

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144477.14 + phase: 1 /pseudo
(190 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
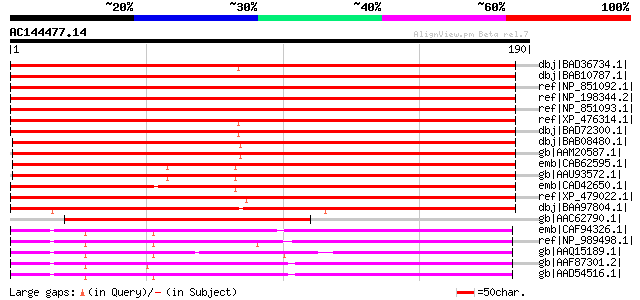
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAD36734.1| putative beta-glucuronidase precursor [Oryza sat... 261 5e-69
dbj|BAB10787.1| unnamed protein product [Arabidopsis thaliana] 250 2e-65
ref|NP_851092.1| glycosyl hydrolase family 79 N-terminal domain-... 250 2e-65
ref|NP_198344.2| glycosyl hydrolase family 79 N-terminal domain-... 250 2e-65
ref|NP_851093.1| glycosyl hydrolase family 79 N-terminal domain-... 250 2e-65
ref|XP_476314.1| contains ESTs AU097236(S3106),D40915(S3106)~sim... 244 9e-64
dbj|BAD72300.1| putative beta-glucuronidase precursor [Oryza sat... 244 9e-64
dbj|BAB08480.1| unnamed protein product [Arabidopsis thaliana] 210 1e-53
gb|AAM20587.1| putative protein [Arabidopsis thaliana] gi|273119... 210 1e-53
emb|CAB62595.1| putative protein [Arabidopsis thaliana] gi|11357... 207 2e-52
gb|AAU93572.1| At5g07830 [Arabidopsis thaliana] gi|51536460|gb|A... 207 2e-52
emb|CAD42650.1| putative heparanase [Hordeum vulgare subsp. vulg... 196 2e-49
ref|XP_479022.1| putative beta-glucuronidase precursor [Oryza sa... 188 8e-47
dbj|BAA97804.1| beta-glucuronidase precursor [Scutellaria baical... 179 5e-44
gb|AAC62790.1| T2L5.7 gene product [Arabidopsis thaliana] gi|748... 96 4e-19
emb|CAF94326.1| unnamed protein product [Tetraodon nigroviridis] 74 2e-12
ref|NP_989498.1| heparanase [Gallus gallus] gi|15081576|gb|AAK82... 74 3e-12
gb|AAQ15189.1| heparanase [Rattus norvegicus] 67 3e-10
gb|AAF87301.2| heparanase [Bos taurus] gi|27806583|ref|NP_776507... 67 3e-10
gb|AAD54516.1| heparanase [Homo sapiens] 66 4e-10
>dbj|BAD36734.1| putative beta-glucuronidase precursor [Oryza sativa (japonica
cultivar-group)] gi|51090574|dbj|BAD36026.1| putative
beta-glucuronidase precursor [Oryza sativa (japonica
cultivar-group)]
Length = 541
Score = 261 bits (668), Expect = 5e-69
Identities = 122/186 (65%), Positives = 145/186 (77%), Gaps = 1/186 (0%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A SFIRYTVSK Y IHGWELGNEL G G+G + QYA D L+ I+ Y+ KP
Sbjct: 186 AASFIRYTVSKGYDIHGWELGNELSGSGVGARVDADQYAQDVLALKQIIDNSYQGHASKP 245
Query: 61 LIIAPGGFFDANWFKKFLNRSE-KLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
L+IAPGGFFDA WF + ++R++ DV+THHIYNLGPGVD H+ +KILDP+YLDG AGT
Sbjct: 246 LVIAPGGFFDAAWFTELISRTKPNQMDVMTHHIYNLGPGVDTHLIDKILDPSYLDGEAGT 305
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
FSSL+ +L+ + T+ AWVGEAGGA+NSGHHLV+DAFV SFWY DQLGMS+ Y TKTYCR
Sbjct: 306 FSSLQGILKSAGTSTVAWVGEAGGAYNSGHHLVTDAFVFSFWYLDQLGMSSKYDTKTYCR 365
Query: 180 QTLIGG 185
QTLIGG
Sbjct: 366 QTLIGG 371
>dbj|BAB10787.1| unnamed protein product [Arabidopsis thaliana]
Length = 536
Score = 250 bits (638), Expect = 2e-65
Identities = 114/185 (61%), Positives = 143/185 (76%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+SFIR+T NYTI GWELGNELCG G+G + QYA D LRNIV VY+ V P
Sbjct: 180 AESFIRFTAENNYTIDGWELGNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMP 239
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
L+I PGGFF+ +WF ++LN++E + T HIY+LGPGVD+H+ EKIL+P+YLD A +F
Sbjct: 240 LVIGPGGFFEVDWFTEYLNKAENSLNATTRHIYDLGPGVDEHLIEKILNPSYLDQEAKSF 299
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
SLKN+++ SST A AWVGE+GGA+NSG +LVS+AFV SFWY DQLGM++ Y TKTYCRQ
Sbjct: 300 RSLKNIIKNSSTKAVAWVGESGGAYNSGRNLVSNAFVYSFWYLDQLGMASLYDTKTYCRQ 359
Query: 181 TLIGG 185
+LIGG
Sbjct: 360 SLIGG 364
>ref|NP_851092.1| glycosyl hydrolase family 79 N-terminal domain-containing protein
[Arabidopsis thaliana]
Length = 401
Score = 250 bits (638), Expect = 2e-65
Identities = 114/185 (61%), Positives = 143/185 (76%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+SFIR+T NYTI GWELGNELCG G+G + QYA D LRNIV VY+ V P
Sbjct: 45 AESFIRFTAENNYTIDGWELGNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMP 104
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
L+I PGGFF+ +WF ++LN++E + T HIY+LGPGVD+H+ EKIL+P+YLD A +F
Sbjct: 105 LVIGPGGFFEVDWFTEYLNKAENSLNATTRHIYDLGPGVDEHLIEKILNPSYLDQEAKSF 164
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
SLKN+++ SST A AWVGE+GGA+NSG +LVS+AFV SFWY DQLGM++ Y TKTYCRQ
Sbjct: 165 RSLKNIIKNSSTKAVAWVGESGGAYNSGRNLVSNAFVYSFWYLDQLGMASLYDTKTYCRQ 224
Query: 181 TLIGG 185
+LIGG
Sbjct: 225 SLIGG 229
>ref|NP_198344.2| glycosyl hydrolase family 79 N-terminal domain-containing protein
[Arabidopsis thaliana]
Length = 382
Score = 250 bits (638), Expect = 2e-65
Identities = 114/185 (61%), Positives = 143/185 (76%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+SFIR+T NYTI GWELGNELCG G+G + QYA D LRNIV VY+ V P
Sbjct: 180 AESFIRFTAENNYTIDGWELGNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMP 239
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
L+I PGGFF+ +WF ++LN++E + T HIY+LGPGVD+H+ EKIL+P+YLD A +F
Sbjct: 240 LVIGPGGFFEVDWFTEYLNKAENSLNATTRHIYDLGPGVDEHLIEKILNPSYLDQEAKSF 299
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
SLKN+++ SST A AWVGE+GGA+NSG +LVS+AFV SFWY DQLGM++ Y TKTYCRQ
Sbjct: 300 RSLKNIIKNSSTKAVAWVGESGGAYNSGRNLVSNAFVYSFWYLDQLGMASLYDTKTYCRQ 359
Query: 181 TLIGG 185
+LIGG
Sbjct: 360 SLIGG 364
>ref|NP_851093.1| glycosyl hydrolase family 79 N-terminal domain-containing protein
[Arabidopsis thaliana]
Length = 536
Score = 250 bits (638), Expect = 2e-65
Identities = 114/185 (61%), Positives = 143/185 (76%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+SFIR+T NYTI GWELGNELCG G+G + QYA D LRNIV VY+ V P
Sbjct: 180 AESFIRFTAENNYTIDGWELGNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMP 239
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
L+I PGGFF+ +WF ++LN++E + T HIY+LGPGVD+H+ EKIL+P+YLD A +F
Sbjct: 240 LVIGPGGFFEVDWFTEYLNKAENSLNATTRHIYDLGPGVDEHLIEKILNPSYLDQEAKSF 299
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
SLKN+++ SST A AWVGE+GGA+NSG +LVS+AFV SFWY DQLGM++ Y TKTYCRQ
Sbjct: 300 RSLKNIIKNSSTKAVAWVGESGGAYNSGRNLVSNAFVYSFWYLDQLGMASLYDTKTYCRQ 359
Query: 181 TLIGG 185
+LIGG
Sbjct: 360 SLIGG 364
>ref|XP_476314.1| contains ESTs AU097236(S3106),D40915(S3106)~similar to
endo-beta-glucuronidase/heparanase [Oryza sativa
(japonica cultivar-group)]
Length = 544
Score = 244 bits (623), Expect = 9e-64
Identities = 112/186 (60%), Positives = 143/186 (76%), Gaps = 1/186 (0%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A S IRYT SK Y IHGWELGNEL G G+G + QYA D L+++V +Y+ KP
Sbjct: 195 AASLIRYTASKGYKIHGWELGNELSGSGVGTKVGADQYAADVIALKSLVDTIYQGNPSKP 254
Query: 61 LIIAPGGFFDANWFKKFLNRSE-KLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
L++APGGFFDA WF + + ++ L +VVTHHIYNLGPGVD H+ EKIL+P+YLDG+ T
Sbjct: 255 LVLAPGGFFDAGWFTEVIVKTRPNLLNVVTHHIYNLGPGVDTHLIEKILNPSYLDGMVST 314
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
FS+L+ +L+ + T+A AWVGE+GGA+NSG HLV+D+FV SFW+ DQLGMSA Y TK+YCR
Sbjct: 315 FSNLQGILKSAGTSAVAWVGESGGAYNSGRHLVTDSFVFSFWFLDQLGMSAKYDTKSYCR 374
Query: 180 QTLIGG 185
Q+LIGG
Sbjct: 375 QSLIGG 380
>dbj|BAD72300.1| putative beta-glucuronidase precursor [Oryza sativa (japonica
cultivar-group)]
Length = 526
Score = 244 bits (623), Expect = 9e-64
Identities = 112/186 (60%), Positives = 143/186 (76%), Gaps = 1/186 (0%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A S IRYT SK Y IHGWELGNEL G G+G + QYA D L+++V +Y+ KP
Sbjct: 177 AASLIRYTASKGYKIHGWELGNELSGSGVGTKVGADQYAADVIALKSLVDTIYQGNPSKP 236
Query: 61 LIIAPGGFFDANWFKKFLNRSE-KLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
L++APGGFFDA WF + + ++ L +VVTHHIYNLGPGVD H+ EKIL+P+YLDG+ T
Sbjct: 237 LVLAPGGFFDAGWFTEVIVKTRPNLLNVVTHHIYNLGPGVDTHLIEKILNPSYLDGMVST 296
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
FS+L+ +L+ + T+A AWVGE+GGA+NSG HLV+D+FV SFW+ DQLGMSA Y TK+YCR
Sbjct: 297 FSNLQGILKSAGTSAVAWVGESGGAYNSGRHLVTDSFVFSFWFLDQLGMSAKYDTKSYCR 356
Query: 180 QTLIGG 185
Q+LIGG
Sbjct: 357 QSLIGG 362
>dbj|BAB08480.1| unnamed protein product [Arabidopsis thaliana]
Length = 516
Score = 210 bits (535), Expect = 1e-53
Identities = 95/185 (51%), Positives = 126/185 (67%), Gaps = 1/185 (0%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKPL 61
+ F+ YTVSK Y I WE GNEL G GI S+S Y D +L+N+++ VY+ KPL
Sbjct: 154 QDFMNYTVSKGYAIDSWEFGNELSGSGIWASVSVELYGKDLIVLKNVIKNVYKNSRTKPL 213
Query: 62 IIAPGGFFDANWFKKFLNRSEK-LADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
++APGGFF+ W+ + L S + DV+THHIYNLGPG D + KILDP YL G++ F
Sbjct: 214 VVAPGGFFEEQWYSELLRLSGPGVLDVLTHHIYNLGPGNDPKLVNKILDPNYLSGISELF 273
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
+++ +Q A AWVGEAGGA+NSG VS+ F+NSFWY DQLG+S+ ++TK YCRQ
Sbjct: 274 ANVNQTIQEHGPWAAAWVGEAGGAFNSGGRQVSETFINSFWYLDQLGISSKHNTKVYCRQ 333
Query: 181 TLIGG 185
L+GG
Sbjct: 334 ALVGG 338
>gb|AAM20587.1| putative protein [Arabidopsis thaliana] gi|27311995|gb|AAO00963.1|
putative protein [Arabidopsis thaliana]
gi|30697502|ref|NP_851238.1| glycosyl hydrolase family
79 N-terminal domain-containing protein [Arabidopsis
thaliana] gi|22327999|ref|NP_200933.2| glycosyl
hydrolase family 79 N-terminal domain-containing protein
[Arabidopsis thaliana]
Length = 539
Score = 210 bits (535), Expect = 1e-53
Identities = 95/185 (51%), Positives = 126/185 (67%), Gaps = 1/185 (0%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKPL 61
+ F+ YTVSK Y I WE GNEL G GI S+S Y D +L+N+++ VY+ KPL
Sbjct: 177 QDFMNYTVSKGYAIDSWEFGNELSGSGIWASVSVELYGKDLIVLKNVIKNVYKNSRTKPL 236
Query: 62 IIAPGGFFDANWFKKFLNRSEK-LADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGTF 120
++APGGFF+ W+ + L S + DV+THHIYNLGPG D + KILDP YL G++ F
Sbjct: 237 VVAPGGFFEEQWYSELLRLSGPGVLDVLTHHIYNLGPGNDPKLVNKILDPNYLSGISELF 296
Query: 121 SSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCRQ 180
+++ +Q A AWVGEAGGA+NSG VS+ F+NSFWY DQLG+S+ ++TK YCRQ
Sbjct: 297 ANVNQTIQEHGPWAAAWVGEAGGAFNSGGRQVSETFINSFWYLDQLGISSKHNTKVYCRQ 356
Query: 181 TLIGG 185
L+GG
Sbjct: 357 ALVGG 361
>emb|CAB62595.1| putative protein [Arabidopsis thaliana] gi|11357400|pir||T45608
hypothetical protein F13G24.30 - Arabidopsis thaliana
Length = 521
Score = 207 bits (526), Expect = 2e-52
Identities = 94/186 (50%), Positives = 127/186 (67%), Gaps = 2/186 (1%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREV-VQKP 60
+ F+ YTVSK Y I WE GNEL G G+G S+S Y D +L++++ +VY+ + KP
Sbjct: 158 QDFLNYTVSKGYVIDSWEFGNELSGSGVGASVSAELYGKDLIVLKDVINKVYKNSWLHKP 217
Query: 61 LIIAPGGFFDANWFKKFLNRS-EKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
+++APGGF++ W+ K L S + DVVTHHIYNLG G D + +KI+DP+YL V+ T
Sbjct: 218 ILVAPGGFYEQQWYTKLLEISGPSVVDVVTHHIYNLGSGNDPALVKKIMDPSYLSQVSKT 277
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
F + +Q A WVGE+GGA+NSG VSD F++SFWY DQLGMSA ++TK YCR
Sbjct: 278 FKDVNQTIQEHGPWASPWVGESGGAYNSGGRHVSDTFIDSFWYLDQLGMSARHNTKVYCR 337
Query: 180 QTLIGG 185
QTL+GG
Sbjct: 338 QTLVGG 343
>gb|AAU93572.1| At5g07830 [Arabidopsis thaliana] gi|51536460|gb|AAU05468.1|
At5g07830 [Arabidopsis thaliana]
gi|10176717|dbj|BAB09947.1| unnamed protein product
[Arabidopsis thaliana] gi|42567736|ref|NP_196400.2|
glycosyl hydrolase family 79 N-terminal
domain-containing protein [Arabidopsis thaliana]
Length = 543
Score = 207 bits (526), Expect = 2e-52
Identities = 94/186 (50%), Positives = 127/186 (67%), Gaps = 2/186 (1%)
Query: 2 KSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREV-VQKP 60
+ F+ YTVSK Y I WE GNEL G G+G S+S Y D +L++++ +VY+ + KP
Sbjct: 180 QDFLNYTVSKGYVIDSWEFGNELSGSGVGASVSAELYGKDLIVLKDVINKVYKNSWLHKP 239
Query: 61 LIIAPGGFFDANWFKKFLNRS-EKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
+++APGGF++ W+ K L S + DVVTHHIYNLG G D + +KI+DP+YL V+ T
Sbjct: 240 ILVAPGGFYEQQWYTKLLEISGPSVVDVVTHHIYNLGSGNDPALVKKIMDPSYLSQVSKT 299
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
F + +Q A WVGE+GGA+NSG VSD F++SFWY DQLGMSA ++TK YCR
Sbjct: 300 FKDVNQTIQEHGPWASPWVGESGGAYNSGGRHVSDTFIDSFWYLDQLGMSARHNTKVYCR 359
Query: 180 QTLIGG 185
QTL+GG
Sbjct: 360 QTLVGG 365
>emb|CAD42650.1| putative heparanase [Hordeum vulgare subsp. vulgare]
Length = 537
Score = 196 bits (499), Expect = 2e-49
Identities = 98/186 (52%), Positives = 121/186 (64%), Gaps = 2/186 (1%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A+ F+ YTVS NY I WE GNEL G GIG S+ QY D L+ IV ++Y E KP
Sbjct: 174 AQEFMEYTVSMNYPIDSWEFGNELSGSGIGASVGAEQYGKDLVELQKIVDQLY-ENPSKP 232
Query: 61 LIIAPGGFFDANWFKKFLNRS-EKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
L++APGGF+D F + L+ S + +THHIYNLG G D + +ILDP YL V+ T
Sbjct: 233 LVLAPGGFYDKQCFAQLLDVSGPNVVRGMTHHIYNLGAGNDPRVANRILDPQYLSRVSDT 292
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
F L+ +QR + WVGEAGGA+NSG VS+ F++SFWY DQLG SA Y TK YCR
Sbjct: 293 FRDLQLTIQRHGPWSAPWVGEAGGAYNSGSRTVSNTFLDSFWYLDQLGQSAKYDTKVYCR 352
Query: 180 QTLIGG 185
QTLIGG
Sbjct: 353 QTLIGG 358
>ref|XP_479022.1| putative beta-glucuronidase precursor [Oryza sativa (japonica
cultivar-group)] gi|34393590|dbj|BAC83217.1| putative
beta-glucuronidase precursor [Oryza sativa (japonica
cultivar-group)]
Length = 529
Score = 188 bits (477), Expect = 8e-47
Identities = 90/186 (48%), Positives = 115/186 (61%), Gaps = 1/186 (0%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKP 60
A +RYT K Y + WELGNEL G G+ ++ QY D +LR V+ VY + P
Sbjct: 185 ALDLMRYTAGKGYRVESWELGNELSGSGVAARVAAAQYGRDVAVLRKAVERVYGGGGEVP 244
Query: 61 LIIAPGGFFDANWFKKFLNRSEKLA-DVVTHHIYNLGPGVDDHITEKILDPTYLDGVAGT 119
++APGGF+D WF + L S + A D VTHHIYNLG G D + K+ DP YLD V T
Sbjct: 245 KVLAPGGFYDGAWFSEMLRVSGRGAVDGVTHHIYNLGSGKDRDLARKMQDPGYLDQVEKT 304
Query: 120 FSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYCR 179
F + ++ S + WVGE+GGA+NSG VSD +VN FWY DQLGMSA + T+ YCR
Sbjct: 305 FRDMAATVRGSGPWSSPWVGESGGAYNSGGKGVSDRYVNGFWYLDQLGMSAAHGTRVYCR 364
Query: 180 QTLIGG 185
Q L+GG
Sbjct: 365 QALVGG 370
>dbj|BAA97804.1| beta-glucuronidase precursor [Scutellaria baicalensis]
Length = 527
Score = 179 bits (453), Expect = 5e-44
Identities = 87/187 (46%), Positives = 120/187 (63%), Gaps = 3/187 (1%)
Query: 1 AKSFIRYTVSKNYT-IHGWELGNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQK 59
+K I Y++ K Y I GW LGNEL G + I +SP YANDA L +V+E+Y++
Sbjct: 189 SKFLIEYSLKKGYKHIRGWTLGNELGGHTLFIGVSPEDYANDAKKLHELVKEIYQDQGTM 248
Query: 60 PLIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLD-GVAG 118
PLIIAPG FD W+ +F++R+ +L V THH+YNLG G DD + + +L ++ D
Sbjct: 249 PLIIAPGAIFDLEWYTEFIDRTPEL-HVATHHMYNLGSGGDDALKDVLLTASFFDEATKS 307
Query: 119 TFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTYC 178
+ L+ ++ R T A AW+GEAGGA+NSG +S+ F+N FWY + LG SA TKT+C
Sbjct: 308 MYEGLQKIVNRPGTKAVAWIGEAGGAFNSGQDGISNTFINGFWYLNMLGYSALLDTKTFC 367
Query: 179 RQTLIGG 185
RQTL GG
Sbjct: 368 RQTLTGG 374
>gb|AAC62790.1| T2L5.7 gene product [Arabidopsis thaliana] gi|7487548|pir||T01954
hypothetical protein T2L5.7 - Arabidopsis thaliana
Length = 196
Score = 96.3 bits (238), Expect = 4e-19
Identities = 43/90 (47%), Positives = 57/90 (62%)
Query: 21 GNELCGKGIGISISPYQYANDATILRNIVQEVYREVVQKPLIIAPGGFFDANWFKKFLNR 80
GNELCG G+G + QYA D LRNIV VY+ V PL+I PGGFF+ +WF ++LN+
Sbjct: 89 GNELCGSGVGARVGANQYAIDTINLRNIVNRVYKNVSPMPLVIGPGGFFEVDWFTEYLNK 148
Query: 81 SEKLADVVTHHIYNLGPGVDDHITEKILDP 110
+E + T HIY+LGPG + + DP
Sbjct: 149 AENSLNATTRHIYDLGPGNPNTQLSFVSDP 178
>emb|CAF94326.1| unnamed protein product [Tetraodon nigroviridis]
Length = 533
Score = 74.3 bits (181), Expect = 2e-12
Identities = 56/188 (29%), Positives = 83/188 (43%), Gaps = 7/188 (3%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCG--KGIGISISPYQYANDATILRNIVQE--VYREV 56
A+S +RY ++ Y + WELGNE K G+ + Q D T+LR I++E YR+
Sbjct: 159 ARSLLRYCEARRYHM-SWELGNEPNSYEKKAGLRLDGRQLGEDFTVLRKILRESRFYRDA 217
Query: 57 VQKPLIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGV 116
+ + FL + D T H Y L + E LDP LD +
Sbjct: 218 GLFGPDVGQPRDHRIDILSGFLQSGAEAVDACTWHHYYLDGR--EASLEDFLDPDVLDTL 275
Query: 117 AGTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKT 176
+ + + S W+GE A+ G +SD FV F + D+LG++AT +
Sbjct: 276 REKIGEVLEEVHQVSPGKPVWLGETSSAYGGGAAGLSDTFVAGFMWLDKLGLAATLGLEL 335
Query: 177 YCRQTLIG 184
RQ LIG
Sbjct: 336 VMRQVLIG 343
>ref|NP_989498.1| heparanase [Gallus gallus] gi|15081576|gb|AAK82648.1| heparanase
[Gallus gallus]
Length = 523
Score = 73.6 bits (179), Expect = 3e-12
Identities = 54/189 (28%), Positives = 80/189 (41%), Gaps = 9/189 (4%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCG--KGIGISISPYQYANDATILRNIVQE--VYREV 56
AK + Y ++Y I WELGNE K GI I +Q D LR ++ + +YR
Sbjct: 183 AKQLLGYCAQRSYNI-SWELGNEPNSFRKKSGICIDGFQLGRDFVHLRQLLSQHPLYRHA 241
Query: 57 VQKPLIIAPGGFFDANWFKKFLNRSEKLADVVT-HHIYNLGPGVDDHITEKILDPTYLDG 115
L + + + F+ K D VT HH Y G E L P LD
Sbjct: 242 ELYGLDVGQPRKHTQHLLRSFMKSGGKAIDSVTWHHYYVNGRSAT---REDFLSPEVLDS 298
Query: 116 VAGTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTK 175
A + +++ + K W+GE G A+ G +S+ +V F + D+LG++A
Sbjct: 299 FATAIHDVLGIVEATVPGKKVWLGETGSAYGGGAPQLSNTYVAGFMWLDKLGLAARRGID 358
Query: 176 TYCRQTLIG 184
RQ G
Sbjct: 359 VVMRQVSFG 367
>gb|AAQ15189.1| heparanase [Rattus norvegicus]
Length = 536
Score = 67.0 bits (162), Expect = 3e-10
Identities = 56/191 (29%), Positives = 81/191 (42%), Gaps = 14/191 (7%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCG--KGIGISISPYQYANDATILRNIVQE--VYREV 56
A+ + Y SK Y I WELGNE K ISI Q D L ++Q+
Sbjct: 197 AQLLLNYCSSKGYNI-SWELGNEPNSFWKKAHISIDGLQLGEDFVELHKLLQKSAFQNAK 255
Query: 57 VQKPLIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGV---DDHITEKILDPTYL 113
+ P I P G + FL ++ D +T H Y L V +D ++ +LD L
Sbjct: 256 LYGPDIGQPRGK-TVKLLRSFLKAGGEVIDSLTWHHYYLNGRVATKEDFLSSDVLDTFIL 314
Query: 114 DGVAGTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYS 173
+ + V + + K W+GE A+ G L+SD F F + D+LG+SA
Sbjct: 315 -----SVQKILKVTKEMTPGKKVWLGETSSAYGGGAPLLSDTFAAGFMWLDKLGLSAQLG 369
Query: 174 TKTYCRQTLIG 184
+ RQ G
Sbjct: 370 IEVVMRQVFFG 380
>gb|AAF87301.2| heparanase [Bos taurus] gi|27806583|ref|NP_776507.1| heparanase
[Bos taurus]
Length = 545
Score = 66.6 bits (161), Expect = 3e-10
Identities = 51/187 (27%), Positives = 78/187 (41%), Gaps = 6/187 (3%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCG--KGIGISISPYQYANDATILRNIV-QEVYREVV 57
A+ + Y SKNY I WELGNE + GI I+ Q D R ++ + ++
Sbjct: 206 AQLLLDYCSSKNYNI-SWELGNEPNSFQRKAGIFINGRQLGEDFIEFRKLLGKSAFKNAK 264
Query: 58 QKPLIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVA 117
I K FL ++ D VT H Y + + E L+P LD
Sbjct: 265 LYGPDIGQPRRNTVKMLKSFLKAGGEVIDSVTWHHYYVNGRIATK--EDFLNPDILDTFI 322
Query: 118 GTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTY 177
+ ++++ K W+GE A+ G +S+ F F + D+LG+SA +
Sbjct: 323 SSVQKTLRIVEKIRPLKKVWLGETSSAFGGGAPFLSNTFAAGFMWLDKLGLSARMGIEVV 382
Query: 178 CRQTLIG 184
RQ L G
Sbjct: 383 MRQVLFG 389
>gb|AAD54516.1| heparanase [Homo sapiens]
Length = 545
Score = 66.2 bits (160), Expect = 4e-10
Identities = 53/187 (28%), Positives = 78/187 (41%), Gaps = 6/187 (3%)
Query: 1 AKSFIRYTVSKNYTIHGWELGNELCG--KGIGISISPYQYANDATILRNIVQE-VYREVV 57
A+ + Y SK Y I WELGNE K I I+ Q D L ++++ ++
Sbjct: 206 AQLLLDYCSSKGYNI-SWELGNEPNSFLKKADIFINGSQLGEDFIQLHKLLRKSTFKNAK 264
Query: 58 QKPLIIAPGGFFDANWFKKFLNRSEKLADVVTHHIYNLGPGVDDHITEKILDPTYLDGVA 117
+ A K FL ++ D VT H Y L E L+P LD
Sbjct: 265 LYGPDVGQPRRKTAKMLKSFLKAGGEVIDSVTWHHYYLNGRTATR--EDFLNPDVLDIFI 322
Query: 118 GTFSSLKNVLQRSSTTAKAWVGEAGGAWNSGHHLVSDAFVNSFWYFDQLGMSATYSTKTY 177
+ + V++ + K W+GE A+ G L+SD F F + D+LG+SA +
Sbjct: 323 SSVQKVFQVVESTRPGKKVWLGETSSAYGGGAPLLSDTFAAGFMWLDKLGLSARMGIEVV 382
Query: 178 CRQTLIG 184
RQ G
Sbjct: 383 MRQVFFG 389
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.320 0.137 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 346,071,073
Number of Sequences: 2540612
Number of extensions: 14017884
Number of successful extensions: 31298
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 31246
Number of HSP's gapped (non-prelim): 47
length of query: 190
length of database: 863,360,394
effective HSP length: 121
effective length of query: 69
effective length of database: 555,946,342
effective search space: 38360297598
effective search space used: 38360297598
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC144477.14


