

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144503.8 - phase: 0
(729 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
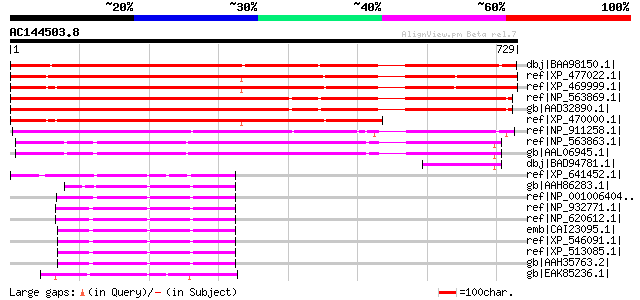
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAA98150.1| unnamed protein product [Arabidopsis thaliana] g... 1008 0.0
ref|XP_477022.1| unknown protein [Oryza sativa (japonica cultiva... 903 0.0
ref|XP_469999.1| expressed protein (with alternative splicing) [... 875 0.0
ref|NP_563869.1| hypothetical protein [Arabidopsis thaliana] 799 0.0
gb|AAD32890.1| F14N23.28 [Arabidopsis thaliana] 794 0.0
ref|XP_470000.1| expressed protein (with alternative splicing) [... 668 0.0
ref|NP_911258.1| unknown protein [Oryza sativa (japonica cultiva... 380 e-103
ref|NP_563863.1| expressed protein [Arabidopsis thaliana] gi|491... 363 1e-98
gb|AAL06945.1| At1g10180/F14N23_6 [Arabidopsis thaliana] 360 9e-98
dbj|BAD94781.1| hypothetical protein [Arabidopsis thaliana] 95 7e-18
ref|XP_641452.1| hypothetical protein DDB0205917 [Dictyostelium ... 74 2e-11
gb|AAH86283.1| Unknown (protein for MGC:83775) [Xenopus laevis] 73 4e-11
ref|NP_001006404.1| similar to Expressed sequence AI414418 [Gall... 71 1e-10
ref|NP_932771.1| exocyst complex component 8 [Mus musculus] gi|3... 68 9e-10
ref|NP_620612.1| exocyst complex component 8 [Rattus norvegicus]... 67 2e-09
emb|CAI23095.1| exocyst complex component 8 [Homo sapiens] 67 3e-09
ref|XP_546091.1| PREDICTED: hypothetical protein XP_546091 [Cani... 67 3e-09
ref|XP_513085.1| PREDICTED: hypothetical protein XP_513085 [Pan ... 67 3e-09
gb|AAH35763.2| Exocyst complex 84-kDa subunit [Homo sapiens] gi|... 67 3e-09
gb|EAK85236.1| hypothetical protein UM04147.1 [Ustilago maydis 5... 66 4e-09
>dbj|BAA98150.1| unnamed protein product [Arabidopsis thaliana]
gi|20334792|gb|AAM16257.1| AT5g49830/K21G20_4
[Arabidopsis thaliana] gi|18700079|gb|AAL77652.1|
AT5g49830/K21G20_4 [Arabidopsis thaliana]
gi|15240560|ref|NP_199794.1| expressed protein
[Arabidopsis thaliana]
Length = 752
Score = 1008 bits (2605), Expect = 0.0
Identities = 528/729 (72%), Positives = 611/729 (83%), Gaps = 45/729 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MRRSVYANY AFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV+ID +SD
Sbjct: 67 MRRSVYANYPAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVNIDDDKVSDES-- 124
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
NG L+ E +SDL+KW EFPD LD LLAERRV+EALAA DEGE +VS+A E +L+
Sbjct: 125 LANGLLNFEDNGLSDLEKWATEFPDHLDALLAERRVDEALAAFDEGEILVSQANEKHTLS 184
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
S+L SLQ +I ER+QKLADQLA+AACQPSTRG ELR++++ALK+LGDGP AH++LL+AH
Sbjct: 185 SSVLSSLQFAIAERKQKLADQLAKAACQPSTRGGELRSAIAALKRLGDGPRAHTVLLDAH 244
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
QRYQYNMQSLRPS+TSYGGAYTAAL+QLVFSA++QA+SDSL IFG+EPAYSSELV WAT
Sbjct: 245 FQRYQYNMQSLRPSSTSYGGAYTAALSQLVFSAISQASSDSLGIFGKEPAYSSELVTWAT 304
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
KQTEAF+LLVKRHALASSAAAGGLRAAAEC QIALGHCSLLEARGL+LCPVLLK F+P V
Sbjct: 305 KQTEAFSLLVKRHALASSAAAGGLRAAAECAQIALGHCSLLEARGLSLCPVLLKHFKPIV 364
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDF 360
EQAL+ANLKRI+E+TAAMAAADDWVLT PP +R ++TAFQ KLTSSAHRFNLMVQDF
Sbjct: 365 EQALEANLKRIEENTAAMAAADDWVLTSPPAGSRH--ASTAFQNKLTSSAHRFNLMVQDF 422
Query: 361 FEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEE-SFEDSGNKIVRMAETE 419
FEDVGPLLSMQLG +ALEGLF+VFNSYV++L++ALP S+EEE+ +FE S NKIV+MAETE
Sbjct: 423 FEDVGPLLSMQLGSKALEGLFRVFNSYVDVLVRALPGSIEEEDPNFESSCNKIVQMAETE 482
Query: 420 AQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLVG 479
A Q+ALLANASLLADELLPRAAMKL SL+Q + D+ RR +RQNR+PEQREW+RRL+
Sbjct: 483 ANQLALLANASLLADELLPRAAMKL-SLDQTGQRTDDLRRPLDRQNRNPEQREWKRRLLS 541
Query: 480 SVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLIF 539
+VD+LKD+FCRQHAL LIFTE+GDSHL+ADMY++++ N ++V++ PSLIFQ
Sbjct: 542 TVDKLKDAFCRQHALDLIFTEEGDSHLSADMYVNIDENGEDVDFFPSLIFQ--------- 592
Query: 540 KNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRLT 599
ELF KLNRMA++AADMFVGRERFA LLMRLT
Sbjct: 593 ----------------------------ELFAKLNRMASLAADMFVGRERFAISLLMRLT 624
Query: 600 ETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEI 659
ETVILW+S DQSFWDDIEEGPRPLGPLGL+Q YLDMKFV+CFAS GRYLSRNL R NEI
Sbjct: 625 ETVILWLSGDQSFWDDIEEGPRPLGPLGLRQLYLDMKFVICFASQGRYLSRNLHRGTNEI 684
Query: 660 IRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPKEINGERELSSPTASVSAQS 719
I KA++AF+ATG+DPYS+LPED+WFN+IC DAMERLSGK K NG ++ SPTASVSAQS
Sbjct: 685 ISKALAAFTATGIDPYSELPEDDWFNDICVDAMERLSGKTKGNNG--DVHSPTASVSAQS 742
Query: 720 ISSVRSHNS 728
+SS RSH S
Sbjct: 743 VSSARSHGS 751
>ref|XP_477022.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|34394796|dbj|BAC84209.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 788
Score = 903 bits (2333), Expect = 0.0
Identities = 467/733 (63%), Positives = 583/733 (78%), Gaps = 44/733 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MRRSVYANYAAFIRTSKEISDLEGEL SIRNLL+TQA LIHGL++GV IDSL+ S+++G
Sbjct: 96 MRRSVYANYAAFIRTSKEISDLEGELLSIRNLLNTQAALIHGLSEGVQIDSLT-SNTEGS 154
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
+ + + E +E S++ KW +FPD+LDVLLAERRV+EAL ALDE ER+ S+AK ++L
Sbjct: 155 AEDDISNVEDQEPSEIQKWSADFPDMLDVLLAERRVDEALDALDEAERLASDAKLKQTLT 214
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
+ + +L+ ++++ RQKLADQLAEAACQ STRG ELRA+ SALK+LGDGP AHSLLLNAH
Sbjct: 215 ATEIAALRRAVSDNRQKLADQLAEAACQSSTRGIELRAAASALKRLGDGPRAHSLLLNAH 274
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
QR Q NMQ++ PS+TSYGGAYTAALAQ VFS VAQA SDS+ +FG+E Y+SELV WAT
Sbjct: 275 NQRLQCNMQTIHPSSTSYGGAYTAALAQQVFSVVAQALSDSVEVFGDESCYASELVTWAT 334
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
KQ +FALLVKRH L+S AAAGGLRAAAECVQI+LGHCSLLEARGL++ VLL+ FRPS+
Sbjct: 335 KQVMSFALLVKRHVLSSCAAAGGLRAAAECVQISLGHCSLLEARGLSVAAVLLRQFRPSL 394
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPN----VNRQTGSTTAFQLKLTSSAHRFNLM 356
EQAL +N++RI+ESTAA+AAADDW+LTYPP + R + + A Q KL++SAHRFN M
Sbjct: 395 EQALYSNIRRIEESTAALAAADDWILTYPPTGIRPLARSSAANLALQPKLSNSAHRFNSM 454
Query: 357 VQDFFEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMA 416
VQ+FFEDV PLLS+QLGG ++ + ++FNSYVN+LI ALP SME+E + + GNKIVRMA
Sbjct: 455 VQEFFEDVAPLLSLQLGGSTMDDITKIFNSYVNLLISALPGSMEDEANIDGLGNKIVRMA 514
Query: 417 ETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRR 476
E+E QQ+ALLANASLLA+ELLPRAAMKLSS+N DD R+R +++QNR PEQREW+R+
Sbjct: 515 ESEEQQLALLANASLLAEELLPRAAMKLSSMNHSS-MDDLRKRGSDKQNRMPEQREWKRK 573
Query: 477 LVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWL 536
L VDRL+DSFCRQHAL LIFT++G++HL+ADMYISM+ +E EW PSLIFQ
Sbjct: 574 LQRMVDRLRDSFCRQHALELIFTDEGETHLSADMYISMDNTVEEPEWAPSLIFQ------ 627
Query: 537 LIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLM 596
EL+ KLNRMA+IAADMFVGRERFATLL+M
Sbjct: 628 -------------------------------ELYAKLNRMASIAADMFVGRERFATLLMM 656
Query: 597 RLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIV 656
RLTETVILW+SEDQ+FW++IE+GP+PLGPLGLQQFYLDM+FV+ F GR+LSR++ +++
Sbjct: 657 RLTETVILWLSEDQAFWEEIEQGPKPLGPLGLQQFYLDMQFVIIF-GQGRFLSRHVHQVI 715
Query: 657 NEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPKEINGERELSSPTASVS 716
+II +AM+AFSATGM+P S LP D+WF ++ Q+ + +SGK + NG+RE++SPTASVS
Sbjct: 716 LDIIDRAMAAFSATGMNPDSVLPGDDWFMDVSQEVVSMISGKGRAANGDREINSPTASVS 775
Query: 717 AQSISSVRSHNSS 729
A S+SS RSH SS
Sbjct: 776 AHSMSSFRSHGSS 788
>ref|XP_469999.1| expressed protein (with alternative splicing) [Oryza sativa
(japonica cultivar-group)] gi|41469399|gb|AAS07222.1|
expressed protein (with alternative splicing) [Oryza
sativa (japonica cultivar-group)]
Length = 771
Score = 875 bits (2262), Expect = 0.0
Identities = 456/733 (62%), Positives = 576/733 (78%), Gaps = 46/733 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MRRSVYANYAAFIRTSKEISDLE EL S+RNLLSTQ+ LI GL++GVHIDSL+ + S+G
Sbjct: 81 MRRSVYANYAAFIRTSKEISDLERELLSVRNLLSTQSALIRGLSEGVHIDSLT-TGSEGS 139
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
+ G E +E S++ W +FP++LDVLLAERRV+EAL ALDE ERVV++ K+ ++L
Sbjct: 140 AEEGT--DEDQEPSEIQNWCTDFPEMLDVLLAERRVDEALDALDEAERVVADEKQKQTLT 197
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
+ +L+++ +I++ R KLA+QLAEAACQ STRG ELRAS SALK+LGDGP AHSLLL+AH
Sbjct: 198 TADILAVKRAISDNRLKLANQLAEAACQSSTRGVELRASASALKRLGDGPRAHSLLLSAH 257
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
QR Q +MQ++ PS+TS+ GAYTA+LA+ VFS +AQA SDSL +FG+EP+Y SEL+ WAT
Sbjct: 258 NQRLQCSMQTIHPSSTSHSGAYTASLARQVFSVIAQALSDSLELFGDEPSYLSELITWAT 317
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
+Q FALLVKRHALA+ AAGGLRAAAEC+QI+LGH SLLE RGL+L VL+K F+PSV
Sbjct: 318 EQAMLFALLVKRHALAACVAAGGLRAAAECIQISLGHSSLLETRGLSLSSVLMKQFKPSV 377
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPN----VNRQTGSTTAFQLKLTSSAHRFNLM 356
EQAL+++L+RI+ESTAA+AAADDWVLTYPP+ R + S+ Q KL++S HRF+ M
Sbjct: 378 EQALESSLRRIEESTAALAAADDWVLTYPPSGIRTFARSSASSLLLQPKLSNSGHRFSSM 437
Query: 357 VQDFFEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMA 416
VQDFFEDVGPL S+QLGG A++GL ++FNSYVN+LI ALP S+++E + E GNKIVR+A
Sbjct: 438 VQDFFEDVGPLHSLQLGGSAMDGLLKIFNSYVNLLISALPHSLDDEANLEGLGNKIVRVA 497
Query: 417 ETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRR 476
ETE QQ+AL ANASLLA+ELLPRAAMKLSS+N +D R+++ +RQNR EQREW+++
Sbjct: 498 ETEEQQLALFANASLLAEELLPRAAMKLSSVNHTGV-NDIRKKSVDRQNRVAEQREWKKK 556
Query: 477 LVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWL 536
L VD+LKDSFCRQHAL LIFTED D+ L+A+MYI+M+ +E EW+PSLIFQ
Sbjct: 557 LQRIVDKLKDSFCRQHALDLIFTEDDDTRLSAEMYINMDNTVEEPEWVPSLIFQ------ 610
Query: 537 LIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLM 596
EL+ KLNRMA+IAAD+FVGRERFAT LLM
Sbjct: 611 -------------------------------ELYAKLNRMASIAADLFVGRERFATFLLM 639
Query: 597 RLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIV 656
RLTETVILW+SEDQSFW++IEEGPR LGPLGLQQFYLDM+FV+ + GR+LSR++ +++
Sbjct: 640 RLTETVILWLSEDQSFWEEIEEGPRALGPLGLQQFYLDMQFVI-LSGRGRFLSRHVHQVI 698
Query: 657 NEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPKEINGERELSSPTASVS 716
+II +AM+AFSATGM+P S LP D+WF ++ D + R+SG P+ NG+RE++SPTASVS
Sbjct: 699 LKIIDRAMAAFSATGMNPDSVLPSDDWFIDVANDTISRISGNPRTANGDREVNSPTASVS 758
Query: 717 AQSISSVRSHNSS 729
AQSISSVRSH SS
Sbjct: 759 AQSISSVRSHGSS 771
>ref|NP_563869.1| hypothetical protein [Arabidopsis thaliana]
Length = 754
Score = 799 bits (2064), Expect = 0.0
Identities = 431/725 (59%), Positives = 545/725 (74%), Gaps = 47/725 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MR+SVYANYAAFIRTSKEIS LEG+L S+RNLLS QA L+HGLADGVHI SL D+D
Sbjct: 73 MRKSVYANYAAFIRTSKEISALEGQLLSMRNLLSAQAALVHGLADGVHISSLCADDADDL 132
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
D ++K++S+++ W+VEF D L+VLLAE+RVEE++AAL+EG RV EA E ++L+
Sbjct: 133 RDEDLYDMDNKQLSNIENWVVEFFDRLEVLLAEKRVEESMAALEEGRRVAVEAHEKRTLS 192
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
P+ LLSL ++I E+RQ+LADQLAEA QPSTRG ELR++V +LKKLGDG AH+LLL ++
Sbjct: 193 PTTLLSLNNAIKEKRQELADQLAEAISQPSTRGGELRSAVLSLKKLGDGSRAHTLLLRSY 252
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
+R Q N+QSLR SNTSYG A+ AAL+QLVFS +AQAASDS A+ GE+PAY+SELV WA
Sbjct: 253 ERRLQANIQSLRASNTSYGVAFAAALSQLVFSTIAQAASDSQAVVGEDPAYTSELVTWAV 312
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
KQ E+FALL+KRH LASSAAAG LR AECVQ+ HCS LE+RGLAL PVLLK FRP V
Sbjct: 313 KQAESFALLLKRHTLASSAAAGSLRVTAECVQLCASHCSSLESRGLALSPVLLKHFRPGV 372
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGST-TAFQLKLTSSAHRFNLMVQD 359
EQAL NLKRI++S+AA+AA+DDW L+Y P +R + +T TA LKL+ SA RFN MVQ+
Sbjct: 373 EQALTGNLKRIEQSSAALAASDDWSLSYTPTGSRASSTTPTAPHLKLSISAQRFNSMVQE 432
Query: 360 FFEDVGPL-LSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMAET 418
F ED GPL ++QL G AL+G+ QVFNSYV++LI ALP S E E E+ ++IV++AET
Sbjct: 433 FLEDAGPLDEALQLDGIALDGVLQVFNSYVDLLINALPGSAENE---ENPVHRIVKVAET 489
Query: 419 EAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLV 478
E+QQ ALL NA LLADEL+PR+A ++ L Q + RR +++RQNR PEQREW+++L
Sbjct: 490 ESQQTALLVNALLLADELIPRSASRI--LPQGTSQSTPRRGSSDRQNR-PEQREWKKKLQ 546
Query: 479 GSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLI 538
SVDRL+DSFCRQHAL LIFTE+G+ L++++YI M+ +E EW PS IFQ
Sbjct: 547 RSVDRLRDSFCRQHALELIFTEEGEVRLSSEIYILMDETTEEPEWFPSPIFQ-------- 598
Query: 539 FKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRL 598
ELF KL R+A I +DMFVGRERFAT+LLMRL
Sbjct: 599 -----------------------------ELFAKLTRIAMIVSDMFVGRERFATILLMRL 629
Query: 599 TETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNE 658
TETVILWIS+DQSFW+++E G +PLGPLGLQQFYLDM+FV+ FAS GRYLSRNL +++
Sbjct: 630 TETVILWISDDQSFWEEMETGDKPLGPLGLQQFYLDMEFVMIFASQGRYLSRNLHQVIKN 689
Query: 659 IIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPK-EINGERELSSPTASVSA 717
II +A+ A SATG+DPYS LPE+EWF E+ Q A++ L GK +GER+++SP+ S SA
Sbjct: 690 IIARAVEAVSATGLDPYSTLPEEEWFAEVAQIAIKMLMGKGNFGGHGERDVTSPSVS-SA 748
Query: 718 QSISS 722
+S +S
Sbjct: 749 KSYTS 753
>gb|AAD32890.1| F14N23.28 [Arabidopsis thaliana]
Length = 824
Score = 794 bits (2051), Expect = 0.0
Identities = 431/727 (59%), Positives = 545/727 (74%), Gaps = 49/727 (6%)
Query: 1 MRRSVYANYAAFIR--TSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSD 58
MR+SVYANYAAFIR TSKEIS LEG+L S+RNLLS QA L+HGLADGVHI SL D+D
Sbjct: 141 MRKSVYANYAAFIRCVTSKEISALEGQLLSMRNLLSAQAALVHGLADGVHISSLCADDAD 200
Query: 59 GFSVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKS 118
D ++K++S+++ W+VEF D L+VLLAE+RVEE++AAL+EG RV EA E ++
Sbjct: 201 DLRDEDLYDMDNKQLSNIENWVVEFFDRLEVLLAEKRVEESMAALEEGRRVAVEAHEKRT 260
Query: 119 LNPSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLN 178
L+P+ LLSL ++I E+RQ+LADQLAEA QPSTRG ELR++V +LKKLGDG AH+LLL
Sbjct: 261 LSPTTLLSLNNAIKEKRQELADQLAEAISQPSTRGGELRSAVLSLKKLGDGSRAHTLLLR 320
Query: 179 AHLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMW 238
++ +R Q N+QSLR SNTSYG A+ AAL+QLVFS +AQAASDS A+ GE+PAY+SELV W
Sbjct: 321 SYERRLQANIQSLRASNTSYGVAFAAALSQLVFSTIAQAASDSQAVVGEDPAYTSELVTW 380
Query: 239 ATKQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRP 298
A KQ E+FALL+KRH LASSAAAG LR AECVQ+ HCS LE+RGLAL PVLLK FRP
Sbjct: 381 AVKQAESFALLLKRHTLASSAAAGSLRVTAECVQLCASHCSSLESRGLALSPVLLKHFRP 440
Query: 299 SVEQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGST-TAFQLKLTSSAHRFNLMV 357
VEQAL NLKRI++S+AA+AA+DDW L+Y P +R + +T TA LKL+ SA RFN MV
Sbjct: 441 GVEQALTGNLKRIEQSSAALAASDDWSLSYTPTGSRASSTTPTAPHLKLSISAQRFNSMV 500
Query: 358 QDFFEDVGPL-LSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMA 416
Q+F ED GPL ++QL G AL+G+ QVFNSYV++LI ALP S E E E+ ++IV++A
Sbjct: 501 QEFLEDAGPLDEALQLDGIALDGVLQVFNSYVDLLINALPGSAENE---ENPVHRIVKVA 557
Query: 417 ETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRR 476
ETE+QQ ALL NA LLADEL+PR+A ++ L Q + RR +++RQNR PEQREW+++
Sbjct: 558 ETESQQTALLVNALLLADELIPRSASRI--LPQGTSQSTPRRGSSDRQNR-PEQREWKKK 614
Query: 477 LVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWL 536
L SVDRL+DSFCRQHAL LIFTE+G+ L++++YI M+ +E EW PS IFQ
Sbjct: 615 LQRSVDRLRDSFCRQHALELIFTEEGEVRLSSEIYILMDETTEEPEWFPSPIFQ------ 668
Query: 537 LIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLM 596
ELF KL R+A I +DMFVGRERFAT+LLM
Sbjct: 669 -------------------------------ELFAKLTRIAMIVSDMFVGRERFATILLM 697
Query: 597 RLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIV 656
RLTETVILWIS+DQSFW+++E G +PLGPLGLQQFYLDM+FV+ FAS GRYLSRNL +++
Sbjct: 698 RLTETVILWISDDQSFWEEMETGDKPLGPLGLQQFYLDMEFVMIFASQGRYLSRNLHQVI 757
Query: 657 NEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPK-EINGERELSSPTASV 715
II +A+ A SATG+DPYS LPE+EWF E+ Q A++ L GK +GER+++SP+ S
Sbjct: 758 KNIIARAVEAVSATGLDPYSTLPEEEWFAEVAQIAIKMLMGKGNFGGHGERDVTSPSVS- 816
Query: 716 SAQSISS 722
SA+S +S
Sbjct: 817 SAKSYTS 823
>ref|XP_470000.1| expressed protein (with alternative splicing) [Oryza sativa
(japonica cultivar-group)] gi|41469398|gb|AAS07221.1|
expressed protein (with alternative splicing) [Oryza
sativa (japonica cultivar-group)]
Length = 641
Score = 668 bits (1724), Expect = 0.0
Identities = 345/540 (63%), Positives = 440/540 (80%), Gaps = 8/540 (1%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDSDGF 60
MRRSVYANYAAFIRTSKEISDLE EL S+RNLLSTQ+ LI GL++GVHIDSL+ + S+G
Sbjct: 81 MRRSVYANYAAFIRTSKEISDLERELLSVRNLLSTQSALIRGLSEGVHIDSLT-TGSEGS 139
Query: 61 SVNGALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLN 120
+ G E +E S++ W +FP++LDVLLAERRV+EAL ALDE ERVV++ K+ ++L
Sbjct: 140 AEEGT--DEDQEPSEIQNWCTDFPEMLDVLLAERRVDEALDALDEAERVVADEKQKQTLT 197
Query: 121 PSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAH 180
+ +L+++ +I++ R KLA+QLAEAACQ STRG ELRAS SALK+LGDGP AHSLLL+AH
Sbjct: 198 TADILAVKRAISDNRLKLANQLAEAACQSSTRGVELRASASALKRLGDGPRAHSLLLSAH 257
Query: 181 LQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWAT 240
QR Q +MQ++ PS+TS+ GAYTA+LA+ VFS +AQA SDSL +FG+EP+Y SEL+ WAT
Sbjct: 258 NQRLQCSMQTIHPSSTSHSGAYTASLARQVFSVIAQALSDSLELFGDEPSYLSELITWAT 317
Query: 241 KQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSV 300
+Q FALLVKRHALA+ AAGGLRAAAEC+QI+LGH SLLE RGL+L VL+K F+PSV
Sbjct: 318 EQAMLFALLVKRHALAACVAAGGLRAAAECIQISLGHSSLLETRGLSLSSVLMKQFKPSV 377
Query: 301 EQALDANLKRIQESTAAMAAADDWVLTYPPN----VNRQTGSTTAFQLKLTSSAHRFNLM 356
EQAL+++L+RI+ESTAA+AAADDWVLTYPP+ R + S+ Q KL++S HRF+ M
Sbjct: 378 EQALESSLRRIEESTAALAAADDWVLTYPPSGIRTFARSSASSLLLQPKLSNSGHRFSSM 437
Query: 357 VQDFFEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMA 416
VQDFFEDVGPL S+QLGG A++GL ++FNSYVN+LI ALP S+++E + E GNKIVR+A
Sbjct: 438 VQDFFEDVGPLHSLQLGGSAMDGLLKIFNSYVNLLISALPHSLDDEANLEGLGNKIVRVA 497
Query: 417 ETEAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRR 476
ETE QQ+AL ANASLLA+ELLPRAAMKLSS+N +D R+++ +RQNR EQREW+++
Sbjct: 498 ETEEQQLALFANASLLAEELLPRAAMKLSSVNHTGV-NDIRKKSVDRQNRVAEQREWKKK 556
Query: 477 LVGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWL 536
L VD+LKDSFCRQHAL LIFTED D+ L+A+MYI+M+ +E EW+PSLIFQ +++L
Sbjct: 557 LQRIVDKLKDSFCRQHALDLIFTEDDDTRLSAEMYINMDNTVEEPEWVPSLIFQVRINFL 616
>ref|NP_911258.1| unknown protein [Oryza sativa (japonica cultivar-group)]
gi|51963482|ref|XP_506414.1| PREDICTED OJ1092_A07.123
gene product [Oryza sativa (japonica cultivar-group)]
gi|27817901|dbj|BAC55667.1| unknown protein [Oryza
sativa (japonica cultivar-group)]
Length = 773
Score = 380 bits (975), Expect = e-103
Identities = 253/735 (34%), Positives = 388/735 (52%), Gaps = 60/735 (8%)
Query: 4 SVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSDGFS 61
++ + Y AF+R S+E+ + E EL ++ +S Q L+ L GV ++ D
Sbjct: 57 NMQSKYHAFLRISEEVVEAEQELIELQKHVSAQGILVQDLMSGVCRELEMWQKHCKDEHV 116
Query: 62 VNGALDSEHKEISDLDKW--LVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSL 119
L +E EI D V F D LD LLAE ++EEAL AL+ E+ +
Sbjct: 117 EEKDLQTELDEILSYDTQDSKVSFLDKLDTLLAEHKIEEALLALETEEKKCMATDDPGKE 176
Query: 120 NPSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNA 179
+ + + ++++++R+ L DQL + QPS ELR S+S L K+G G AH +LL A
Sbjct: 177 LDAEISTYKTALSKRKSILEDQLVRYSEQPSLSITELRKSLSGLIKIGKGSLAHQVLLKA 236
Query: 180 HLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWA 239
+ R Q N+++ P+ + Y Y+A L+++VFSA+++ + +S ++FG+ P + ++ WA
Sbjct: 237 YGSRLQKNVEAFLPTCSIYTETYSATLSKIVFSAISKVSKESSSLFGDSPMNLNRIIQWA 296
Query: 240 TKQTEAFALLVKRHA-LASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRP 298
+ E FA LVK ++ L S +A LR+A C+Q +L HCS LE+ GL +L+ L P
Sbjct: 297 EYEIETFARLVKENSPLPESVSA--LRSACICIQTSLTHCSYLESYGLKFSNLLMVLLHP 354
Query: 299 SVEQALDANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQ 358
VE+ L+ N +R++ A DD +L P +R S+ A + LTSS +F +V
Sbjct: 355 YVEEVLELNFRRLRRKIVDSAKNDDILLPSPQEGSR-LSSSVAPNIMLTSSGKKFMSIVN 413
Query: 359 DFFEDVGPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMAET 418
D + + P+ + GG L Q+F+ YV LI+ LP + E++ E S I AE+
Sbjct: 414 DVLDQITPMTIVHFGGTILNKFVQLFDKYVEALIEVLPGASEDDHLVE-SKEPIEFKAES 472
Query: 419 EAQQIALLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHP-EQREWRRRL 477
+AQQI L+ A +ADELLP A K + + + + + E +EW+R L
Sbjct: 473 DAQQIQLIGTAYTVADELLPAAVSKFFDIQTEKKRIGGTGEGLGSGSIYSIEYKEWKRSL 532
Query: 478 VGSVDRLKDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEW----IPSLIFQSVV 533
S+D+L+D FC Q+ LS I+ E G S L A MY +E D++ W PSL FQ+
Sbjct: 533 QHSLDKLRDHFCLQYVLSFIYLE-GKSRLDARMY--LELKTDDLLWECDPSPSLPFQA-- 587
Query: 534 SWLLIFKNYRIRQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATL 593
LF+KL ++A++A D+ +G+E+ +
Sbjct: 588 -----------------------------------LFVKLRQLASVAGDVLLGKEKIQKV 612
Query: 594 LLMRLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQ 653
LL RLTETV++W+S +Q FWD E+ L P GLQQ LDM FVV A GRY R +Q
Sbjct: 613 LLSRLTETVVMWLSNEQEFWDVFEDQSIQLRPSGLQQLILDMHFVVEIAVCGRYPHRPVQ 672
Query: 654 RIVNEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERLSGKPKEINGERELSSPTA 713
++V+ II +A++AFS +DP S LPEDEWF ++ + A+ + G E EL P
Sbjct: 673 QLVSVIITRAIAAFSVRNVDPQSSLPEDEWFLDMAKVAINKQLGTS---GSESELEEPVV 729
Query: 714 ---SVSAQSISSVRS 725
+S SS+ S
Sbjct: 730 VHDEISDSEESSISS 744
>ref|NP_563863.1| expressed protein [Arabidopsis thaliana] gi|4914320|gb|AAD32868.1|
F14N23.6 [Arabidopsis thaliana]
Length = 769
Score = 363 bits (931), Expect = 1e-98
Identities = 236/709 (33%), Positives = 372/709 (52%), Gaps = 59/709 (8%)
Query: 9 YAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSDGFSVNGAL 66
Y AF+R S+E ++E EL +R +S+Q L+ L GV +D + D
Sbjct: 59 YLAFLRISEEAVEMEHELVELRKHISSQGILVQDLMAGVCREMDDWNRLPGDVHDAEVEE 118
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
D E++D EF + +D+LLAE +V+EAL A+D ER + K ++ S
Sbjct: 119 DPLPNEVTDPKS---EFLEKIDLLLAEHKVDEALEAMDAEERSSPDLKGSVEMS-----S 170
Query: 127 LQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAHLQRYQY 186
+S+ ER+ L DQL A QPS AEL+ ++ L +LG GP AH LLL + +
Sbjct: 171 YKSAFMERKAVLEDQLLRIAKQPSICVAELKHALIGLIRLGKGPSAHQLLLKFYATSLRR 230
Query: 187 NMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEE--PAYSSELVMWATKQTE 244
+++ PS + + A L++LVFS ++ A +S A+FG++ PAYS+++V WA ++ E
Sbjct: 231 RIEAFLPSCLTCPNTFPATLSKLVFSNISVATKESAAMFGDDDNPAYSNKVVQWAEREVE 290
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
LVK +A + S A LRAA+ C+Q L +C +LE +GL L + L LFRP VE+ L
Sbjct: 291 YLVRLVKENA-SPSETASALRAASICLQDCLNYCKVLEPQGLFLSKLFLVLFRPYVEEVL 349
Query: 305 DANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDFFEDV 364
+ N +R + + D+ + + V + A +T + RF +VQD E +
Sbjct: 350 ELNFRRARRVIFDLNETDEGLESPSDFVTILSEFAIASDTMMTDCSIRFMQIVQDILEQL 409
Query: 365 GPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMAETEAQQIA 424
L+ + G L + Q+++ Y++ LIKALP +E+ E N ++ AET+++Q+A
Sbjct: 410 THLVVLHFGESVLTRILQLYDKYIDFLIKALPGHSDEDGLPELQDNTVLARAETDSEQLA 469
Query: 425 LLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLVGSVDRL 484
LL A + DELLPR+ +K+ L + + + PE +EW+R +V + D+L
Sbjct: 470 LLGAAFTILDELLPRSLVKVWKLQIENGGGEGENSAALNSSAAPELKEWKRHMVQAFDKL 529
Query: 485 KDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLIFKNYRI 544
++ FC Q LS I++ +G + L A +Y++ D++ +PSL FQ+
Sbjct: 530 RNYFCLQFVLSFIYSREGLTRLDALIYLT--ETPDDLH-LPSLPFQA------------- 573
Query: 545 RQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRLTETVIL 604
LF KL ++A IA D+ +G+E+ +LL RLTETVI+
Sbjct: 574 ------------------------LFSKLQQLAIIAGDVLLGKEKLQKILLARLTETVII 609
Query: 605 WISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEIIRKAM 664
W+S +Q FW E+ PL P GLQQ LDM F V A Y + +Q + +I +A+
Sbjct: 610 WLSNEQEFWSAFEDESNPLQPSGLQQLILDMNFTVEIARFAGYPFKVVQNHASVVINRAI 669
Query: 665 SAFSATGMDPYSDLPEDEWFNEICQDAMERL------SGKPKEINGERE 707
+ FS G++P S LP+ EWF E + A+ RL + +P+E E E
Sbjct: 670 NIFSERGINPQSSLPKTEWFTEAAKSAINRLLMGSEDASEPEEYECEEE 718
>gb|AAL06945.1| At1g10180/F14N23_6 [Arabidopsis thaliana]
Length = 769
Score = 360 bits (924), Expect = 9e-98
Identities = 235/709 (33%), Positives = 371/709 (52%), Gaps = 59/709 (8%)
Query: 9 YAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV--HIDSLSISDSDGFSVNGAL 66
Y AF+R S+E ++E EL +R +S+Q L+ L GV +D + D
Sbjct: 59 YLAFLRISEEAVEMEHELVELRKHISSQGILVQDLMAGVCREMDDWNRLPGDVHDAEVEE 118
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
D E++D EF + +D+LLAE +V+EAL A+D ER + K ++ S
Sbjct: 119 DPLPNEVTDPKS---EFLEKIDLLLAEHKVDEALEAMDAEERSSPDLKGSVEMS-----S 170
Query: 127 LQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLNAHLQRYQY 186
+S+ ER+ L DQL A QPS AEL+ ++ L +LG GP AH LLL + +
Sbjct: 171 YKSAFMERKAVLEDQLLRIAKQPSICVAELKHALIGLIRLGKGPSAHQLLLKFYATSLRR 230
Query: 187 NMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEE--PAYSSELVMWATKQTE 244
+++ PS + + A L++LVFS ++ A +S A+FG++ PAYS+++V WA ++ E
Sbjct: 231 RIEAFLPSCLTCPNTFPATLSKLVFSNISVATKESAAMFGDDDNPAYSNKVVQWAEREVE 290
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
LVK +A + S A LRAA+ C+Q L +C +LE +GL L + L LFRP VE+ L
Sbjct: 291 YLVRLVKENA-SPSETASALRAASICLQDCLNYCKVLEPQGLFLSKLFLVLFRPYVEEVL 349
Query: 305 DANLKRIQESTAAMAAADDWVLTYPPNVNRQTGSTTAFQLKLTSSAHRFNLMVQDFFEDV 364
+ N +R + + D+ + + V + A +T + RF +VQD E +
Sbjct: 350 ELNFRRARRVIFDLNETDEGLESPSDFVTILSEFAIASDTMMTDCSIRFMQIVQDILEQL 409
Query: 365 GPLLSMQLGGQALEGLFQVFNSYVNMLIKALPESMEEEESFEDSGNKIVRMAETEAQQIA 424
L+ + G L + Q+++ Y++ LIKALP +E+ E N ++ AET+++Q+A
Sbjct: 410 THLVVLHFGESVLTRILQLYDKYIDFLIKALPGHSDEDGLPELQDNTVLARAETDSEQLA 469
Query: 425 LLANASLLADELLPRAAMKLSSLNQDPYKDDNRRRTTERQNRHPEQREWRRRLVGSVDRL 484
LL A + DELLPR+ +K+ L + + + PE +EW+R +V + D+L
Sbjct: 470 LLGAAFTILDELLPRSLVKVWKLQIENGGGEGENSAALNSSAAPELKEWKRHMVQAFDKL 529
Query: 485 KDSFCRQHALSLIFTEDGDSHLTADMYISMERNADEVEWIPSLIFQSVVSWLLIFKNYRI 544
++ FC Q LS I++ +G + L A +Y++ D++ +PSL FQ+
Sbjct: 530 RNYFCLQFVLSFIYSREGLTRLDALIYLT--ETPDDLH-LPSLPFQA------------- 573
Query: 545 RQQIDDLSIHIFLIVECLVHSFEELFIKLNRMANIAADMFVGRERFATLLLMRLTETVIL 604
LF KL ++A IA D+ +G+E+ +LL RLTETVI+
Sbjct: 574 ------------------------LFSKLQQLAIIAGDVLLGKEKLQKILLARLTETVII 609
Query: 605 WISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQRIVNEIIRKAM 664
W+S +Q FW E+ PL P GLQQ L M F V A Y + +Q + +I +A+
Sbjct: 610 WLSNEQEFWSAFEDESNPLQPSGLQQLILGMNFTVEIARFAGYPFKVVQNHASVVINRAI 669
Query: 665 SAFSATGMDPYSDLPEDEWFNEICQDAMERL------SGKPKEINGERE 707
+ FS G++P S LP+ EWF E + A+ RL + +P+E E E
Sbjct: 670 NIFSERGINPQSSLPKTEWFTEAAKSAINRLLMGSEDASEPEEYECEEE 718
>dbj|BAD94781.1| hypothetical protein [Arabidopsis thaliana]
Length = 171
Score = 95.1 bits (235), Expect = 7e-18
Identities = 50/120 (41%), Positives = 69/120 (56%), Gaps = 6/120 (5%)
Query: 594 LLMRLTETVILWISEDQSFWDDIEEGPRPLGPLGLQQFYLDMKFVVCFASNGRYLSRNLQ 653
LL RLTETVI+W+S +Q FW E+ PL P GLQQ LDM F V A Y + +Q
Sbjct: 1 LLARLTETVIIWLSNEQEFWSAFEDESNPLQPSGLQQLILDMNFTVEIARFAGYPFKVVQ 60
Query: 654 RIVNEIIRKAMSAFSATGMDPYSDLPEDEWFNEICQDAMERL------SGKPKEINGERE 707
+ +I +A++ FS G++P S LP+ EWF E + A+ RL + +P+E E E
Sbjct: 61 NHASVVINRAINIFSERGINPQSSLPKTEWFTEAAKSAINRLLMGSEDASEPEEYECEEE 120
>ref|XP_641452.1| hypothetical protein DDB0205917 [Dictyostelium discoideum]
gi|60469474|gb|EAL67467.1| hypothetical protein
DDB0205917 [Dictyostelium discoideum]
Length = 815
Score = 73.9 bits (180), Expect = 2e-11
Identities = 68/327 (20%), Positives = 146/327 (43%), Gaps = 20/327 (6%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSIS-DSDG 59
+++ VY N+ FI SKEI++ E ++ RNL+S ++ L ++SIS D
Sbjct: 151 LKKDVYKNHLIFIGASKEIANSEVDMLDFRNLISDYGNVMSSL------QNISISWDHYK 204
Query: 60 FSVNGALDSEH-KEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKS 118
+G +D E ++ +WL P+ L V + +R E A+ +++ ++ +++
Sbjct: 205 VKKSGKIDFEPLSPATEPIQWLTTAPNELSVSIEQREFEVAVGLVEKINKIYESNPKVEI 264
Query: 119 LNPSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRASVSALKKLGDGPHAHSLLLN 178
+ + L+ I + + L D+L P + +++ ++S L +L A S+ L
Sbjct: 265 VMQT--HPLKDQIENKVKILTDKLMNELRSPLLKANQIKDTISLLVRLSQNDKAKSIFLE 322
Query: 179 AHLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAY-SSELVM 237
+ ++ + S + LA+++F+++ +D F P+Y +S LV
Sbjct: 323 SRSHSINQAIKKIVFSGDL--NRFIGELARVIFNSINSTCNDFTNSF---PSYMNSGLVS 377
Query: 238 WATKQTEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFR 297
W ++ + + R + ++ ++I HC +++ GL++ L +
Sbjct: 378 WIIEELVLISDIFNRQVF----ILDNFYSISQAIRIIESHCEMMDQTGLSIGFYWNLLLQ 433
Query: 298 PSVEQALDANLKRIQESTAAMAAADDW 324
P VEQ + +I++S + W
Sbjct: 434 PHVEQLIVNYEIKIRDSMLHQLMDEKW 460
>gb|AAH86283.1| Unknown (protein for MGC:83775) [Xenopus laevis]
Length = 685
Score = 72.8 bits (177), Expect = 4e-11
Identities = 66/249 (26%), Positives = 109/249 (43%), Gaps = 13/249 (5%)
Query: 79 WLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQSSITERRQKL 138
W+ E P+ LDV +A+R E A+ LD ++ S ++ +P + L+S + ER ++L
Sbjct: 297 WIQELPEDLDVCIAQRNFEGAVDLLD---KLNSYLEDKPLTHP--VKELKSKVDERVRQL 351
Query: 139 ADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQRYQYNMQSLRPSN 195
D L E + S RG R +VS L +LG A L L Q ++ LR
Sbjct: 352 TDVLVFELSPDRSLRGGPKATRRAVSQLVRLGQSTKACELFLKNQAAAVQTAIRQLRIEG 411
Query: 196 TSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWATKQTEAFALLVKRHAL 255
+ Y L + F+++ + A + F E S ++W+ + F +
Sbjct: 412 ATL--LYIHKLCNVFFTSLLETAKEFEMDFAENHGCYSAFIVWSRLALKMFVDAFSKQVF 469
Query: 256 ASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQALDANLKRIQEST 315
S + L AECV++A HC L GL L +L L ++ AL + + E+T
Sbjct: 470 DSKES---LSTVAECVKVAKEHCKQLSEIGLDLTFILHTLLVKDIKAALQSYKDIVIEAT 526
Query: 316 AAMAAADDW 324
+ + W
Sbjct: 527 KHRNSEEMW 535
Score = 43.1 bits (100), Expect = 0.032
Identities = 18/47 (38%), Positives = 32/47 (67%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV 47
++R+VY NY FI T+KEIS LEGE+ + ++L+ Q +++ + +
Sbjct: 57 LKRNVYQNYRQFIETAKEISYLEGEMYQLSHILTEQKSIMESVTQAL 103
>ref|NP_001006404.1| similar to Expressed sequence AI414418 [Gallus gallus]
gi|53133842|emb|CAG32250.1| hypothetical protein [Gallus
gallus]
Length = 708
Score = 70.9 bits (172), Expect = 1e-10
Identities = 68/261 (26%), Positives = 111/261 (42%), Gaps = 14/261 (5%)
Query: 68 SEHKEISDLD-KWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
S +E DL +W+ E P+ LDV +A+R E A+ LD+ +++ + +
Sbjct: 308 SAEEEAVDLSLEWIQELPEDLDVCIAQRDFEGAVDLLDKLNEYLADKPVSQPVK-----E 362
Query: 127 LQSSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQR 183
L++ + ER ++L D L E + S RG R +VS L +LG A L L
Sbjct: 363 LRAKVDERVRQLTDVLVFELSPDRSLRGGPRATRRAVSQLIRLGQSTKACELFLKNRAAA 422
Query: 184 YQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIFGEEPAYSSELVMWATKQT 243
++ LR + Y L + F+++ + A + F S V+WA
Sbjct: 423 VHTAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFETDFAGNNGCYSAFVVWARSSM 480
Query: 244 EAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQA 303
F + S + L AAECV++A HC L GL L ++ L ++ A
Sbjct: 481 RMFVDAFSKQVFDSKES---LSTAAECVKVAKEHCKQLSDIGLDLTFIIHALLVKDIKGA 537
Query: 304 LDANLKRIQESTAAMAAADDW 324
L + I E+T + + W
Sbjct: 538 LQSYKDIIIEATKHRNSEEMW 558
Score = 38.5 bits (88), Expect = 0.79
Identities = 16/47 (34%), Positives = 30/47 (63%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGV 47
++R+VY NY FI T++EIS LE E+ + ++L+ Q ++ + +
Sbjct: 60 LKRNVYQNYRQFIETAREISYLESEMYQLSHILTEQKGIMEAVTQAL 106
>ref|NP_932771.1| exocyst complex component 8 [Mus musculus]
gi|34784278|gb|AAH57052.1| Exoc8 protein [Mus musculus]
Length = 716
Score = 68.2 bits (165), Expect = 9e-10
Identities = 66/262 (25%), Positives = 115/262 (43%), Gaps = 14/262 (5%)
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
++E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P +
Sbjct: 315 EAEEEKVDLSMEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPPVKE 369
Query: 127 LQSSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQR 183
L++ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 370 LRAKVDERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAA 429
Query: 184 YQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIF-GEEPAYSSELVMWATKQ 242
++ LR + Y L + F+++ + A + F G + S V+WA
Sbjct: 430 VHTAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFETDFAGTDSGCYSAFVVWARSA 487
Query: 243 TEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQ 302
F + S + L AAECV++A HC L GL L ++ L ++
Sbjct: 488 MGMFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGEIGLDLTFIIHALLVKDIQG 544
Query: 303 ALDANLKRIQESTAAMAAADDW 324
AL + + I E+T + + W
Sbjct: 545 ALHSYKEIIIEATKHRNSEEMW 566
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 55 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 97
>ref|NP_620612.1| exocyst complex component 8 [Rattus norvegicus]
gi|2827164|gb|AAC01581.1| exo84 [Rattus norvegicus]
gi|7513969|pir||T09222 exocyst complex protein Exo84 -
rat
Length = 716
Score = 67.4 bits (163), Expect = 2e-09
Identities = 66/262 (25%), Positives = 115/262 (43%), Gaps = 14/262 (5%)
Query: 67 DSEHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLS 126
++E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P +
Sbjct: 315 EAEEEKVDLSMEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPSVKE 369
Query: 127 LQSSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQR 183
L++ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 370 LRAKVDERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAA 429
Query: 184 YQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIF-GEEPAYSSELVMWATKQ 242
++ LR + Y L + F+++ + A + F G + S V+WA
Sbjct: 430 VHTAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFETDFAGTDSGCYSAFVVWARSA 487
Query: 243 TEAFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQ 302
F + S + L AAECV++A HC L GL L ++ L ++
Sbjct: 488 MGMFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGEIGLDLTFIIHALLVKDIQG 544
Query: 303 ALDANLKRIQESTAAMAAADDW 324
AL + + I E+T + + W
Sbjct: 545 ALLSYKEIIIEATKHRNSEEMW 566
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 55 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 97
>emb|CAI23095.1| exocyst complex component 8 [Homo sapiens]
Length = 721
Score = 66.6 bits (161), Expect = 3e-09
Identities = 65/260 (25%), Positives = 113/260 (43%), Gaps = 14/260 (5%)
Query: 69 EHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQ 128
E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P + L+
Sbjct: 322 EEEKVDLSMEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPPVKELR 376
Query: 129 SSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQRYQ 185
+ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 377 AKVEERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAAVH 436
Query: 186 YNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASD-SLAIFGEEPAYSSELVMWATKQTE 244
++ LR + Y L + F+++ + A + + G + S V+WA
Sbjct: 437 TAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFEIDFAGTDSGCYSAFVVWARSAMG 494
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
F + S + L AAECV++A HC L GL L ++ L ++ AL
Sbjct: 495 MFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGDIGLDLTFIIHALLVKDIQGAL 551
Query: 305 DANLKRIQESTAAMAAADDW 324
+ + I E+T + + W
Sbjct: 552 HSYKEIIIEATKHRNSEEMW 571
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 55 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 97
>ref|XP_546091.1| PREDICTED: hypothetical protein XP_546091 [Canis familiaris]
Length = 668
Score = 66.6 bits (161), Expect = 3e-09
Identities = 66/260 (25%), Positives = 113/260 (43%), Gaps = 14/260 (5%)
Query: 69 EHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQ 128
E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P + L+
Sbjct: 269 EEEKVDLSVEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPPVKELR 323
Query: 129 SSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQRYQ 185
+ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 324 AKVDERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAAVH 383
Query: 186 YNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASDSLAIF-GEEPAYSSELVMWATKQTE 244
++ LR + Y L + F+++ + A + F G + S V+WA
Sbjct: 384 TAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFETDFAGTDSGCYSAFVVWARSAMG 441
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
F + S + L AAECV++A HC L GL L ++ L ++ AL
Sbjct: 442 MFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGDIGLDLTFIIHALLVKDIQGAL 498
Query: 305 DANLKRIQESTAAMAAADDW 324
+ + I E+T + + W
Sbjct: 499 HSYKEIIIEATKHRNSEEMW 518
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 55 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 97
>ref|XP_513085.1| PREDICTED: hypothetical protein XP_513085 [Pan troglodytes]
Length = 725
Score = 66.6 bits (161), Expect = 3e-09
Identities = 65/260 (25%), Positives = 113/260 (43%), Gaps = 14/260 (5%)
Query: 69 EHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQ 128
E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P + L+
Sbjct: 326 EEEKVDLSMEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPPVKELR 380
Query: 129 SSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQRYQ 185
+ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 381 AKVEERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAAVH 440
Query: 186 YNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASD-SLAIFGEEPAYSSELVMWATKQTE 244
++ LR + Y L + F+++ + A + + G + S V+WA
Sbjct: 441 TAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFEIDFAGTDSGCYSAFVVWARSAMG 498
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
F + S + L AAECV++A HC L GL L ++ L ++ AL
Sbjct: 499 MFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGDIGLDLTFIIHALLVKDIQGAL 555
Query: 305 DANLKRIQESTAAMAAADDW 324
+ + I E+T + + W
Sbjct: 556 HSYKEIIIEATKHRNSEEMW 575
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 59 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 101
>gb|AAH35763.2| Exocyst complex 84-kDa subunit [Homo sapiens]
gi|44921615|ref|NP_787072.2| exocyst complex 84-kDa
subunit [Homo sapiens]
Length = 725
Score = 66.6 bits (161), Expect = 3e-09
Identities = 65/260 (25%), Positives = 113/260 (43%), Gaps = 14/260 (5%)
Query: 69 EHKEISDLDKWLVEFPDLLDVLLAERRVEEALAALDEGERVVSEAKEMKSLNPSLLLSLQ 128
E +++ +W+ E P+ LDV +A+R E A+ LD+ + + +P + L+
Sbjct: 326 EEEKVDLSMEWIQELPEDLDVCIAQRDFEGAVDLLDKLNHYLEDKP-----SPPPVKELR 380
Query: 129 SSITERRQKLADQLA-EAACQPSTRGAE--LRASVSALKKLGDGPHAHSLLLNAHLQRYQ 185
+ + ER ++L + L E + S RG R +VS L +LG A L L
Sbjct: 381 AKVEERVRQLTEVLVFELSPDRSLRGGPKATRRAVSQLIRLGQCTKACELFLRNRAAAVH 440
Query: 186 YNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAASD-SLAIFGEEPAYSSELVMWATKQTE 244
++ LR + Y L + F+++ + A + + G + S V+WA
Sbjct: 441 TAIRQLRIEGATL--LYIHKLCHVFFTSLLETAREFEIDFAGTDSGCYSAFVVWARSAMG 498
Query: 245 AFALLVKRHALASSAAAGGLRAAAECVQIALGHCSLLEARGLALCPVLLKLFRPSVEQAL 304
F + S + L AAECV++A HC L GL L ++ L ++ AL
Sbjct: 499 MFVDAFSKQVFDSKES---LSTAAECVKVAKEHCQQLGDIGLDLTFIIHALLVKDIQGAL 555
Query: 305 DANLKRIQESTAAMAAADDW 324
+ + I E+T + + W
Sbjct: 556 HSYKEIIIEATKHRNSEEMW 575
Score = 39.3 bits (90), Expect = 0.46
Identities = 17/43 (39%), Positives = 29/43 (66%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGL 43
++R+VY NY FI T++EIS LE E+ + +LL+ Q + + +
Sbjct: 59 LKRNVYQNYRQFIETAREISYLESEMYQLSHLLTEQKSSLESI 101
>gb|EAK85236.1| hypothetical protein UM04147.1 [Ustilago maydis 521]
gi|49075386|ref|XP_401762.1| hypothetical protein
UM04147.1 [Ustilago maydis 521]
Length = 809
Score = 66.2 bits (160), Expect = 4e-09
Identities = 67/294 (22%), Positives = 126/294 (42%), Gaps = 17/294 (5%)
Query: 45 DGVHIDSLS-ISDSDGFSVNG-----ALDSEHKEISDLDKWLVEFPDLLDVLLAERRVEE 98
DG + S+S D D V G A S E D +W +F D L V +A R ++
Sbjct: 479 DGGLLTSVSEAGDGDAMQVGGGMMAAAAASAASERKDPGRWNNDFADELSVCIALREWDQ 538
Query: 99 ALAALDEGERVVSEAKEMKSLNPSLLLSLQSSITERRQKLADQLAEAACQPSTRGAELRA 158
A+ +++G V+S S N + L + +T +L ++ + + + +
Sbjct: 539 AVTLIEKGRAVLS---TYTSTNDASWRDLSAKLTSLTSELVSAISLDFVRQHLKKSLVTR 595
Query: 159 SVSALKKLGDGPHAHSLLLNAHLQRYQYNMQSLRPSNTSYGGAYTAALAQLVFSAVAQAA 218
+ S L +L G A L L+A + + + ++ + G Y + LA + F+ + +
Sbjct: 596 NASYLLRLDQGEKARQLFLDARTESLKKRTRQIKFEGDT--GLYISELAMVHFALIKNTS 653
Query: 219 SDSLAIFGEEPAYSSELVMWATKQTEAFALLVKRHALA-----SSAAAGGLRAAAECVQI 273
+A F ++ + +S V WA+++ E +A+L +R A +E +I
Sbjct: 654 EWYMAAF-KDGSMASGFVEWASERVEEYAVLFRRQVFGVQEQQDEAGTTNAEMVSEMREI 712
Query: 274 ALGHCSLLEARGLALCPVLLKLFRPSVEQALDANLKRIQESTAAMAAADDWVLT 327
+L + L GL +L +L +P D + + E+ + A+ VLT
Sbjct: 713 SLRLATQLNEVGLDFTFLLDRLLQPEPRTMADKHEQEEAEAGESDASVPTLVLT 766
Score = 37.0 bits (84), Expect = 2.3
Identities = 20/57 (35%), Positives = 31/57 (54%)
Query: 1 MRRSVYANYAAFIRTSKEISDLEGELSSIRNLLSTQATLIHGLADGVHIDSLSISDS 57
++R V+ NY+ FI SKEI+ LE ++ ++ LLS L L + S SD+
Sbjct: 225 LKRQVFKNYSEFITISKEIATLENDMLELKELLSEWKQLPQALELDDSSSAASFSDA 281
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.318 0.132 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,104,575,070
Number of Sequences: 2540612
Number of extensions: 43302941
Number of successful extensions: 139912
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 139762
Number of HSP's gapped (non-prelim): 169
length of query: 729
length of database: 863,360,394
effective HSP length: 136
effective length of query: 593
effective length of database: 517,837,162
effective search space: 307077437066
effective search space used: 307077437066
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144503.8


