CAB89048.1| pectinesterase-like protein [Arabidopsis thaliana]
gi|20465719|gb|AAM20328.1| putative pectinesterase
[Arabidopsis thaliana] gi|17979141|gb|AAL49828.1|
putative pectinesterase [Arabidopsis thaliana]
gi|30690925|ref|NP_189913.3| pectinesterase family
protein [Arabidopsis thaliana] gi|11279193|pir||T49241
pectinesterase-like protein - Arabidopsis thaliana
Length = 527
Score = 477 bits (1227), Expect = e-133
Identities = 239/480 (49%), Positives = 328/480 (67%), Gaps = 17/480 (3%)
Query: 76 ALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGD 135
A +T ++ A+ ++K + +R AI DC +LLD + EL+W + + D
Sbjct: 50 AAKTVVDAITKAVAIVSKFDKKAGKSRVSNAIVDCVDLLDSAAEELSWIISASQSPNGKD 109
Query: 136 R-TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQL 194
T +L W+SAALSNQDTC++GFEGT+ ++ ++G +++V + N+L T +
Sbjct: 110 NSTGDVGSDLRTWISAALSNQDTCLDGFEGTNGIIKKIVAGGLSKVGTTVRNLL---TMV 166
Query: 195 NRLPFRPPRNTTLHETSTDESL---EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQY 251
+ P +P +T T +FP W+ D++LL++ + +ADAVVA DG+G +
Sbjct: 167 HSPPSKPKPKPIKAQTMTKAHSGFSKFPSWVKPGDRKLLQTD-NITVADAVVAADGTGNF 225
Query: 252 RTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQ 311
TI++AV AAP +S +R+VI+VK+G+Y EN+++KKK NIMMVGDGI T++T NR+F+
Sbjct: 226 TTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATVITGNRSFID 285
Query: 312 GWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLY 371
GWTTFR+ATFAVSG+GFIA+D+TF+NTAGP HQAVA+R D+D F+RC++ G QDTLY
Sbjct: 286 GWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLY 345
Query: 372 AHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGF 431
AHS+RQF+REC I GT+DFIFG+ AV Q+C+I + LP QK +ITAQGRK P++ TGF
Sbjct: 346 AHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEPTGF 405
Query: 432 TIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALD 482
TIQ S + A + TYLGRPWK YSRTV++ YMS + P GWLEW GNFALD
Sbjct: 406 TIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWNGNFALD 465
Query: 483 TLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
TL+YGEY N GPG+SL RVKWPGYHV+ ++ A FTV + + G WLP TG+ F AGL
Sbjct: 466 TLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFIAGL 525
>gb|AAK81875.1| pectin methylesterase PME1 [Vitis vinifera]
Length = 531
Score = 469 bits (1206), Expect = e-130
Identities = 234/485 (48%), Positives = 330/485 (67%), Gaps = 15/485 (3%)
Query: 69 PTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
P+S ++L++TI+ G ++ +++ + + R AI DC ELLDF+ +L+WSL +
Sbjct: 51 PSSAFISSLKSTIDVLRGTMSVVSQFTKVFNDFRLSNAISDCLELLDFAADDLSWSLSAI 110
Query: 129 RRIRAGDR-TAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNV 187
+ + D T +L+ WLS+ +NQDTCIEGF GT+ +++ ++ S++QV L+
Sbjct: 111 QNPKGKDNGTGDLGSDLKTWLSSTFTNQDTCIEGFVGTNGIVKTVVAESLSQVASLVH-- 168
Query: 188 LSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDG 247
SL T ++ + N S +FP W+ + ++LL++ D VA DG
Sbjct: 169 -SLLTMVHDPAPKGKSNGGGGGVKHVGSGDFPSWVGKHSRKLLQASSVSP--DVTVAADG 225
Query: 248 SGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNR 307
+G Y T+ +AV AAP +S +VIY+K+G+Y+EN+++KKK N+MMVGDG+G T++T NR
Sbjct: 226 TGNYTTVMDAVQAAPDYSQNHYVIYIKQGIYRENVEIKKKKWNLMMVGDGMGATVITGNR 285
Query: 308 NFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQ 367
+++ GWTT+ +ATFAV GKGFIA+DMTF NTAGP HQAVALR DSD S ++RCS+ G Q
Sbjct: 286 SYIDGWTTYASATFAVKGKGFIARDMTFENTAGPEKHQAVALRSDSDLSVYYRCSMRGYQ 345
Query: 368 DTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQ 427
DTLY H+ RQFYREC I GT+DFIFG+ V QNC+I + LP QK TITAQGRK P Q
Sbjct: 346 DTLYPHTNRQFYRECRISGTVDFIFGDATVVFQNCQILVKKGLPNQKNTITAQGRKDPAQ 405
Query: 428 STGFTIQ------DSYVLA---SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
TGF+IQ DS +LA S +YLGRPWK+YSRT+ + +Y+S ++P GWLEW G+
Sbjct: 406 PTGFSIQFSNISADSDLLASVNSTLSYLGRPWKQYSRTIIMKSYISDAIRPEGWLEWNGD 465
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
FALDTL+YGEY NYGP + L RV+WPG+H++ +++ A FTV F+ G WLP TGVK+
Sbjct: 466 FALDTLYYGEYMNYGPSAGLGSRVQWPGFHLLNNSAQAANFTVTEFIAGNLWLPSTGVKY 525
Query: 539 TAGLS 543
+AGL+
Sbjct: 526 SAGLA 530
>emb|CAA65237.1| pectinesterase [Prunus persica] gi|6093744|sp|Q43062|PME_PRUPE
Pectinesterase PPE8B precursor (Pectin methylesterase)
(PE)
Length = 522
Score = 465 bits (1196), Expect = e-129
Identities = 233/478 (48%), Positives = 317/478 (65%), Gaps = 35/478 (7%)
Query: 80 TINEAIGAINNMTKISTFSVNN----REQLAIEDCKELLDFSVSELAWSLGEMRRIRA-G 134
++ + I A+ + I + N R AI DC +LLDFS EL WSL + +
Sbjct: 59 SLKDTIDAVQQVASILSQFANAFGDFRLANAISDCLDLLDFSADELNWSLSASQNQKGKN 118
Query: 135 DRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQL 194
+ T + +L WLSAAL NQDTC GFEGT+ ++ IS + QVT L+ +L
Sbjct: 119 NSTGKLSSDLRTWLSAALVNQDTCSNGFEGTNSIVQGLISAGLGQVTSLVQELL------ 172
Query: 195 NRLPFRPPRNTTLHETSTDESL--EFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYR 252
T +H S + + P W+ D++LL++ G DA+VA DG+G +
Sbjct: 173 ----------TQVHPNSNQQGPNGQIPSWVKTKDRKLLQAD--GVSVDAIVAQDGTGNFT 220
Query: 253 TINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQG 312
+ +AV AAP +S RR+VIY+K+G YKEN+++KKK N+MM+GDG+ TI++ NR+F+ G
Sbjct: 221 NVTDAVLAAPDYSMRRYVIYIKRGTYKENVEIKKKKWNLMMIGDGMDATIISGNRSFVDG 280
Query: 313 WTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYA 372
WTTFR+ATFAVSG+GFIA+D+TF NTAGP HQAVALR DSD S F+RC+I G QDTLY
Sbjct: 281 WTTFRSATFAVSGRGFIARDITFENTAGPEKHQAVALRSDSDLSVFYRCNIRGYQDTLYT 340
Query: 373 HSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFT 432
H++RQFYR+C+I GT+DFIFG+ V QNC+I + LP QK +ITAQGRK P++ TG +
Sbjct: 341 HTMRQFYRDCKISGTVDFIFGDATVVFQNCQILAKKGLPNQKNSITAQGRKDPNEPTGIS 400
Query: 433 IQDSYVLA----------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALD 482
IQ + A S PTYLGRPWK YSRTV + +++S++++P GWLEW G+FAL+
Sbjct: 401 IQFCNITADSDLEAASVNSTPTYLGRPWKLYSRTVIMQSFLSNVIRPEGWLEWNGDFALN 460
Query: 483 TLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTA 540
+L+YGEY NYGPG+ L RVKWPGY V +++ A +TV +F+ G WLP TGVK+TA
Sbjct: 461 SLFYGEYMNYGPGAGLGSRVKWPGYQVFNESTQAKNYTVAQFIEGNLWLPSTGVKYTA 518
>ref|XP_482697.1| putative pectinesterase [Oryza sativa (japonica cultivar-group)]
gi|42407616|dbj|BAD08731.1| putative pectinesterase
[Oryza sativa (japonica cultivar-group)]
Length = 557
Score = 454 bits (1168), Expect = e-126
Identities = 241/516 (46%), Positives = 324/516 (62%), Gaps = 53/516 (10%)
Query: 73 INAALRTTINEAIGAINNMTKI-STFSVNN-------REQLAIEDCKELLDFSVSELAWS 124
I+ L +T+ E + AI N+ I S+F + R AI DC +LLD S EL+WS
Sbjct: 51 ISPVLVSTLRETLDAIKNVASIISSFPIGGILGGGDLRLSSAIADCLDLLDLSSDELSWS 110
Query: 125 LG-----EMRRIRAGDRTAQYEG------NLEAWLSAALSNQDTCIEGFEGTDRRLESYI 173
+ + AG T+ + G +L +WL ALSNQDTC EG + T L S +
Sbjct: 111 MSTTSSSSYQPTNAGAATSSHVGTGDARSDLRSWLGGALSNQDTCKEGLDDTGSVLGSLV 170
Query: 174 SGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLE----FPEWMTEADQEL 229
++ VT L+++ L ++ +S+ L P W+ ++ L
Sbjct: 171 GTALQTVTSLLTDGLGQVAA---------GEASIAWSSSRRGLAEGGGAPHWLGARERRL 221
Query: 230 LKSK--PHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKK 287
L+ P G DAVVA DGSG Y T++ AV+AAP+ S R+VIYVKKG+YKE +D+KKK
Sbjct: 222 LQMPLGPGGMPVDAVVAKDGSGNYTTVSAAVDAAPTESASRYVIYVKKGVYKETVDIKKK 281
Query: 288 MTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAV 347
N+M+VGDG+G T+++ +RN++ G+TTFR+AT AV+GKGF+A+D+TF NTAGP HQAV
Sbjct: 282 KWNLMLVGDGMGVTVISGHRNYVDGYTTFRSATVAVNGKGFMARDVTFENTAGPSKHQAV 341
Query: 348 ALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTR 407
ALR DSD S F+RC EG QDTLYAHSLRQFYR+C + GT+DF+FGN AAV QNC + R
Sbjct: 342 ALRCDSDLSVFYRCGFEGYQDTLYAHSLRQFYRDCRVSGTVDFVFGNAAAVFQNCTLAAR 401
Query: 408 VPLPLQKVTITAQGRKSPHQSTGFTIQ-------------------DSYVLASQPTYLGR 448
+PLP QK ++TAQGR + +TGF Q S A TYLGR
Sbjct: 402 LPLPDQKNSVTAQGRLDGNMTTGFAFQFCNVTADDDLQRALAGGGNQSSAAAVTQTYLGR 461
Query: 449 PWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYH 508
PWK+YSR V++ +Y+ ++V+P GWL W G FALDTL+YGEY N GPG+ + GRVKWPG+H
Sbjct: 462 PWKQYSRVVFMQSYIGAVVRPEGWLAWDGQFALDTLYYGEYMNTGPGAGVGGRVKWPGFH 521
Query: 509 VIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLSN 544
V+ + AG FTV +F+ G WLP TGVK+TAGL++
Sbjct: 522 VMTSPAQAGNFTVAQFIEGNMWLPPTGVKYTAGLTS 557
>gb|AAQ21124.1| pectinesterase [Fragaria x ananassa]
Length = 514
Score = 449 bits (1156), Expect = e-125
Identities = 229/485 (47%), Positives = 313/485 (64%), Gaps = 27/485 (5%)
Query: 69 PTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEM 128
P S ++ TIN ++ ++ ++ + R A++DC EL+D S +L+W+L
Sbjct: 42 PNSEFGGSIINTINVLQQVVSILSDVAKGFGDFRLSNAVDDCLELMDDSTDQLSWTLSAT 101
Query: 129 RRIRAG-DRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLISNV 187
+ + T +L WLSA L NQDTC EG +GT+ ++S +SGS+ Q+T L V
Sbjct: 102 QNKNGKHNSTGNLSSDLRTWLSATLVNQDTCNEGLDGTNSIVKSLVSGSLNQITSL---V 158
Query: 188 LSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDG 247
L L Q++ + HE+S ++ P W D++LL++ +G D VVA DG
Sbjct: 159 LELLGQVHP-------TSDQHESSNGQT---PAWFKAEDRKLLQA--NGVPVDVVVAQDG 206
Query: 248 SGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNR 307
+G + I A+ +AP +S +R+VIYVKKGLYKE +++KKK NIMM+GDG+ T+++ N
Sbjct: 207 TGNFTNITAAILSAPDYSLKRYVIYVKKGLYKEYVEIKKKKWNIMMIGDGMDATVISGNH 266
Query: 308 NFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQ 367
NF+ GWTTFR+ATFAVSG+GFIA+D+TF NTAGP H AVALR DSD SAF+RC G Q
Sbjct: 267 NFVDGWTTFRSATFAVSGRGFIARDITFENTAGPEKHMAVALRSDSDLSAFYRCEFRGYQ 326
Query: 368 DTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQ 427
DTLY HS+RQFYR+C+I GT+DFIFG+G + QNC+I R LP QK +ITA GRK +
Sbjct: 327 DTLYTHSMRQFYRDCKISGTVDFIFGDGTVMFQNCQILARKALPNQKNSITAHGRKYKDE 386
Query: 428 STGFTIQDSYVLA-----------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWL 476
TGF+ Q + A S PTYLGRPWKEYSRT+ + ++MS+M++P GWLEW
Sbjct: 387 PTGFSFQFCNISAHPDLLATPVNSSTPTYLGRPWKEYSRTIIMQSFMSNMIKPAGWLEWN 446
Query: 477 GNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGV 536
G+ L TL+YGE+ NYGPG+ L RV WPGY + A +TV F+ G WLP TGV
Sbjct: 447 GDMFLKTLFYGEHMNYGPGAGLGSRVTWPGYQKFNQSGQAKNYTVAEFIEGNLWLPSTGV 506
Query: 537 KFTAG 541
K+T+G
Sbjct: 507 KYTSG 511
>ref|NP_567917.1| pectinesterase family protein [Arabidopsis thaliana]
Length = 404
Score = 432 bits (1111), Expect = e-119
Identities = 214/405 (52%), Positives = 284/405 (69%), Gaps = 24/405 (5%)
Query: 159 IEGFEGTDRRLESYISGSVTQVTQLISNVLSLYTQ------------LNRLPFRPPRNTT 206
+EGF+GT ++S ++GS+ Q+ ++ +L L + + P PP
Sbjct: 1 MEGFDGTSGLVKSLVAGSLDQLYSMLRELLPLVQPEQKPKAVSKPGPIAKGPKAPP-GRK 59
Query: 207 LHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSN 266
L +T DESL+FP+W+ D++LL+S +G+ D VALDG+G + I +A+ AP +S+
Sbjct: 60 LRDTDEDESLQFPDWVRPDDRKLLES--NGRTYDVSVALDGTGNFTKIMDAIKKAPDYSS 117
Query: 267 RRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGK 326
R VIY+KKGLY EN+++KKK NI+M+GDGI T+++ NR+F+ GWTTFR+ATFAVSG+
Sbjct: 118 TRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNRSFIDGWTTFRSATFAVSGR 177
Query: 327 GFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYG 386
GF+A+D+TF+NTAGP HQAVALR DSD S FFRC++ G QDTLY H++RQFYREC I G
Sbjct: 178 GFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYTHTMRQFYRECTITG 237
Query: 387 TIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYV-------- 438
T+DFIFG+G V QNC+I + LP QK TITAQGRK +Q +GF+IQ S +
Sbjct: 238 TVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQPSGFSIQFSNISADADLVP 297
Query: 439 -LASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSS 497
L + TYLGRPWK YSRTV+I MS +V+P GWLEW +FALDTL+YGE+ NYGPGS
Sbjct: 298 YLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNADFALDTLFYGEFMNYGPGSG 357
Query: 498 LAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
L+ RVKWPGYHV ++ A FTV +F+ G WLP TGV F+ GL
Sbjct: 358 LSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTGVTFSDGL 402
>gb|AAK93754.1| putative pectinesterase [Arabidopsis thaliana]
gi|13507549|gb|AAK28637.1| putative pectinesterase
[Arabidopsis thaliana] gi|9759184|dbj|BAB09799.1|
pectinesterase [Arabidopsis thaliana]
gi|15238729|ref|NP_200149.1| pectinesterase family
protein [Arabidopsis thaliana]
Length = 587
Score = 431 bits (1108), Expect = e-119
Identities = 224/485 (46%), Positives = 305/485 (62%), Gaps = 17/485 (3%)
Query: 72 VINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRI 131
+I+ + T+ + A+ + I+ + R + A + C ELLD SV L +L + +
Sbjct: 106 LIHISFNATLQKFSKALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALTRALSSVVVV 165
Query: 132 RAGDRTAQYEGNLEAWLSAALSNQDTCIEGF---EGTDRRLESYISGSVTQVTQLISNVL 188
+GD + ++ WLS+A++N DTC +GF EG ++ + G+V +++++SN L
Sbjct: 166 -SGDES---HSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDLSEMVSNCL 221
Query: 189 SLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGS 248
+++ + P T+E+ E P W+ D+ELL + AD V+ DGS
Sbjct: 222 AIFAGKVKDLSGVPVVNNRKLLGTEETEELPNWLKREDRELLGTPTSAIQADITVSKDGS 281
Query: 249 GQYRTINEAVNAAPSHSNRRHVIYVKKGLYKE-NIDMKKKMTNIMMVGDGIGQTIVTSNR 307
G ++TI EA+ AP HS+RR VIYVK G Y+E N+ + +K TN+M +GDG G+T++T +
Sbjct: 282 GTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGRKKTNLMFIGDGKGKTVITGGK 341
Query: 308 NFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQ 367
+ TTF TATFA +G GFI +DMTF N AGP HQAVALRV D + +RC+I G Q
Sbjct: 342 SIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQAVALRVGGDHAVVYRCNIIGYQ 401
Query: 368 DTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQ 427
D LY HS RQF+RECEIYGT+DFIFGN A +LQ+C IY R P+ QK+TITAQ RK P+Q
Sbjct: 402 DALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYARKPMAQQKITITAQNRKDPNQ 461
Query: 428 STGFTIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGN 478
+TG +I +LA S PTYLGRPWK YSR VY+ + M + PRGWLEW G
Sbjct: 462 NTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVVYMMSDMGDHIDPRGWLEWNGP 521
Query: 479 FALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKF 538
FALD+L+YGEY N G GS + RVKWPGYHVI A FTV +F++G SWLP TGV F
Sbjct: 522 FALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEASKFTVAQFISGSSWLPSTGVSF 581
Query: 539 TAGLS 543
+GLS
Sbjct: 582 FSGLS 586
>gb|AAP37714.1| At3g49220 [Arabidopsis thaliana] gi|6723392|emb|CAB66401.1|
pectinesterase-like protein [Arabidopsis thaliana]
gi|16648929|gb|AAL24316.1| pectinesterase-like protein
[Arabidopsis thaliana] gi|15229105|ref|NP_190491.1|
pectinesterase family protein [Arabidopsis thaliana]
gi|11279196|pir||T45827 pectinesterase-like protein -
Arabidopsis thaliana
Length = 598
Score = 423 bits (1088), Expect = e-117
Identities = 228/518 (44%), Positives = 317/518 (61%), Gaps = 21/518 (4%)
Query: 40 IAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSV 99
I++AC C+ ++ + S +I+ + T++ A+ + +S +
Sbjct: 87 ISKACELTRFPELCVDSLMDFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFVDM 146
Query: 100 NNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCI 159
R + A + C ELLD SV L+ +L + A + ++ WLSAAL+N DTC
Sbjct: 147 PPRARSAYDSCVELLDDSVDALSRALSSVVSSSAKPQ------DVTTWLSAALTNHDTCT 200
Query: 160 EGFEGTDRR-LESYISGSVTQVTQLISNVLSLYTQLNR---LPFRPPRNTTLHETSTDES 215
EGF+G D ++ +++ ++ +++L+SN L++++ + P +N L E
Sbjct: 201 EGFDGVDDGGVKDHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREE 260
Query: 216 LEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKK 275
+FP WM ++E+L+ AD +V+ DG+G +TI+EA+ AP +S RR +IYVK
Sbjct: 261 -KFPRWMRPKEREILEMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKA 319
Query: 276 GLYKEN-IDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMT 334
G Y+EN + + +K N+M VGDG G+T+++ ++ TTF TA+FA +G GFIA+D+T
Sbjct: 320 GRYEENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDIT 379
Query: 335 FRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGN 394
F N AGP HQAVALR+ +D + +RC+I G QDTLY HS RQF+REC+IYGT+DFIFGN
Sbjct: 380 FENWAGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGN 439
Query: 395 GAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTY 445
A VLQNC IY R P+ QK TITAQ RK P+Q+TG +I S VLA S TY
Sbjct: 440 AAVVLQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTY 499
Query: 446 LGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWP 505
LGRPWK +SRTVY+ +Y+ V RGWLEW FALDTL+YGEY N GPGS L RV WP
Sbjct: 500 LGRPWKLFSRTVYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWP 559
Query: 506 GYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
GY VI + A FTV F+ G SWLP TGV F AGLS
Sbjct: 560 GYRVINSTAEANRFTVAEFIYGSSWLPSTGVSFLAGLS 597
>ref|XP_479497.1| putative pectinesterase [Oryza sativa (japonica cultivar-group)]
gi|50509816|dbj|BAD31979.1| putative pectinesterase
[Oryza sativa (japonica cultivar-group)]
gi|34393863|dbj|BAC83543.1| putative pectinesterase
[Oryza sativa (japonica cultivar-group)]
Length = 579
Score = 416 bits (1070), Expect = e-115
Identities = 226/523 (43%), Positives = 322/523 (61%), Gaps = 47/523 (8%)
Query: 53 CLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFS----------VNNR 102
C +H ++ + + ++ LR I+E +G + + S+ + + R
Sbjct: 72 CAGTLHRDVCVSTLSTIPNLARKPLRDVISEVVGRAASAVRASSSNCTSYLQRPRQLRTR 131
Query: 103 EQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEG-NLEAWLSAALSNQDTCIEG 161
++LA+ DC EL ++ L + E+ AG+ TA+ ++ LSAA++NQ TC++G
Sbjct: 132 DRLALSDCLELFGHTLDLLGTAAAELS---AGNSTAEESAAGVQTVLSAAMTNQYTCLDG 188
Query: 162 FEGT----DRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLE 217
F G D R+ +I G + V L+SN L++ + RLP + R + +E LE
Sbjct: 189 FAGPSASEDGRVRPFIQGRIYHVAHLVSNSLAM---VRRLPTQRRRG------AEEEPLE 239
Query: 218 --------FPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRH 269
FP W++ +D+ L+ + AD VVA DGSG++ T++EAV AAP++S R+
Sbjct: 240 GYGRVRRGFPSWVSASDRRRLQQQV---AADVVVAKDGSGKFTTVSEAVAAAPNNSETRY 296
Query: 270 VIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFI 329
VIY+K G Y EN+++ + TNIM VGDG +T++ ++RN + TTFR+AT AV G GF+
Sbjct: 297 VIYIKAGGYFENVEVGSEKTNIMFVGDGTWKTVIKASRNVVDNSTTFRSATLAVVGTGFL 356
Query: 330 AKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTID 389
A+D+T N AGP HQAVALRV++D SAF+RCS G QDTLYAHSLRQFYR+C+IYGT+D
Sbjct: 357 ARDITVENAAGPSKHQAVALRVNADLSAFYRCSFAGYQDTLYAHSLRQFYRDCDIYGTVD 416
Query: 390 FIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQ------- 442
FIFG+ A VLQNC +Y R P P QK TAQGR+ P+Q+TG IQ V A+
Sbjct: 417 FIFGDAAVVLQNCNLYARRPDPNQKNVFTAQGREDPNQNTGIAIQGCKVAAAADLVPVQA 476
Query: 443 --PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAG 500
+YLGRPWK YSRTV++ + + S++ PRGWLEW G+FALDTL+Y EY N G G+ +
Sbjct: 477 NFSSYLGRPWKTYSRTVFLQSKIDSLIHPRGWLEWNGSFALDTLYYAEYMNRGDGADTSA 536
Query: 501 RVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGLS 543
RV WPGYHV+ +A+ A FTV F+ G WL + + GLS
Sbjct: 537 RVSWPGYHVLTNATDAANFTVLNFVQGDLWLNSSSFPYILGLS 579
>sp|O04887|PME2_CITSI Pectinesterase-2 precursor (Pectin methylesterase) (PE)
gi|2098709|gb|AAB57669.1| pectinesterase [Citrus
sinensis]
Length = 510
Score = 416 bits (1070), Expect = e-115
Identities = 236/509 (46%), Positives = 302/509 (58%), Gaps = 40/509 (7%)
Query: 44 CMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSVNNRE 103
C N C + + T T +L+ + A A + + + N RE
Sbjct: 32 CGKTPNPQPCEYFLTQKTDVTSIKQDTDFYKISLQLALERATTAQSRTYTLGSKCRNERE 91
Query: 104 QLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFE 163
+ A EDC+EL + +V +L + D+ + WLS AL+N +TC E
Sbjct: 92 KAAWEDCRELYELTVLKLNQTSNSSPGCTKVDK--------QTWLSTALTNLETCRASLE 143
Query: 164 GTDRRLESYISGSVTQ-VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWM 222
D + Y+ ++ VT+LISN LSL N++P+ P FP W+
Sbjct: 144 --DLGVPEYVLPLLSNNVTKLISNTLSL----NKVPYNEPSYKD----------GFPTWV 187
Query: 223 TEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENI 282
D++LL++ P I VVA DGSG +TI EAV AA R+VIY+K G Y ENI
Sbjct: 188 KPGDRKLLQTTPRANI---VVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAGTYNENI 244
Query: 283 DMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPV 342
++K K NIM VGDGIG+TI+T +++ G TTF++AT AV G FIA+D+T RNTAGP
Sbjct: 245 EVKLK--NIMFVGDGIGKTIITGSKSVGGGATTFKSATVAVVGDNFIARDITIRNTAGPN 302
Query: 343 NHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNC 402
NHQAVALR SD S F+RCS EG QDTLY HS RQFYREC+IYGT+DFIFGN A VLQNC
Sbjct: 303 NHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNAAVVLQNC 362
Query: 403 KIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTYLGRPWKEY 453
I+ R P P + T+TAQGR P+QSTG I + V A S T+LGRPWK+Y
Sbjct: 363 NIFARKP-PNRTNTLTAQGRTDPNQSTGIIIHNCRVTAASDLKPVQSSVKTFLGRPWKQY 421
Query: 454 SRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDA 513
SRTVYI T++ S++ P GW+EW G+FAL+TL+Y EY N GPGSS A RVKW GYHV+
Sbjct: 422 SRTVYIKTFLDSLINPAGWMEWSGDFALNTLYYAEYMNTGPGSSTANRVKWRGYHVLTSP 481
Query: 514 SAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
S FTV F+ G SWLP T V FT+GL
Sbjct: 482 SQVSQFTVGNFIAGNSWLPATNVPFTSGL 510
>dbj|BAD45460.1| putative pectinesterase [Oryza sativa (japonica cultivar-group)]
Length = 426
Score = 414 bits (1063), Expect = e-114
Identities = 210/423 (49%), Positives = 280/423 (65%), Gaps = 28/423 (6%)
Query: 148 LSAALSNQDTCIEGFEGTD-RRLESYISGSVTQVTQLISNVLSLYTQL------NRLPFR 200
LSAA++NQ TC++GF+ D R+ Y+ S+ V++++SN L++ +L P
Sbjct: 4 LSAAMTNQYTCLDGFDYKDGERVRHYMESSIHHVSRMVSNSLAMAKKLPGAGGGGMTPSS 63
Query: 201 PPRNTTLHETSTDESLE------------FPEWMTEADQELLKSKPHGKIADAVVALDGS 248
+T S++ + FP+W+ D+ LL++ DAVVA DGS
Sbjct: 64 SSPDTATQSESSETTQRQPFMGYGQMANGFPKWVRPGDRRLLQAPASSITPDAVVAKDGS 123
Query: 249 GQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRN 308
G Y T++ AV AAP++SN+R+VI++K G Y EN+++ K N+M +GDGIG+T++ ++RN
Sbjct: 124 GGYTTVSAAVAAAPANSNKRYVIHIKAGAYMENVEVGKSKKNLMFIGDGIGKTVIKASRN 183
Query: 309 FMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQD 368
+ G TTFR+AT AV G F+A+D+T N+AGP HQAVALRV +D SAF+RCS G QD
Sbjct: 184 VVDGSTTFRSATVAVVGNNFLARDLTIENSAGPSKHQAVALRVGADLSAFYRCSFVGYQD 243
Query: 369 TLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQS 428
TLY HSLRQF+REC+IYGTIDFIFGN A V Q+C +Y R PLP Q TAQGR+ P+Q+
Sbjct: 244 TLYVHSLRQFFRECDIYGTIDFIFGNSAVVFQSCNLYARRPLPNQSNVYTAQGREDPNQN 303
Query: 429 TGFTIQDSYVLA---------SQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNF 479
TG +IQ V A S TYLGRPWK+YSRTV++ + + S+V P GWLEW GNF
Sbjct: 304 TGISIQKCKVAAASDLLAVQSSFKTYLGRPWKQYSRTVFMQSELDSVVNPAGWLEWSGNF 363
Query: 480 ALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFT 539
ALDTL+YGEY+N GPG+S + RVKW GY VI AS A FTV F++G WL T V FT
Sbjct: 364 ALDTLYYGEYQNTGPGASTSNRVKWKGYRVITSASEASTFTVGNFIDGDVWLAGTSVPFT 423
Query: 540 AGL 542
GL
Sbjct: 424 VGL 426
>gb|AAB57671.1| pectinesterase [Citrus sinensis] gi|7447381|pir||T10494
pectinesterase (EC 3.1.1.11) PECS-c2 - sweet orange
Length = 510
Score = 413 bits (1061), Expect = e-114
Identities = 234/509 (45%), Positives = 302/509 (58%), Gaps = 40/509 (7%)
Query: 44 CMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEAIGAINNMTKISTFSVNNRE 103
C N C + + T T +L+ + A A + + + N RE
Sbjct: 32 CGKTPNPQPCEYFLTQKTDVTSIKQDTDFYKISLQLALERATTAQSRTYTLGSKCRNERE 91
Query: 104 QLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFE 163
+ A EDC+EL + +V +L + D+ + WLS+AL+N +TC E
Sbjct: 92 KAAWEDCRELYELTVLKLNQTSNSSPGCTKVDK--------QTWLSSALTNLETCRASLE 143
Query: 164 GTDRRLESYISGSVTQ-VTQLISNVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWM 222
D + Y+ ++ VT+LISN LSL N++P+ P FP W+
Sbjct: 144 --DLGVPEYVLPLLSNNVTKLISNALSL----NKVPYNEPSYKD----------GFPTWV 187
Query: 223 TEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENI 282
D++LL++ P I VVA DGSG +TI EAV AA R+VIY+K G Y ENI
Sbjct: 188 KPGDRKLLQTTPRANI---VVAQDGSGNVKTIQEAVAAASRAGGSRYVIYIKAGTYNENI 244
Query: 283 DMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPV 342
++K K NIM VGDGIG+TI+T +++ G TTF++AT AV G FIA+D+T RNTAGP
Sbjct: 245 EVKLK--NIMFVGDGIGKTIITGSKSVGGGATTFKSATVAVVGDNFIARDITIRNTAGPN 302
Query: 343 NHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNC 402
NHQAVALR SD S F+RCS EG QDTLY HS RQFYREC+IYGT+DFIFGN A VLQNC
Sbjct: 303 NHQAVALRSGSDLSVFYRCSFEGYQDTLYVHSQRQFYRECDIYGTVDFIFGNAAVVLQNC 362
Query: 403 KIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQPTYLGRPWKEY 453
I+ R P P + T+TAQGR P+Q+TG I + V A S T+LGRPWK+Y
Sbjct: 363 NIFARXP-PNRTNTLTAQGRTDPNQNTGIIIHNCRVTAASDLKPVQSSVKTFLGRPWKQY 421
Query: 454 SRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDA 513
SRTV I T++ S++ P GW+EW G+FAL+TL+Y EY N GPGSS A RVKW GYHV+
Sbjct: 422 SRTVXIKTFLDSLINPAGWMEWSGDFALNTLYYAEYMNTGPGSSTANRVKWRGYHVLTSP 481
Query: 514 SAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
S FTV F+ G SWLP T V FT+GL
Sbjct: 482 SQVSQFTVGNFIAGNSWLPATNVPFTSGL 510
>gb|AAG17110.1| putative pectin methylesterase 3 [Linum usitatissimum]
Length = 555
Score = 400 bits (1029), Expect = e-110
Identities = 219/520 (42%), Positives = 311/520 (59%), Gaps = 20/520 (3%)
Query: 38 ALIAQACMDIENQNSCLTNIHNELTRTGPPSPTSVINAALRTTINEA---IGAINNMTKI 94
A++ +C + + + C +++ + + VI A++ T I A+N
Sbjct: 41 AILTSSCSNTRHPDLCFSSLASAPVHVSLNTQMDVIKASINVTCTSVLRNIAAVNKALST 100
Query: 95 STFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSN 154
T + R + A++DC E + S+ EL +L E+ ++ +L+ LSAA +N
Sbjct: 101 RT-DLTPRSRSALKDCVETMSTSLDELHVALAELDEYPNKKSITRHADDLKTLLSAATTN 159
Query: 155 QDTCIEGF--EGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETST 212
Q+TC++GF + +++++ + +V ++ N L + + N T
Sbjct: 160 QETCLDGFSHDDSEKKVRKTLETGPVRVEKMCGNALGMIVNMTETDMASATNAV--NTEG 217
Query: 213 DESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVIY 272
S +P WM D+ LL++ + VVA DGSG+YR ++EAV AAPS S++R+VI
Sbjct: 218 GSSGSWPIWMKGGDRRLLQAGTT-VTPNVVVAADGSGKYRRVSEAVAAAPSKSSKRYVIR 276
Query: 273 VKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAKD 332
+K G+Y+EN+++ K TNIM VGDG TI+T N+N + G TTF +AT AV G+GF+A+D
Sbjct: 277 IKAGIYRENVEVPKDKTNIMFVGDGRSNTIITGNKNVVDGSTTFNSATVAVVGQGFLARD 336
Query: 333 MTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFIF 392
+TF+NTAGP HQAVALRV +D +AF+RC QDTLY HS RQF+ C + GT+DFIF
Sbjct: 337 ITFQNTAGPSKHQAVALRVGADLAAFYRCDFLAYQDTLYVHSNRQFFINCLVVGTVDFIF 396
Query: 393 GNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLASQP--------- 443
GN AAV QNC I+ R P P QK +TA GR P+Q+TG IQ S + A+
Sbjct: 397 GNSAAVFQNCDIHARRPNPGQKNMLTAHGRTDPNQNTGIVIQKSRIAATSDLQSVKGSFG 456
Query: 444 TYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRVK 503
TYLGRPWK Y+RTV + + +S +V P GW EW GNFAL+TL+YGE++N G GS + GRVK
Sbjct: 457 TYLGRPWKAYARTVIMQSTISDVVHPAGWHEWDGNFALNTLFYGEHKNSGAGSGVNGRVK 516
Query: 504 WPGYHVI-KDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
W G+ VI DA AAG FT RF+ GGSWL T FT GL
Sbjct: 517 WKGHKVISSDAEAAG-FTPGRFIAGGSWLGSTTFPFTLGL 555
>gb|AAF23892.1| pectin methyl esterase [Solanum tuberosum]
Length = 576
Score = 400 bits (1027), Expect = e-110
Identities = 215/520 (41%), Positives = 316/520 (60%), Gaps = 18/520 (3%)
Query: 38 ALIAQACMDIENQNSCLTNIHNELTRTGP-PSPTSVINAALRTTINEA---IGAINNMTK 93
A++ AC + + C + I N + S VI +L T+ A+ + K
Sbjct: 60 AIVKSACSNTLHPELCYSAIVNVTDFSKKVTSQKDVIELSLNITVKAVRRNYYAVKELIK 119
Query: 94 ISTFSVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALS 153
+ RE++A+ DC E +D ++ EL ++ ++ ++ +L+ +S+A++
Sbjct: 120 TRK-GLTPREKVALHDCLETMDETLDELHTAVADLELYPNKKSLKEHAEDLKTLISSAIT 178
Query: 154 NQDTCIEGF--EGTDRRLESYISGSVTQVTQLISNVLSLYTQLNRLPFRPPRNTTLHETS 211
NQ+TC++GF + D+++ + V ++ SN L++ + +
Sbjct: 179 NQETCLDGFSHDEADKKVRKVLLKGQKHVEKMCSNALAMICNMTNTDIANEMKLSGSRKL 238
Query: 212 TDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTINEAVNAAPSHSNRRHVI 271
+++ E+PEW++ D+ LL+S D VVA DGSG Y+T++EAV AP S++R+VI
Sbjct: 239 VEDNGEWPEWLSAGDRRLLQSST--VTPDVVVAADGSGDYKTVSEAVAKAPEKSSKRYVI 296
Query: 272 YVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGWTTFRTATFAVSGKGFIAK 331
+K G+Y+EN+D+ KK TNIM +GDG TI+T++RN G TTF +AT A G+ F+A+
Sbjct: 297 RIKAGVYRENVDVPKKKTNIMFMGDGRSNTIITASRNVQDGSTTFHSATVAAVGEKFLAR 356
Query: 332 DMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAHSLRQFYRECEIYGTIDFI 391
D+TF+NTAG HQAVALRV SD SAF++C I QDTLY HS RQF+ +C + GT+DFI
Sbjct: 357 DITFQNTAGASKHQAVALRVGSDLSAFYKCDILAYQDTLYVHSNRQFFVQCLVAGTVDFI 416
Query: 392 FGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTIQDSYVLA---------SQ 442
FGNGAAVLQ+C I+ R P QK +TAQGR P+Q+TG IQ + A S
Sbjct: 417 FGNGAAVLQDCDIHARRPGSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLRPVQKSF 476
Query: 443 PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYGPGSSLAGRV 502
PTYLGRPWKEYSRTV + + ++ ++QP GW EW GNFAL+TL+YGEY N G G++ +GRV
Sbjct: 477 PTYLGRPWKEYSRTVIMQSSITDVIQPAGWHEWNGNFALNTLFYGEYANTGAGAATSGRV 536
Query: 503 KWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
KW G+ VI ++ A +T F+ GGSWL TG F+ GL
Sbjct: 537 KWKGHKVITSSTEAQAYTPGSFIAGGSWLSSTGFPFSLGL 576
>ref|NP_915736.1| putative pectin esterase [Oryza sativa (japonica cultivar-group)]
Length = 563
Score = 399 bits (1026), Expect = e-110
Identities = 216/487 (44%), Positives = 294/487 (60%), Gaps = 28/487 (5%)
Query: 66 PPSPTSVINAALRTTINEAIGAINNMTKISTFSVNNREQLAIEDCKELLDFSVSELAWSL 125
P P V+ A L T+++ A + + + + R + A+EDC +L+ + LA +
Sbjct: 95 PSRPAHVLRAILATSLDRHDAAAEAVAGMRRRASDPRHRAALEDCVQLMGLARDRLADAA 154
Query: 126 GEMRRIRAGDRTAQYEGNLEAWLSAALSNQDTCIEGFEGTDRRLESYISGSVTQVTQLIS 185
G A D + + WLSA L++ TC++G + D L + + + L S
Sbjct: 155 G------APDVDVDVD-DARTWLSAVLTDHVTCLDGLD--DGPLRDSVGAHLEPLKSLAS 205
Query: 186 NVLSLYTQLNRLPFRPPRNTTLHETSTDESLEFPEWMTEADQELLKSKPHGKIADAVVAL 245
L++ + R + + FP W+T D+ LL + AD VVA
Sbjct: 206 ASLAVLSAAGR---------GARDVLAEAVDRFPSWLTARDRTLLDAGAGAVQADVVVAK 256
Query: 246 DGSGQYRTINEAVNAAPSHSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTS 305
DGSG+Y TI EAV+AAP R+VIYVKKG+YKEN+++ K +M+VGDG+ QT++T
Sbjct: 257 DGSGKYTTIKEAVDAAPDGGKSRYVIYVKKGVYKENLEVGKTKRVLMIVGDGMDQTVITG 316
Query: 306 NRNFMQGWTTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEG 365
+RN + G TTF +AT A+SG G I +D+ NTAG QAVALRV +D++ RC ++G
Sbjct: 317 SRNVVDGSTTFNSATLALSGDGIILQDLKVENTAGAEKQQAVALRVSADRAVINRCRLDG 376
Query: 366 NQDTLYAHSLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSP 425
QDTLYAH LRQFYR+C + GT+DF+FGN AAVLQ C + R P QK +TAQGR P
Sbjct: 377 YQDTLYAHQLRQFYRDCAVSGTVDFVFGNAAAVLQGCVLTARRPAQAQKNAVTAQGRTDP 436
Query: 426 HQSTGFTIQDSYVLASQ---------PTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWL 476
+Q+TG +I V+ + PT+LGRPWKEYSRTVY+ +Y+ S V PRGWLEW
Sbjct: 437 NQNTGTSIHRCRVVPAPDLAPAAKQFPTFLGRPWKEYSRTVYMLSYLDSHVDPRGWLEWN 496
Query: 477 G-NFALDTLWYGEYRNYGPGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTG 535
G +FAL TL+YGEY+N GPG+S AGRV WPGYHVI D S A FTV +F+ GG+WL TG
Sbjct: 497 GADFALKTLFYGEYQNQGPGASTAGRVNWPGYHVITDQSVAMQFTVGQFIQGGNWLKATG 556
Query: 536 VKFTAGL 542
V + GL
Sbjct: 557 VNYNEGL 563
>gb|AAM14264.1| putative pectinesterase [Arabidopsis thaliana]
gi|17529058|gb|AAL38739.1| putative pectinesterase
[Arabidopsis thaliana] gi|15220671|ref|NP_173733.1|
pectinesterase family protein [Arabidopsis thaliana]
gi|25313428|pir||C86366 protein F26F24.2 [imported] -
Arabidopsis thaliana gi|9295687|gb|AAF86993.1| F26F24.2
[Arabidopsis thaliana] gi|2829892|gb|AAC00600.1|
putative pectinesterase [Arabidopsis thaliana]
Length = 554
Score = 399 bits (1026), Expect = e-110
Identities = 222/469 (47%), Positives = 284/469 (60%), Gaps = 33/469 (7%)
Query: 98 SVNNREQLAIEDCKELLDFSVSELAWSLGEMRRIRAGDRTAQYEGNLEAWLSAALSNQDT 157
S++ A+ DC EL + ++ +L S R G ++ ++ LSAA++NQDT
Sbjct: 95 SLHKHATSALFDCLELYEDTIDQLNHS-----RRSYGQYSSPHDRQTS--LSAAIANQDT 147
Query: 158 CIEGFEGTD--------------RRLESYISGS--VTQVTQLISNVLSLYTQLNRLPFRP 201
C GF R L IS S VT+ V Y F
Sbjct: 148 CRNGFRDFKLTSSYSKYFPVQFHRNLTKSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSK 207
Query: 202 PRNTTLHETS------TDESLEFPEWMTEADQELLKSKPHGKIADAVVALDGSGQYRTIN 255
R++ + +DE +FP W +D++LL+ AD VVA DGSG Y +I
Sbjct: 208 QRSSAGGGSHRRLLLFSDE--KFPSWFPLSDRKLLEDSKTTAKADLVVAKDGSGHYTSIQ 265
Query: 256 EAVNAAPS--HSNRRHVIYVKKGLYKENIDMKKKMTNIMMVGDGIGQTIVTSNRNFMQGW 313
+AVNAA N+R VIYVK G+Y+EN+ +KK + N+M++GDGI TIVT NRN G
Sbjct: 266 QAVNAAAKLPRRNQRLVIYVKAGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNVQDGT 325
Query: 314 TTFRTATFAVSGKGFIAKDMTFRNTAGPVNHQAVALRVDSDQSAFFRCSIEGNQDTLYAH 373
TTFR+ATFAVSG GFIA+ +TF NTAGP HQAVALR SD S F+ CS +G QDTLY H
Sbjct: 326 TTFRSATFAVSGNGFIAQGITFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLH 385
Query: 374 SLRQFYRECEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPHQSTGFTI 433
S RQF R C IYGT+DFIFG+ A+LQNC IY R P+ QK TITAQ RK P ++TGF I
Sbjct: 386 SSRQFLRNCNIYGTVDFIFGDATAILQNCNIYARKPMSGQKNTITAQSRKEPDETTGFVI 445
Query: 434 QDSYVLASQPTYLGRPWKEYSRTVYINTYMSSMVQPRGWLEWLGNFALDTLWYGEYRNYG 493
Q S V + TYLGRPW+ +SRTV++ + ++V P GWL W G+FAL TL+YGEY N G
Sbjct: 446 QSSTVATASETYLGRPWRSHSRTVFMKCNLGALVSPAGWLPWSGSFALSTLYYGEYGNTG 505
Query: 494 PGSSLAGRVKWPGYHVIKDASAAGYFTVQRFLNGGSWLPRTGVKFTAGL 542
G+S++GRVKWPGYHVIK + A FTV+ FL+G W+ TGV GL
Sbjct: 506 AGASVSGRVKWPGYHVIKTVTEAEKFTVENFLDGNYWITATGVPVNDGL 554
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.319 0.133 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 908,211,053
Number of Sequences: 2540612
Number of extensions: 37244059
Number of successful extensions: 88135
Number of sequences better than 10.0: 405
Number of HSP's better than 10.0 without gapping: 332
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 86854
Number of HSP's gapped (non-prelim): 451
length of query: 544
length of database: 863,360,394
effective HSP length: 133
effective length of query: 411
effective length of database: 525,458,998
effective search space: 215963648178
effective search space used: 215963648178
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC144513.13


