

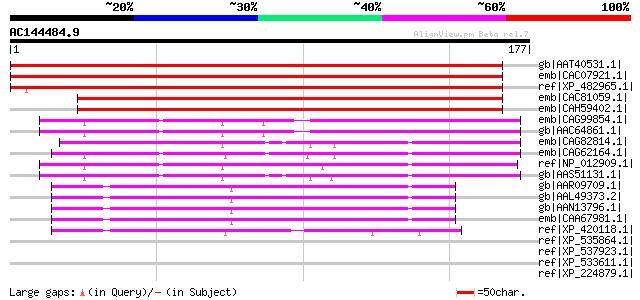
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144484.9 + phase: 0
(177 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAT40531.1| putative mitochondrial ATP synthase [Solanum demi... 297 1e-79
emb|CAC07921.1| putative protein [Arabidopsis thaliana] gi|21555... 290 2e-77
ref|XP_482965.1| putative mitochondrial F0 ATP synthase D chain ... 278 5e-74
emb|CAC81059.1| mitochondrial F0 ATP synthase D chain [Arabidops... 256 3e-67
emb|CAH59402.1| mitochondrial F0 ATP synthase delta chain [Plant... 227 1e-58
emb|CAG99854.1| ATP7_KLULA [Kluyveromyces lactis NRRL Y-1140] gi... 57 2e-07
gb|AAC64861.1| F1Fo-ATP synthase subunit 7 [Kluyveromyces lactis] 57 2e-07
emb|CAG82814.1| unnamed protein product [Yarrowia lipolytica CLI... 55 9e-07
emb|CAG62164.1| unnamed protein product [Candida glabrata CBS138... 54 1e-06
ref|NP_012909.1| Subunit d of the stator stalk of mitochondrial ... 52 7e-06
gb|AAS51131.1| ACL097Cp [Ashbya gossypii ATCC 10895] gi|45185591... 47 2e-04
gb|AAR09709.1| similar to Drosophila melanogaster ATPsyn-d [Dros... 47 3e-04
gb|AAL49373.2| RH59211p [Drosophila melanogaster] 47 3e-04
gb|AAN13796.1| CG6030-PB, isoform B [Drosophila melanogaster] gi... 47 3e-04
emb|CAA67981.1| mitochondrial ATP synthase [Drosophila melanogas... 46 4e-04
ref|XP_420118.1| PREDICTED: similar to Hypothetical protein MGC7... 46 5e-04
ref|XP_535864.1| PREDICTED: similar to H+-transporting two-secto... 45 0.001
ref|XP_537923.1| PREDICTED: similar to Chemokine receptor-like 2... 44 0.002
ref|XP_533611.1| PREDICTED: similar to myosin, heavy polypeptide... 44 0.002
ref|XP_224879.1| PREDICTED: similar to ATP synthase D chain, mit... 44 0.002
>gb|AAT40531.1| putative mitochondrial ATP synthase [Solanum demissum]
Length = 168
Score = 297 bits (760), Expect = 1e-79
Identities = 141/168 (83%), Positives = 158/168 (93%)
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSGTVKK+ DV FKAG+ IDW+GMAKLLVTDEAR+EF NLRRAFD+VN+QL+TKFSQEPE
Sbjct: 1 MSGTVKKIADVTFKAGRTIDWEGMAKLLVTDEARKEFSNLRRAFDDVNSQLQTKFSQEPE 60
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKE 120
PI+WEYYRKGIG+RLVDMYK+ YESIEIPKF DTVTPQYKPKFDALLVELKEAE++SLKE
Sbjct: 61 PINWEYYRKGIGSRLVDMYKEAYESIEIPKFEDTVTPQYKPKFDALLVELKEAEKQSLKE 120
Query: 121 SERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
SERLEKE+ VQ LKK++STMTA+EYFA+HPELKKKFDDEIRND WGY
Sbjct: 121 SERLEKEVAEVQELKKKLSTMTAEEYFAKHPELKKKFDDEIRNDYWGY 168
>emb|CAC07921.1| putative protein [Arabidopsis thaliana] gi|21555349|gb|AAM63838.1|
mitochondrial F0 ATP synthase D chain [Arabidopsis
thaliana] gi|20334872|gb|AAM16192.1| AT3g52300/T25B15_70
[Arabidopsis thaliana] gi|15215604|gb|AAK91347.1|
AT3g52300/T25B15_70 [Arabidopsis thaliana]
gi|15231176|ref|NP_190798.1| ATP synthase D
chain-related [Arabidopsis thaliana]
gi|11358239|pir||T46100 hypothetical protein T25B15.70 -
Arabidopsis thaliana gi|25089786|sp|Q9FT52|ATPQ_ARATH
ATP synthase D chain, mitochondrial
Length = 168
Score = 290 bits (741), Expect = 2e-77
Identities = 137/168 (81%), Positives = 153/168 (90%)
Query: 1 MSGTVKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPE 60
MSG KK+ DVAFKA + IDWDGMAK+LVTDEARREF NLRRAFDEVNTQL+TKFSQEPE
Sbjct: 1 MSGAGKKIADVAFKASRTIDWDGMAKVLVTDEARREFSNLRRAFDEVNTQLQTKFSQEPE 60
Query: 61 PIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKE 120
PIDW+YYRKGIG +VD YK+ Y+SIEIPK+VD VTP+YKPKFDALLVELKEAE+KSLKE
Sbjct: 61 PIDWDYYRKGIGAGIVDKYKEAYDSIEIPKYVDKVTPEYKPKFDALLVELKEAEQKSLKE 120
Query: 121 SERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
SERLEKEI +VQ + K++STMTADEYF +HPELKKKFDDEIRNDNWGY
Sbjct: 121 SERLEKEIADVQEISKKLSTMTADEYFEKHPELKKKFDDEIRNDNWGY 168
>ref|XP_482965.1| putative mitochondrial F0 ATP synthase D chain [Oryza sativa
(japonica cultivar-group)] gi|42407865|dbj|BAD09007.1|
putative mitochondrial F0 ATP synthase D chain [Oryza
sativa (japonica cultivar-group)]
gi|32352148|dbj|BAC78567.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 169
Score = 278 bits (711), Expect = 5e-74
Identities = 132/169 (78%), Positives = 155/169 (91%), Gaps = 1/169 (0%)
Query: 1 MSGT-VKKVTDVAFKAGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEP 59
MSG VKKV +VA KAGK IDW+GMAK+LV+DEAR+EF LRR F++VN QL+TKFSQEP
Sbjct: 1 MSGNGVKKVAEVAAKAGKAIDWEGMAKMLVSDEARKEFNTLRRTFEDVNHQLQTKFSQEP 60
Query: 60 EPIDWEYYRKGIGTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLK 119
+PIDWEYYRKGIG+++VDMYK+ YESIEIPK+VDTVTPQYKPKFDALLVELKEAE++SLK
Sbjct: 61 QPIDWEYYRKGIGSKVVDMYKEAYESIEIPKYVDTVTPQYKPKFDALLVELKEAEKESLK 120
Query: 120 ESERLEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
ESER+EKE+ +Q +KK ISTMTADEYFA+HPE+K+KFDDEIRNDNWGY
Sbjct: 121 ESERIEKELAELQEMKKNISTMTADEYFAKHPEVKQKFDDEIRNDNWGY 169
>emb|CAC81059.1| mitochondrial F0 ATP synthase D chain [Arabidopsis thaliana]
Length = 145
Score = 256 bits (653), Expect = 3e-67
Identities = 121/145 (83%), Positives = 135/145 (92%)
Query: 24 MAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHY 83
MAK+LVTDEARREF NLRRAFDEVNTQL+TKFSQEPEPIDW+YYRKGIG +VD YK+ Y
Sbjct: 1 MAKVLVTDEARREFSNLRRAFDEVNTQLQTKFSQEPEPIDWDYYRKGIGAGIVDKYKEAY 60
Query: 84 ESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTA 143
+SIEIPK+VD VTP+YKPKFDALLVELKEAE+KSLKESERLEKEI +VQ + K++STMTA
Sbjct: 61 DSIEIPKYVDKVTPEYKPKFDALLVELKEAEQKSLKESERLEKEIADVQEISKKLSTMTA 120
Query: 144 DEYFAEHPELKKKFDDEIRNDNWGY 168
DEYF +HPELKKKFDDEIRNDNWGY
Sbjct: 121 DEYFEKHPELKKKFDDEIRNDNWGY 145
>emb|CAH59402.1| mitochondrial F0 ATP synthase delta chain [Plantago major]
Length = 145
Score = 227 bits (578), Expect = 1e-58
Identities = 104/145 (71%), Positives = 132/145 (90%)
Query: 24 MAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTRLVDMYKQHY 83
MAK+LV+DEAR+EFF LRRAFD+VNTQL+TKFSQEP PIDWEYYRKG+G+RLVDMYKQ +
Sbjct: 1 MAKVLVSDEARKEFFALRRAFDDVNTQLQTKFSQEPVPIDWEYYRKGLGSRLVDMYKQAH 60
Query: 84 ESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLKKRISTMTA 143
+ I+IPK+VD VTP+Y KFDAL VE+++AE++SL+ ++RLEKEI +V+ LK ++STMTA
Sbjct: 61 DEIKIPKYVDNVTPEYTVKFDALSVEMQQAEQESLEVTKRLEKEIADVEELKNKVSTMTA 120
Query: 144 DEYFAEHPELKKKFDDEIRNDNWGY 168
DEYFA+HPE+K+KFD+EIRND WGY
Sbjct: 121 DEYFAKHPEVKEKFDEEIRNDYWGY 145
>emb|CAG99854.1| ATP7_KLULA [Kluyveromyces lactis NRRL Y-1140]
gi|50309515|ref|XP_454767.1| ATP7_KLULA [Kluyveromyces
lactis] gi|52788215|sp|O13350|ATP7_KLULA ATP synthase D
chain, mitochondrial
Length = 174
Score = 57.4 bits (137), Expect = 2e-07
Identities = 40/171 (23%), Positives = 79/171 (45%), Gaps = 13/171 (7%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + + L +T + + + ++ DE QL + +P +D+ +YR
Sbjct: 3 LAKSAANKLDWAKVISSLKLTGKTATQLSSFKKRNDEARRQL-LELQSQPTSVDFSHYRS 61
Query: 70 GI-GTRLVDMYKQHYES-----IEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESER 123
+ T +VD +Q Y+S +++ K + T+ F++ +E EK + + +
Sbjct: 62 VLKNTEVVDKIEQFYKSYKPVSVDVSKQLSTIEA-----FESQAIENAAETEKLVAQELK 116
Query: 124 LEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
KE +N + +T DE PE+ K ++ ++ W YKE+
Sbjct: 117 DLKETLNNIESARPFDQLTVDELTKARPEIDAKVEEMVKKGRWDVPGYKEK 167
>gb|AAC64861.1| F1Fo-ATP synthase subunit 7 [Kluyveromyces lactis]
Length = 174
Score = 57.4 bits (137), Expect = 2e-07
Identities = 40/171 (23%), Positives = 79/171 (45%), Gaps = 13/171 (7%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + + L +T + + + ++ DE QL + +P +D+ +YR
Sbjct: 3 LAKSAANKLDWAKVISSLKLTGKTATQLSSFKKRNDEARIQL-LELQSQPTSVDFSHYRS 61
Query: 70 GI-GTRLVDMYKQHYES-----IEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESER 123
+ T +VD +Q Y+S +++ K + T+ F++ +E EK + + +
Sbjct: 62 VLKNTEVVDKIEQFYKSYKPVSVDVSKQLSTIEA-----FESQAIENAAETEKLVAQELK 116
Query: 124 LEKEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
KE +N + +T DE PE+ K ++ ++ W YKE+
Sbjct: 117 DLKETLNNIESARPFDQLTVDELTKARPEIDAKVEEMVKKGRWDVPGYKEK 167
>emb|CAG82814.1| unnamed protein product [Yarrowia lipolytica CLIB99]
gi|50546112|ref|XP_500583.1| hypothetical protein
[Yarrowia lipolytica]
Length = 176
Score = 55.1 bits (131), Expect = 9e-07
Identities = 42/161 (26%), Positives = 76/161 (47%), Gaps = 7/161 (4%)
Query: 18 KIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGI-GTRLV 76
K+DW + L A R E + + +P +D+ +YRK + ++V
Sbjct: 12 KVDWGKIVSSLGLTGATVSSLQAFRKRHEEAKKNAYELQNQPTTVDFAHYRKVLKNQKVV 71
Query: 77 DMYKQHYESIEIPKFVDTVTPQYKP--KFDALLVE-LKEAEEKSLKESERLEKEIVNVQS 133
D +QH++S + P D V+ Q K F+A +E K E K +E L+K + N++S
Sbjct: 72 DEIEQHFKSFK-PVTYD-VSKQLKTIDAFEAKAIEDAKATEGKVNQEIGDLQKTLENIES 129
Query: 134 LKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
+ ++ D+ F P+L+KK ++ ++ W Y E+
Sbjct: 130 ARP-FDQLSVDDVFKARPDLEKKIEEMVKKGRWSVPGYNEK 169
>emb|CAG62164.1| unnamed protein product [Candida glabrata CBS138]
gi|50293565|ref|XP_449194.1| unnamed protein product
[Candida glabrata]
Length = 176
Score = 54.3 bits (129), Expect = 1e-06
Identities = 42/164 (25%), Positives = 80/164 (48%), Gaps = 7/164 (4%)
Query: 15 AGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG- 72
A K+DW + + L +T + + ++ DE QL + Q+P +D+ +YR +
Sbjct: 9 AANKLDWAKVISSLKLTGKTATQLSTFKKRNDEARRQL-FELEQQPVEVDFAHYRSVLNN 67
Query: 73 TRLVDMYKQHYESIEIPKFVDTVTPQYK-PKFDALLVE-LKEAEEKSLKESERLEKEIVN 130
+ +VD +++ +S P VDT F+ L +E KE EE KE + L++ + N
Sbjct: 68 SEVVDKIERYVKSYS-PVTVDTSKQLGAIASFEKLALENAKETEEIVSKELKALKETLAN 126
Query: 131 VQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
++S + +T DE PE+ ++ + ++N W Y ++
Sbjct: 127 IESARP-FDQLTVDELVKAKPEIDERVAELVKNGKWEIPSYYQK 169
>ref|NP_012909.1| Subunit d of the stator stalk of mitochondrial F1F0 ATP synthase,
which is a large, evolutionarily conserved enzyme
complex required for ATP synthesis; Atp7p [Saccharomyces
cerevisiae] gi|486005|emb|CAA81851.1| ATP7
[Saccharomyces cerevisiae] gi|395254|emb|CAA52265.1|
unnamed protein product [Saccharomyces cerevisiae]
gi|399076|sp|P30902|ATP7_YEAST ATP synthase D chain,
mitochondrial gi|45270588|gb|AAS56675.1| YKL016C
[Saccharomyces cerevisiae] gi|171112|gb|AAA34445.1| ATP
synthase d subunit
Length = 174
Score = 52.0 bits (123), Expect = 7e-06
Identities = 37/166 (22%), Positives = 74/166 (44%), Gaps = 5/166 (3%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + + L +T + + ++ DE QL + +P +D+ +YR
Sbjct: 3 LAKSAANKLDWAKVISSLRITGSTATQLSSFKKRNDEARRQL-LELQSQPTEVDFSHYRS 61
Query: 70 GI-GTRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDA-LLVELKEAEEKSLKESERLEKE 127
+ T ++D + + + + K + Q F+ + KE E KE + L+
Sbjct: 62 VLKNTSVIDKIESYVKQYKPVKIDASKQLQVIESFEKHAMTNAKETESLVSKELKDLQST 121
Query: 128 IVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKE 173
+ N+QS + +T D+ PE+ K ++ ++ W YK+
Sbjct: 122 LDNIQSARP-FDELTVDDLTKIKPEIDAKVEEMVKKGKWDVPGYKD 166
>gb|AAS51131.1| ACL097Cp [Ashbya gossypii ATCC 10895] gi|45185591|ref|NP_983307.1|
ACL097Cp [Eremothecium gossypii]
Length = 174
Score = 47.0 bits (110), Expect = 2e-04
Identities = 42/169 (24%), Positives = 79/169 (45%), Gaps = 9/169 (5%)
Query: 11 VAFKAGKKIDWDGM-AKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRK 69
+A A K+DW + A L +T + + ++ DE +L + +P +D+ +YR
Sbjct: 3 LARSAANKLDWAKVIASLKLTGKTAAQLTQFKKRNDEARRRL-FELESQPAEVDFAHYRA 61
Query: 70 GI-GTRLVDMYKQHYESIEIPKFVDTVTPQYKP--KFDALLV-ELKEAEEKSLKESERLE 125
+ T +VD + + P VD V+ Q F+A V + +E E +E + L+
Sbjct: 62 VLKNTAVVDQIEASCRAYR-PVTVD-VSKQISSLEAFEAQAVRDAQETEAVVGRELKALK 119
Query: 126 KEIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGYSHYKEE 174
+ + N+++ + +T DE A PE+ K + ++ W YKE+
Sbjct: 120 ETLANIEAARP-FDELTVDELAAARPEIDTKVAEMVKRGKWEVPGYKEK 167
>gb|AAR09709.1| similar to Drosophila melanogaster ATPsyn-d [Drosophila yakuba]
Length = 178
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 7 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 64
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + + E+ ++ S + + +KEI +++S
Sbjct: 65 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKSSQSEIDAYKKASEQRIQNYQKEIAHLKS 124
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y PE
Sbjct: 125 LLP-YDQMTMEDYRDAFPE 142
>gb|AAL49373.2| RH59211p [Drosophila melanogaster]
Length = 190
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 19 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 76
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + A E+ ++ S + + +KEI +++S
Sbjct: 77 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKASQSEIDAYKKASEQRIQNYQKEIAHLKS 136
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y P+
Sbjct: 137 LLP-YDQMTMEDYRDAFPD 154
>gb|AAN13796.1| CG6030-PB, isoform B [Drosophila melanogaster]
gi|7300478|gb|AAF55633.1| CG6030-PA, isoform A
[Drosophila melanogaster] gi|24648125|ref|NP_732400.1|
CG6030-PB, isoform B [Drosophila melanogaster]
gi|17738043|ref|NP_524402.1| CG6030-PA, isoform A
[Drosophila melanogaster]
gi|14286117|sp|Q24251|ATP5H_DROME ATP synthase D chain,
mitochondrial
Length = 178
Score = 46.6 bits (109), Expect = 3e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 7 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 64
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + A E+ ++ S + + +KEI +++S
Sbjct: 65 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKASQSEIDAYKKASEQRIQNYQKEIAHLKS 124
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y P+
Sbjct: 125 LLP-YDQMTMEDYRDAFPD 142
>emb|CAA67981.1| mitochondrial ATP synthase [Drosophila melanogaster]
Length = 178
Score = 46.2 bits (108), Expect = 4e-04
Identities = 32/139 (23%), Positives = 67/139 (48%), Gaps = 4/139 (2%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIGTR 74
A I+W +A+ + ++ + F + ++ + + P IDW Y+K +
Sbjct: 7 AQSSINWSALAERVPANQ--KSSFGAFKTKSDIYVRAVLANPECPPQIDWANYKKLVPVA 64
Query: 75 -LVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQS 133
LVD +++ YE++++P D V+ Q + A E+ ++ S + + +KEI +++S
Sbjct: 65 GLVDSFQKQYEALKVPYPQDKVSSQVDAEIKASQNEIDAYKKASEQRIQNYQKEIAHLKS 124
Query: 134 LKKRISTMTADEYFAEHPE 152
L MT ++Y P+
Sbjct: 125 LLP-YDQMTMEDYRDAFPD 142
>ref|XP_420118.1| PREDICTED: similar to Hypothetical protein MGC73071 [Gallus gallus]
Length = 161
Score = 45.8 bits (107), Expect = 5e-04
Identities = 36/144 (25%), Positives = 68/144 (47%), Gaps = 10/144 (6%)
Query: 15 AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-T 73
A K IDW A+ + ++ R FN + + + ++P IDW YY+ +
Sbjct: 7 AVKAIDWAAFAERVPANQ--RAMFNALKTRSDALSARLAALPEKPPAIDWTYYKTAVAKA 64
Query: 74 RLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESE-RLEKEIVNVQ 132
+VD +++ + ++++P+ VDT T K DA E + + +K S+ R+ + +Q
Sbjct: 65 GMVDEFQKKFSALKVPEPVDTQT----AKIDAQEQEAAKGIVEYVKASKARIAEYEQQLQ 120
Query: 133 SLKKRI--STMTADEYFAEHPELK 154
L+ I MT ++ PE +
Sbjct: 121 KLRSMIPFEQMTFEDLHEAFPETR 144
>ref|XP_535864.1| PREDICTED: similar to H+-transporting two-sector ATPase (EC
3.6.3.14) chain d - bovine [Canis familiaris]
Length = 161
Score = 44.7 bits (104), Expect = 0.001
Identities = 32/139 (23%), Positives = 63/139 (45%), Gaps = 4/139 (2%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-TRL 75
K IDW +++ ++ + F N +++E+ T ++P IDW YY+ + L
Sbjct: 9 KTIDWVAFGEIIPQNQ--KAFANSLESWNEMLTSRLATLPEKPPAIDWAYYKANVAKAGL 66
Query: 76 VDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLK 135
VD +++ + ++++P D T Q + + E S E EKE+ ++++
Sbjct: 67 VDDFEKKFNALKVPVPEDKYTVQVDAEEKEDVKSCAEFLSLSKTRIEEYEKELEKMKNII 126
Query: 136 KRISTMTADEYFAEHPELK 154
MT ++ PE K
Sbjct: 127 P-FDQMTIEDLNEVFPETK 144
>ref|XP_537923.1| PREDICTED: similar to Chemokine receptor-like 2 (G protein-coupled
receptor 30) (IL8-related receptor DRY12) (Flow-induced
endothelial G protein-coupled receptor) (FEG-1)
(GPCR-BR) [Canis familiaris]
Length = 535
Score = 44.3 bits (103), Expect = 0.002
Identities = 32/139 (23%), Positives = 63/139 (45%), Gaps = 4/139 (2%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-TRL 75
K IDW +++ ++ + N ++++E+ T ++P IDW YY+ + T L
Sbjct: 384 KTIDWVAFGEIIPRNQ--KAIANSLKSWNEMLTSRLATLPEKPPAIDWAYYKANVAKTGL 441
Query: 76 VDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSLKESERLEKEIVNVQSLK 135
VD +++ + ++++P D Q K + E S E EKE+ ++++
Sbjct: 442 VDDFEKKFNALKVPVPEDKYIVQMDAKEKEGVKSCAEFLSLSKARIEEYEKELEKMKNII 501
Query: 136 KRISTMTADEYFAEHPELK 154
MT ++ PE K
Sbjct: 502 P-FDQMTIEDLNEVFPETK 519
>ref|XP_533611.1| PREDICTED: similar to myosin, heavy polypeptide 14 [Canis familiaris]
Length = 1025
Score = 44.3 bits (103), Expect = 0.002
Identities = 37/156 (23%), Positives = 72/156 (45%), Gaps = 11/156 (7%)
Query: 1 MSGTVKKVTDVAFK-AGKKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEP 59
+ G+VK + K A K IDW +++ ++ + N ++++E+ T ++P
Sbjct: 862 LPGSVKDLKMAGRKLALKTIDWVAFGEIIPQNQ--KAIANSLKSWNEMLTSRLATLPEKP 919
Query: 60 EPIDWEYYRKGIG-TRLVDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEEKSL 118
IDW YY+ + LVD +++ + ++++P D T Q A + L +A
Sbjct: 920 PAIDWAYYKANVAKAGLVDDFEKKFNALKVPVPEDKYTVQVDAGSCAQFLSLSKA----- 974
Query: 119 KESERLEKEIVNVQSLKKRISTMTADEYFAEHPELK 154
E EKE+ ++++ MT ++ PE K
Sbjct: 975 -RIEEYEKELEKMKNIIP-FDQMTIEDLNEVFPETK 1008
>ref|XP_224879.1| PREDICTED: similar to ATP synthase D chain, mitochondrial [Rattus
norvegicus]
Length = 161
Score = 43.9 bits (102), Expect = 0.002
Identities = 40/162 (24%), Positives = 70/162 (42%), Gaps = 25/162 (15%)
Query: 17 KKIDWDGMAKLLVTDEARREFFNLRRAFDEVNTQLETKFSQEPEPIDWEYYRKGIG-TRL 75
K IDW +++ ++ + N +++++ S++P IDW YYR +G L
Sbjct: 9 KTIDWVSFVEIMPQNQ--KAIGNALKSWNKTFHTRLASMSEKPPAIDWAYYRTNVGKAGL 66
Query: 76 VDMYKQHYESIEIPKFVDTVTPQYKPKFDALLVELKEAEE---------KSLKESERLEK 126
VD +++ Y ++IP D T LV+ +E EE S + EK
Sbjct: 67 VDDFEKKYNVLKIPVPEDKYT---------ALVDAEEKEEVKTCAQFVSGSQARVQEYEK 117
Query: 127 EIVNVQSLKKRISTMTADEYFAEHPELKKKFDDEIRNDNWGY 168
++ ++S+ MT D+ PE K DE + W +
Sbjct: 118 QLEKIKSMIP-FDQMTIDDLNEIFPETKL---DEKKYPYWSH 155
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.313 0.132 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 294,102,585
Number of Sequences: 2540612
Number of extensions: 11873433
Number of successful extensions: 48190
Number of sequences better than 10.0: 301
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 270
Number of HSP's that attempted gapping in prelim test: 47928
Number of HSP's gapped (non-prelim): 426
length of query: 177
length of database: 863,360,394
effective HSP length: 119
effective length of query: 58
effective length of database: 561,027,566
effective search space: 32539598828
effective search space used: 32539598828
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144484.9


