

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144502.3 + phase: 0 /pseudo
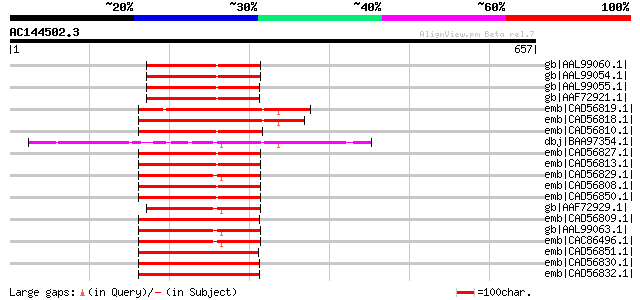
(657 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAL99060.1| NBS-LRR-Toll resistance gene analog protein [Medi... 250 1e-64
gb|AAL99054.1| NBS-LRR-Toll resistance gene analog protein [Medi... 223 2e-56
gb|AAL99055.1| NBS-LRR-Toll resistance gene analog protein [Medi... 222 3e-56
gb|AAF72921.1| resistance gene analog protein [Medicago sativa] 222 3e-56
emb|CAD56819.1| putative resistance gene analogue protein [Lens ... 209 2e-52
emb|CAD56818.1| putative resistance gene analogue protein [Lens ... 208 5e-52
emb|CAD56810.1| putative resistance gene analogue protein [Lens ... 199 2e-49
dbj|BAA97354.1| disease resistance protein-like [Arabidopsis tha... 198 5e-49
emb|CAD56827.1| putative resistance gene analogue protein [Lens ... 197 1e-48
emb|CAD56813.1| putative resistance gene analogue protein [Lens ... 196 3e-48
emb|CAD56829.1| putative resistance gene analogue protein [Lens ... 191 5e-47
emb|CAD56808.1| putative resistance gene analogue protein [Lens ... 191 8e-47
emb|CAD56850.1| putative resistance gene analogue protein [Lens ... 188 5e-46
gb|AAF72929.1| resistance gene analog protein [Medicago sativa] 180 1e-43
emb|CAD56809.1| putative resistance gene analogue protein [Lens ... 180 1e-43
gb|AAL99063.1| NBS-LRR-Toll resistance gene analog protein [Medi... 179 3e-43
emb|CAC86496.1| RGA-G protein [Cicer arietinum] 178 4e-43
emb|CAD56851.1| putative resistance gene analogue protein [Lens ... 177 7e-43
emb|CAD56830.1| putative resistance gene analogue protein [Lens ... 175 4e-42
emb|CAD56832.1| putative resistance gene analogue protein [Lens ... 174 8e-42
>gb|AAL99060.1| NBS-LRR-Toll resistance gene analog protein [Medicago edgeworthii]
Length = 171
Score = 250 bits (638), Expect = 1e-64
Identities = 128/143 (89%), Positives = 133/143 (92%), Gaps = 3/143 (2%)
Query: 172 LYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRR 231
LYDRISHQFGA CFIDDVSKMFRLHDGPLGVQKQILYQT EEHNQICNLSTASNLI+RR
Sbjct: 12 LYDRISHQFGARCFIDDVSKMFRLHDGPLGVQKQILYQTLSEEHNQICNLSTASNLIQRR 71
Query: 232 LCRQRVLLIFDNVDKVEQLEKIGVC-VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPL 290
LCRQRVLLI DNVDKVEQLEKIGVC W + GSKI+IISRDEHILKFFGVD+VYKVPL
Sbjct: 72 LCRQRVLLILDNVDKVEQLEKIGVCSEW--LGGGSKIVIISRDEHILKFFGVDKVYKVPL 129
Query: 291 LDRTNSLQLLCRKAFKLDHILSS 313
LDRTNSLQLLCRKAFKLDHIL+S
Sbjct: 130 LDRTNSLQLLCRKAFKLDHILNS 152
>gb|AAL99054.1| NBS-LRR-Toll resistance gene analog protein [Medicago sativa]
Length = 171
Score = 223 bits (568), Expect = 2e-56
Identities = 113/143 (79%), Positives = 127/143 (88%), Gaps = 3/143 (2%)
Query: 172 LYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRR 231
LYDRIS QFGACCFIDDVSK FRLHDGPLGVQKQIL QT GE+H+QICNL +ASNLIRRR
Sbjct: 12 LYDRISLQFGACCFIDDVSKTFRLHDGPLGVQKQILLQTLGEDHHQICNLYSASNLIRRR 71
Query: 232 LCRQRVLLIFDNVDKVEQLEKIGVC-VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPL 290
LCRQRVL+I DNVDKVEQ++KIGVC W + GS+I++ISRDEHILK +GVD VYKVPL
Sbjct: 72 LCRQRVLIILDNVDKVEQMKKIGVCREW--LGAGSRIVVISRDEHILKEYGVDVVYKVPL 129
Query: 291 LDRTNSLQLLCRKAFKLDHILSS 313
LD TNS +LLC+KAFKLDHI+SS
Sbjct: 130 LDWTNSRRLLCQKAFKLDHIMSS 152
>gb|AAL99055.1| NBS-LRR-Toll resistance gene analog protein [Medicago sativa]
Length = 170
Score = 222 bits (565), Expect = 3e-56
Identities = 112/142 (78%), Positives = 126/142 (87%), Gaps = 3/142 (2%)
Query: 172 LYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRR 231
LYDRIS QFGACCFIDDVSK FRLHDGPLGVQKQIL QT GE+H+QICNL +ASNLIRRR
Sbjct: 12 LYDRISLQFGACCFIDDVSKTFRLHDGPLGVQKQILLQTLGEDHHQICNLCSASNLIRRR 71
Query: 232 LCRQRVLLIFDNVDKVEQLEKIGVC-VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPL 290
LCRQRVL+I DNVDKVEQ++KIGVC W + GS+I++ISRDEHILK +GVD VYKVPL
Sbjct: 72 LCRQRVLIILDNVDKVEQMKKIGVCREW--LGAGSRIVVISRDEHILKEYGVDVVYKVPL 129
Query: 291 LDRTNSLQLLCRKAFKLDHILS 312
LD TNS +LLC+KAFKLDHI+S
Sbjct: 130 LDWTNSRRLLCQKAFKLDHIMS 151
>gb|AAF72921.1| resistance gene analog protein [Medicago sativa]
Length = 158
Score = 222 bits (565), Expect = 3e-56
Identities = 112/142 (78%), Positives = 126/142 (87%), Gaps = 3/142 (2%)
Query: 172 LYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRR 231
LYDRIS QFGACCFIDDVSK FRLHDGPLGVQKQIL QT GE+H+QICNL +ASNLIRRR
Sbjct: 5 LYDRISLQFGACCFIDDVSKTFRLHDGPLGVQKQILLQTLGEDHHQICNLCSASNLIRRR 64
Query: 232 LCRQRVLLIFDNVDKVEQLEKIGVC-VW*MVRWGSKIIIISRDEHILKFFGVDEVYKVPL 290
LCRQRVL+I DNVDKVEQ++KIGVC W + GS+I++ISRDEHILK +GVD VYKVPL
Sbjct: 65 LCRQRVLIILDNVDKVEQMKKIGVCREW--LGAGSRIVVISRDEHILKEYGVDVVYKVPL 122
Query: 291 LDRTNSLQLLCRKAFKLDHILS 312
LD TNS +LLC+KAFKLDHI+S
Sbjct: 123 LDWTNSRRLLCQKAFKLDHIMS 144
>emb|CAD56819.1| putative resistance gene analogue protein [Lens culinaris]
Length = 215
Score = 209 bits (532), Expect = 2e-52
Identities = 120/218 (55%), Positives = 149/218 (68%), Gaps = 7/218 (3%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDV+ +DGP+GVQKQIL+QT GEEHNQI NL
Sbjct: 2 GMGKTTLAGVLYGRISHQFNACCLIDDVASF--KYDGPIGVQKQILHQTLGEEHNQIYNL 59
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+III+SRD HILK +G
Sbjct: 60 HDGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNPK-SLGSGSRIIIVSRDAHILKEYG 118
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYR---AHF 338
V+ +YKV L+ TNSLQL CRKAFK D+I++ + Y I+ A L + +
Sbjct: 119 VNTLYKVQPLNHTNSLQLFCRKAFKCDNIMNDAYK-ELTYGILDIACGLPLAIKVIGSFL 177
Query: 339 CGRDIFEWKSALARLRDSPDKDVMDVLRLSFDGLEESE 376
GR I EW+SAL RLR+SP+KD+MD L+ SF GLE+ E
Sbjct: 178 FGRSISEWRSALTRLRESPNKDIMDALQFSFYGLEKME 215
>emb|CAD56818.1| putative resistance gene analogue protein [Lens culinaris]
Length = 217
Score = 208 bits (529), Expect = 5e-52
Identities = 118/211 (55%), Positives = 148/211 (69%), Gaps = 6/211 (2%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDVSK+F+ +DGP+GVQKQIL+QT GEEHNQI NL
Sbjct: 2 GVGKTTLAGVLYGRISHQFNACCLIDDVSKVFK-YDGPIGVQKQILHQTLGEEHNQIYNL 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+III+SRD HILK +G
Sbjct: 61 HDGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNPK-SLGSGSRIIIVSRDAHILKEYG 119
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMAYYIMLRAHR*QLKYR---AHF 338
V+ ++KV L+ TNSLQL CRKAFK D+I++ + Y I+ A L + +
Sbjct: 120 VNTLHKVQPLNHTNSLQLFCRKAFKCDNIMNDAYK-ELTYGILDIACGLPLAIKVIGSFL 178
Query: 339 CGRDIFEWKSALARLRDSPDKDVMDVLRLSF 369
GR I EW+SAL RLR+SP+KD+MD L+ SF
Sbjct: 179 FGRSISEWRSALTRLRESPNKDIMDALQFSF 209
>emb|CAD56810.1| putative resistance gene analogue protein [Lens culinaris]
Length = 171
Score = 199 bits (506), Expect = 2e-49
Identities = 107/156 (68%), Positives = 123/156 (78%), Gaps = 3/156 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFIDDVSK +RLHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GVGKTTLATVLYDRISQQFVACCFIDDVSKNYRLHDGPLGVQKQILDQTLGQDNHQIHNH 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILKFF 280
A+NLIR RLC Q+ L+I DNVD +EQLEKI V W + GS+IIIISRDEHILK +
Sbjct: 62 YNATNLIRCRLCHQKALMILDNVDHIEQLEKIAVHREW--LGAGSRIIIISRDEHILKQY 119
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKG 316
GVD VY+VPLLD TNSLQLLCRKAFKLDHIL+S +G
Sbjct: 120 GVDAVYQVPLLDSTNSLQLLCRKAFKLDHILNSYEG 155
>dbj|BAA97354.1| disease resistance protein-like [Arabidopsis thaliana]
Length = 1152
Score = 198 bits (503), Expect = 5e-49
Identities = 141/437 (32%), Positives = 229/437 (52%), Gaps = 45/437 (10%)
Query: 24 YDAFVTFRGEDTRNNFTYHLFDAFNREGILAFRDDTNLPKGESIASELLRAIEDSYIFVA 83
YD F +F GED R F HL +R+ I +F+D+ + + +SIA EL++ I+DS I +
Sbjct: 12 YDVFPSFSGEDVRKTFLSHLQLVLDRKLITSFKDN-EIERSQSIAPELVQGIKDSRIAIV 70
Query: 84 VLSRNYASSIWCLQELEKILECVHVSKKHVLPVFYDVDPPVVRKQSGIYCEAFVKHEQIF 143
+ S+NYASS WCL EL +I+ C + V+PVFY +DP VRKQ+G + AF +
Sbjct: 71 IFSKNYASSSWCLNELLEIVSCKEDKGQLVIPVFYALDPTHVRKQTGDFGMAFERTCLNK 130
Query: 144 QQDSQMVLRWREALTQVAGLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQ 203
+D + + WR ALT VA + G A C + +LH +Q
Sbjct: 131 TEDEKNL--WRVALTHVANILG-------------YHSAQCRANPDDYNMKLH-----LQ 170
Query: 204 KQILYQTHGEEHNQICNLSTASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRW 263
+ L G+++ +I +L + RL Q+VLL D++D+ L + ++W
Sbjct: 171 ETFLSTILGKQNIKIDHLGA----LGERLKHQKVLLFIDDLDQQVVLNALAG----QIQW 222
Query: 264 ---GSKIIIISRDEHILKFFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSSMKGWSMA 320
GS+II+++ D+H+L G++ +Y+V L + +L++LCR AF+ + K +A
Sbjct: 223 FGSGSRIIVVTNDKHLLISHGIENIYQVCLPSKELALEMLCRYAFRQNTPPDGFK--KLA 280
Query: 321 YYIMLRAHR*QLKYR---AHFCGRDIFEWKSALARLRDSPDKDVMDVLRLSFDGLEESEK 377
++ A L ++ GR+ W L RLR D + LR+ +DGL+ +
Sbjct: 281 VEVVRHAGILPLGLNVLGSYLRGRNKRYWMDMLPRLRKGLDGKIQKALRVGYDGLDNKKD 340
Query: 378 E-IFLHIACFFNPSMEKYVKNVLNCCGFHADIGLRVLIDKSLISIDESFSSLKEESISMH 436
E IF HIAC FN ++ +L + +IGL L+DKSL+++ + + MH
Sbjct: 341 EAIFRHIACLFNFEKVNDIRLLLADSDLNFNIGLENLVDKSLVNV-------RSNIVEMH 393
Query: 437 GLLEELGRKIVQENSSK 453
LL+E+GR+IV+ S++
Sbjct: 394 CLLQEMGREIVRAQSNE 410
>emb|CAD56827.1| putative resistance gene analogue protein [Lens culinaris subsp.
orientalis]
Length = 171
Score = 197 bits (500), Expect = 1e-48
Identities = 106/153 (69%), Positives = 121/153 (78%), Gaps = 3/153 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFIDDVSK +RLHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GVGKTTLATVLYDRISQQFVACCFIDDVSKNYRLHDGPLGVQKQILDQTLGQDNHQIHNH 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILKFF 280
A+NLIR RLC Q+ L+I DNVD +EQLEKI V W + GS+IIIISRDEHILK +
Sbjct: 62 YNATNLIRCRLCHQKALMILDNVDHIEQLEKIAVHREW--LGAGSRIIIISRDEHILKQY 119
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
GVD VY+VPLLD TNSLQLLCRKAFKLDHIL+S
Sbjct: 120 GVDAVYQVPLLDSTNSLQLLCRKAFKLDHILNS 152
>emb|CAD56813.1| putative resistance gene analogue protein [Lens culinaris]
Length = 171
Score = 196 bits (497), Expect = 3e-48
Identities = 106/153 (69%), Positives = 122/153 (79%), Gaps = 3/153 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFIDDVSK +RLHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GMGKTTLATVLYDRISQQFVACCFIDDVSKNYRLHDGPLGVQKQILDQTLGQDNHQIHNH 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILKFF 280
+A+NLIR RL RQ+ L+I DNVD +EQLEKI V W + GS+IIIISRDEHILK +
Sbjct: 62 YSATNLIRCRLSRQKALMILDNVDHIEQLEKIAVHREW--LGAGSRIIIISRDEHILKQY 119
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
GVD VY+VPLLD TNSLQLLCRKAFKLDHIL+S
Sbjct: 120 GVDAVYQVPLLDSTNSLQLLCRKAFKLDHILNS 152
>emb|CAD56829.1| putative resistance gene analogue protein [Lens ervoides]
Length = 171
Score = 191 bits (486), Expect = 5e-47
Identities = 104/155 (67%), Positives = 118/155 (76%), Gaps = 7/155 (4%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFIDDVSK + LHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GVGKTTLATVLYDRISQQFVACCFIDDVSKNYTLHDGPLGVQKQILDQTLGQDNHQIHNN 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRW---GSKIIIISRDEHILK 278
A+ LIR RLC Q+ L+I DNVD +EQLEKI V W GS+IIIISRDEHILK
Sbjct: 62 YNATYLIRCRLCHQKALMILDNVDHIEQLEKIAV----HREWLGPGSRIIIISRDEHILK 117
Query: 279 FFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
+GVD VY+VPLLD TNSLQLLCRKAFKLDHIL+S
Sbjct: 118 QYGVDAVYQVPLLDSTNSLQLLCRKAFKLDHILNS 152
>emb|CAD56808.1| putative resistance gene analogue protein [Lens culinaris]
Length = 171
Score = 191 bits (484), Expect = 8e-47
Identities = 104/153 (67%), Positives = 120/153 (77%), Gaps = 3/153 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFI+DVSK +RLHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GMGKTTLATVLYDRISQQFVACCFIEDVSKNYRLHDGPLGVQKQILDQTLGQDNHQIHNH 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILKFF 280
A+NLIR RL Q+ L+I DNVD +EQLEKI V W + GS+IIIISRDEHILK +
Sbjct: 62 YNATNLIRCRLSVQKALMILDNVDHIEQLEKIAVHREW--LGAGSRIIIISRDEHILKQY 119
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
GVD VY+VPLLD TNSLQLLCRKAFKLDHIL+S
Sbjct: 120 GVDAVYQVPLLDSTNSLQLLCRKAFKLDHILNS 152
>emb|CAD56850.1| putative resistance gene analogue protein [Lens odemensis]
Length = 171
Score = 188 bits (477), Expect = 5e-46
Identities = 103/153 (67%), Positives = 119/153 (77%), Gaps = 3/153 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LYDRIS QF ACCFI+DVSK +RLHDGPLGVQKQIL QT G++++QI N
Sbjct: 2 GMGKTTLATVLYDRISQQFVACCFIEDVSKNYRLHDGPLGVQKQILDQTLGQDNHQIHNH 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGV-CVW*MVRWGSKIIIISRDEHILKFF 280
A+NLIR RL Q+ L+I DNVD +EQLEKI V W + GS+IIIISRDEHILK +
Sbjct: 62 YNATNLIRCRLSVQKALMILDNVDHIEQLEKIAVHREW--LGAGSRIIIISRDEHILKQY 119
Query: 281 GVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
GVD VY+VPLLD TNS QLLCRKAFKLDHIL+S
Sbjct: 120 GVDAVYQVPLLDSTNSPQLLCRKAFKLDHILNS 152
>gb|AAF72929.1| resistance gene analog protein [Medicago sativa]
Length = 157
Score = 180 bits (456), Expect = 1e-43
Identities = 96/145 (66%), Positives = 114/145 (78%), Gaps = 8/145 (5%)
Query: 172 LYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNLSTASNLIRRR 231
LY RIS QF A CFIDD+SK++R HDGP+G QKQIL+QT G+EH QICNL +NLI+ R
Sbjct: 5 LYRRISSQFDARCFIDDLSKIYR-HDGPIGAQKQILHQTLGKEHFQICNLYDTTNLIQSR 63
Query: 232 LCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRW---GSKIIIISRDEHILKFFGVDEVYKV 288
L R R L++ DNVDKVEQL+K+GV +W GS+IIIISRDEHILK +G D VYKV
Sbjct: 64 LRRLRALIVVDNVDKVEQLDKLGV----NRQWLGAGSRIIIISRDEHILKEYGTDVVYKV 119
Query: 289 PLLDRTNSLQLLCRKAFKLDHILSS 313
PLL+ TNSLQL C+KAFKLDH +SS
Sbjct: 120 PLLNGTNSLQLFCQKAFKLDHTISS 144
>emb|CAD56809.1| putative resistance gene analogue protein [Lens culinaris]
Length = 171
Score = 180 bits (456), Expect = 1e-43
Identities = 96/151 (63%), Positives = 116/151 (76%), Gaps = 2/151 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDVSK+F+ +DGP+GVQKQIL+QT GEEHNQI NL
Sbjct: 2 GVGKTTLAGVLYGRISHQFNACCLIDDVSKVFK-YDGPIGVQKQILHQTLGEEHNQIYNL 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+III+SRD HILK +G
Sbjct: 61 HGGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNPK-SLGSGSRIIIVSRDAHILKEYG 119
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHILS 312
V+ +YKV L+ TNSLQLLCRKAFK D+I++
Sbjct: 120 VNTLYKVQPLNHTNSLQLLCRKAFKCDNIMN 150
>gb|AAL99063.1| NBS-LRR-Toll resistance gene analog protein [Medicago ruthenica]
Length = 172
Score = 179 bits (453), Expect = 3e-43
Identities = 96/155 (61%), Positives = 117/155 (74%), Gaps = 7/155 (4%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L +LY +ISH+F A CFIDDVSK+ RLHDGPL QKQIL+QT G EH+QICN
Sbjct: 2 GVGKTTLALDLYGQISHRFDASCFIDDVSKICRLHDGPLEAQKQILFQTLGIEHHQICNR 61
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRW---GSKIIIISRDEHILK 278
+A+ LIRRRLC +R L+IFDNVD+VEQLEKI + W GS+IIIISRDEHILK
Sbjct: 62 YSATYLIRRRLCHERALVIFDNVDQVEQLEKIAM----HREWFGAGSRIIIISRDEHILK 117
Query: 279 FFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
+GVDEVY+VPLL+ S +L C+KAFK + I+SS
Sbjct: 118 EYGVDEVYRVPLLNWAESHKLFCQKAFKHEKIISS 152
>emb|CAC86496.1| RGA-G protein [Cicer arietinum]
Length = 169
Score = 178 bits (452), Expect = 4e-43
Identities = 92/155 (59%), Positives = 117/155 (75%), Gaps = 8/155 (5%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY++ISHQF C I D+SK++R HDGP+G QKQIL QT GEE+ Q CNL
Sbjct: 2 GVGKTTLATVLYNKISHQFPVYCRIADLSKIYR-HDGPIGAQKQILLQTLGEEYLQTCNL 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRW---GSKIIIISRDEHILK 278
ASNLI+ RLCR++ ++I DNVD++EQLEK+ V W GS+++IISRDEHILK
Sbjct: 61 HNASNLIQNRLCREKAIIIIDNVDQIEQLEKLDV----RREWLGAGSRVVIISRDEHILK 116
Query: 279 FFGVDEVYKVPLLDRTNSLQLLCRKAFKLDHILSS 313
+GVD VYKVPLL++++SLQ CRKAFKLDHI+SS
Sbjct: 117 EYGVDVVYKVPLLNKSDSLQFFCRKAFKLDHIMSS 151
>emb|CAD56851.1| putative resistance gene analogue protein [Lens odemensis]
Length = 170
Score = 177 bits (450), Expect = 7e-43
Identities = 95/150 (63%), Positives = 114/150 (75%), Gaps = 2/150 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDVSK+F+ +DGP+GVQKQIL+QT GEEHNQI NL
Sbjct: 2 GVGKTTLAGVLYGRISHQFNACCLIDDVSKVFK-YDGPIGVQKQILHQTLGEEHNQIYNL 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+III+SRD HILK +G
Sbjct: 61 HGGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNRK-SLGSGSRIIIVSRDAHILKEYG 119
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHIL 311
V+ +YKV L+ TNSLQL CRKAFK D+I+
Sbjct: 120 VNTLYKVQPLNHTNSLQLFCRKAFKCDNIM 149
>emb|CAD56830.1| putative resistance gene analogue protein [Lens odemensis]
Length = 171
Score = 175 bits (444), Expect = 4e-42
Identities = 93/151 (61%), Positives = 114/151 (74%), Gaps = 2/151 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDVSK+F+ +DGP+GVQKQIL+QT GEEHNQI N
Sbjct: 2 GVGKTTLAGVLYGRISHQFNACCLIDDVSKVFK-YDGPIGVQKQILHQTLGEEHNQIYNP 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+II++SRD HILK +G
Sbjct: 61 HGGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNPK-SLGSGSRIIVVSRDAHILKEYG 119
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHILS 312
V+ +YKV L+ TNSLQL CRKAFK D+I++
Sbjct: 120 VNTLYKVQPLNHTNSLQLFCRKAFKCDNIMN 150
>emb|CAD56832.1| putative resistance gene analogue protein [Lens nigricans]
Length = 171
Score = 174 bits (441), Expect = 8e-42
Identities = 94/151 (62%), Positives = 114/151 (75%), Gaps = 2/151 (1%)
Query: 162 GLSGCDLRDNLYDRISHQFGACCFIDDVSKMFRLHDGPLGVQKQILYQTHGEEHNQICNL 221
G+ L LY RISHQF ACC IDDVSK+F+ +DGP+GVQKQIL+QT GEEHNQI NL
Sbjct: 2 GVGKTTLAGVLYGRISHQFNACCLIDDVSKVFK-YDGPIGVQKQILHQTLGEEHNQIYNL 60
Query: 222 STASNLIRRRLCRQRVLLIFDNVDKVEQLEKIGVCVW*MVRWGSKIIIISRDEHILKFFG 281
+NLI+ RLCR+R LLIFDNVD EQLEK+ V + GS+III+SRD HILK +G
Sbjct: 61 HDGTNLIQNRLCRRRALLIFDNVDHNEQLEKLAVNPK-SLGSGSRIIIVSRDAHILKEYG 119
Query: 282 VDEVYKVPLLDRTNSLQLLCRKAFKLDHILS 312
V+ +YKV L+ TNSLQL RKAFK D+I++
Sbjct: 120 VNTLYKVQPLNHTNSLQLFRRKAFKCDNIMN 150
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.333 0.145 0.451
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,032,272,554
Number of Sequences: 2540612
Number of extensions: 41385615
Number of successful extensions: 109183
Number of sequences better than 10.0: 1107
Number of HSP's better than 10.0 without gapping: 867
Number of HSP's successfully gapped in prelim test: 240
Number of HSP's that attempted gapping in prelim test: 104902
Number of HSP's gapped (non-prelim): 2513
length of query: 657
length of database: 863,360,394
effective HSP length: 135
effective length of query: 522
effective length of database: 520,377,774
effective search space: 271637198028
effective search space used: 271637198028
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 79 (35.0 bits)
Medicago: description of AC144502.3


