

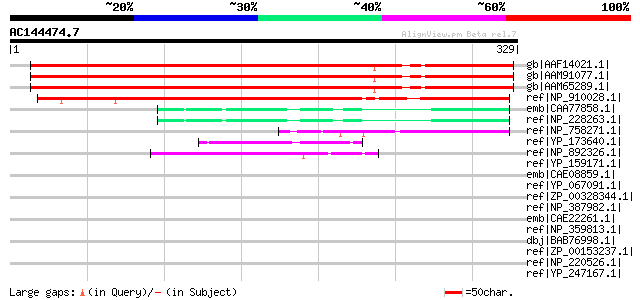
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144474.7 - phase: 0
(329 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF14021.1| unknown protein [Arabidopsis thaliana] 342 1e-92
gb|AAM91077.1| AT3g09210/F3L24_8 [Arabidopsis thaliana] gi|15146... 342 1e-92
gb|AAM65289.1| unknown [Arabidopsis thaliana] 338 1e-91
ref|NP_910028.1| putative transcription factor [Oryza sativa] gi... 243 6e-63
emb|CAA77858.1| NusG [Thermotoga maritima] gi|128916|sp|P29397|N... 58 3e-07
ref|NP_228263.1| N utilization substance protein G [Thermotoga m... 58 3e-07
ref|NP_758271.1| transcription antitermination factor [Mycoplasm... 48 3e-04
ref|YP_173640.1| transcriptional antitermination factor [Bacillu... 47 6e-04
ref|NP_892326.1| transcription antitermination protein, NusG [Pr... 47 8e-04
ref|YP_159171.1| transcription antitermination protein [Azoarcus... 47 0.001
emb|CAE08859.1| transcription antitermination protein, NusG [Syn... 46 0.001
ref|YP_067091.1| transcription antitermination protein NusG (N u... 46 0.001
ref|ZP_00328344.1| COG0250: Transcription antiterminator [Tricho... 46 0.002
ref|NP_387982.1| transcription antitermination factor [Bacillus ... 46 0.002
emb|CAE22261.1| transcription antitermination protein, NusG [Pro... 45 0.004
ref|NP_359813.1| transcription antitermination protein NusG [Ric... 45 0.004
dbj|BAB76998.1| transcription antitermination protein [Nostoc sp... 45 0.004
ref|ZP_00153237.1| COG0250: Transcription antiterminator [Ricket... 45 0.004
ref|NP_220526.1| TRANSCRIPTION ANTITERMINATION PROTEIN NUSG (nus... 44 0.005
ref|YP_247167.1| Transcription antitermination protein NusG [Ric... 44 0.005
>gb|AAF14021.1| unknown protein [Arabidopsis thaliana]
Length = 332
Score = 342 bits (876), Expect = 1e-92
Identities = 177/319 (55%), Positives = 228/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 17 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 76
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 77 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 136
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFIGSKVGNTK+QINKPR
Sbjct: 137 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIGSKVGNTKRQINKPR 196
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 197 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 256
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 257 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 310
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 311 TLFGKETLVEIDINELVPE 329
>gb|AAM91077.1| AT3g09210/F3L24_8 [Arabidopsis thaliana] gi|15146210|gb|AAK83588.1|
AT3g09210/F3L24_8 [Arabidopsis thaliana]
gi|18398425|ref|NP_566346.1| KOW domain-containing
transcription factor family protein [Arabidopsis
thaliana]
Length = 333
Score = 342 bits (876), Expect = 1e-92
Identities = 177/319 (55%), Positives = 228/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 18 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 77
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 78 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 137
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFIGSKVGNTK+QINKPR
Sbjct: 138 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIGSKVGNTKRQINKPR 197
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 198 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 257
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 258 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 311
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 312 TLFGKETLVEIDINELVPE 330
>gb|AAM65289.1| unknown [Arabidopsis thaliana]
Length = 333
Score = 338 bits (867), Expect = 1e-91
Identities = 176/319 (55%), Positives = 227/319 (70%), Gaps = 11/319 (3%)
Query: 14 PRFYTPSLSTKPPLSISATSSPELLTAKERRRLRNERRESN-ATNWKEEVENKLIQKTKK 72
P YTP T+ ++ LTAKERR+LRNERRES +W+EEVE KLI+K KK
Sbjct: 18 PSIYTPINKTQFSIAACVIERNHQLTAKERRQLRNERRESKLGYSWREEVEEKLIKKPKK 77
Query: 73 VNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHE 132
+W +ELNLD L + GPQWW VRVSR++G TA+ LAR+LA+ FP++EF VYAP++
Sbjct: 78 RYATWTEELNLDTLAESGPQWWAVRVSRLRGHETAQILARALARQFPEMEFTVYAPSVQV 137
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
K++LK+GSISVK KP+FPGCIF+RC LNK +HD I++ +GVGGFI SKVGNTK+QINKPR
Sbjct: 138 KRKLKNGSISVKPKPVFPGCIFIRCILNKEIHDSIRDVDGVGGFIVSKVGNTKRQINKPR 197
Query: 193 PVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKEL----ESDVSKAIVDSKP 248
PV + D+EAIFKQAK QE AD FEE ++ S ++EL SDV + + +SKP
Sbjct: 198 PVDDSDLEAIFKQAKEAQEKADSEFEEADRAEEEASILASQELLALSNSDVIETVAESKP 257
Query: 249 KRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHF 308
KR RK T+ E + KK KL GSTV+++SG+F F G LKKLN KT ATV F
Sbjct: 258 KRAPRKA----TLATETKA--KKKKLAAGSTVRVLSGTFAEFVGNLKKLNRKTAKATVGF 311
Query: 309 TMFGKENIVDLDVSEIVPE 327
T+FGKE +V++D++E+VPE
Sbjct: 312 TLFGKETLVEIDINELVPE 330
>ref|NP_910028.1| putative transcription factor [Oryza sativa]
gi|15042831|gb|AAK82454.1| putative transcription factor
[Oryza sativa]
Length = 330
Score = 243 bits (620), Expect = 6e-63
Identities = 131/313 (41%), Positives = 194/313 (61%), Gaps = 19/313 (6%)
Query: 19 PSLSTKPPLSISAT--SSPELLTAKERRRLRNERRESNATNWKEEVENKLI-----QKTK 71
P S P +S+S + LT +ERR+ R ERRE A +WKEEV+ +LI ++ K
Sbjct: 25 PRASPGPTISVSMSVDGGEGELTGRERRKQRGERRELRARDWKEEVQERLIHEPARRRKK 84
Query: 72 KVNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIH 131
++W++ LNLD L + GPQWW VRVS G + L +++++ +P++ FK+Y P+I
Sbjct: 85 PPKRTWRENLNLDFLAEHGPQWWLVRVSMAPGTDYVDLLTKAISRRYPELSFKIYNPSIQ 144
Query: 132 EKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKP 191
KKRLK+GSIS KSKPL PG +FL C LNK +HD+I++ EG GFIG+ VG+ K+QI KP
Sbjct: 145 VKKRLKNGSISTKSKPLHPGLVFLYCTLNKEVHDFIRDTEGCYGFIGATVGSIKRQIKKP 204
Query: 192 RPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRG 251
+P+ +++E+I ++ K EQE D+ FEE E V S NK +E S+ ++ +K KR
Sbjct: 205 KPIPVEEVESIIREEKEEQERVDREFEEMENGGIVESF--NKPVED--SELMLMNKIKRQ 260
Query: 252 SRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMF 311
+K ++ G++V ++SG F GF G+L ++N K K AT+ T+F
Sbjct: 261 FKKPISK--------GGSNHNAFTPGASVHVLSGPFEGFTGSLLEVNRKNKKATLQLTLF 312
Query: 312 GKENIVDLDVSEI 324
GKE+ VDLD +I
Sbjct: 313 GKESFVDLDFDQI 325
>emb|CAA77858.1| NusG [Thermotoga maritima] gi|128916|sp|P29397|NUSG_THEMA
Transcription antitermination protein nusG
Length = 353
Score = 58.2 bits (139), Expect = 3e-07
Identities = 54/228 (23%), Positives = 92/228 (39%), Gaps = 61/228 (26%)
Query: 97 RVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLR 156
R+ + K E LA + KFF + +V +K ++ K + LFPG +F+
Sbjct: 183 RIKKGKEVKQGEMLAEA-RKFFAKVSGRVEVVDYSTRKEIRI--YKTKRRKLFPGYVFVE 239
Query: 157 CELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKA 216
+N +++++ V GF+ S +P PV + +M I + A +E+
Sbjct: 240 MIMNDEAYNFVRSVPYVMGFVSSG--------GQPVPVKDREMRPILRLAGLEE------ 285
Query: 217 FEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVT 276
+EE++K V G
Sbjct: 286 YEEKKKPVKVELG--------------------------------------------FKV 301
Query: 277 GSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
G V+IISG F FAG +K+++ + + V+ T+FG+E V L VSE+
Sbjct: 302 GDMVKIISGPFEDFAGVIKEIDPERQELKVNVTIFGRETPVVLHVSEV 349
>ref|NP_228263.1| N utilization substance protein G [Thermotoga maritima MSB8]
gi|4980959|gb|AAD35536.1| N utilization substance
protein G [Thermotoga maritima MSB8]
gi|7443529|pir||F72375 transcription antitermination
factor nusG - Thermotoga maritima (strain MSB8)
Length = 354
Score = 58.2 bits (139), Expect = 3e-07
Identities = 54/228 (23%), Positives = 92/228 (39%), Gaps = 61/228 (26%)
Query: 97 RVSRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLR 156
R+ + K E LA + KFF + +V +K ++ K + LFPG +F+
Sbjct: 184 RIKKGKEVKQGEMLAEA-RKFFAKVSGRVEVVDYSTRKEIRI--YKTKRRKLFPGYVFVE 240
Query: 157 CELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKA 216
+N +++++ V GF+ S +P PV + +M I + A +E+
Sbjct: 241 MIMNDEAYNFVRSVPYVMGFVSSG--------GQPVPVKDREMRPILRLAGLEE------ 286
Query: 217 FEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVT 276
+EE++K V G
Sbjct: 287 YEEKKKPVKVELG--------------------------------------------FKV 302
Query: 277 GSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
G V+IISG F FAG +K+++ + + V+ T+FG+E V L VSE+
Sbjct: 303 GDMVKIISGPFEDFAGVIKEIDPERQELKVNVTIFGRETPVVLHVSEV 350
>ref|NP_758271.1| transcription antitermination factor [Mycoplasma penetrans HF-2]
gi|26454346|dbj|BAC44675.1| transcription
antitermination factor [Mycoplasma penetrans HF-2]
Length = 348
Score = 48.1 bits (113), Expect = 3e-04
Identities = 40/154 (25%), Positives = 71/154 (45%), Gaps = 12/154 (7%)
Query: 175 GFIGSKVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENA--DKAFEEEEKKAAVNS--GN 230
GF S + Q NK + + E + + + VE+ + D++ E N N
Sbjct: 199 GFFNSNLN----QENKGEEASSESTENV-ESSNVEEVSVSMDQSTNVEVDTNVENQYLSN 253
Query: 231 PNKELESDVSKAIVDSKPKRGSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGF 290
+ E+E VD+K + +T ++ + + +S K P G+TV+I+ S G+
Sbjct: 254 HDGEVEEVTINEDVDNKQEE---ETEEEVELFHKGNSDSKSPLFAVGNTVEILVESMQGY 310
Query: 291 AGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
G ++ ++ + A V + GKE IVD+ SEI
Sbjct: 311 EGVIESIDLEKDKAKVLVDILGKETIVDVHFSEI 344
>ref|YP_173640.1| transcriptional antitermination factor [Bacillus clausii KSM-K16]
gi|56908152|dbj|BAD62679.1| transcriptional
antitermination factor [Bacillus clausii KSM-K16]
Length = 178
Score = 47.4 bits (111), Expect = 6e-04
Identities = 31/107 (28%), Positives = 51/107 (46%), Gaps = 7/107 (6%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P + E+ +K+G S+ +FPG + + + ++ GV GF+GS
Sbjct: 36 FRVLVP-VEEETEIKNGKTKQVSRKVFPGYVLVEMVMTDDSWYVVRNTPGVTGFVGSAGA 94
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSG 229
+KP P+ D++E I KQ V + + FE +E V SG
Sbjct: 95 G-----SKPTPLMPDEVERILKQMGVVEAQEEVDFELKE-SVKVKSG 135
Score = 38.1 bits (87), Expect = 0.36
Identities = 15/46 (32%), Positives = 29/46 (62%)
Query: 279 TVQIISGSFLGFAGTLKKLNSKTKMATVHFTMFGKENIVDLDVSEI 324
+V++ SG F F GT++++ + + VH MFG+E V+L+ ++
Sbjct: 129 SVKVKSGPFADFVGTIEEIQVEKRKLKVHVNMFGRETPVELEFGQV 174
>ref|NP_892326.1| transcription antitermination protein, NusG [Prochlorococcus
marinus subsp. pastoris str. CCMP1986]
gi|33633707|emb|CAE18664.1| transcription
antitermination protein, NusG [Prochlorococcus marinus
subsp. pastoris str. CCMP1986]
Length = 203
Score = 47.0 bits (110), Expect = 8e-04
Identities = 40/152 (26%), Positives = 72/152 (47%), Gaps = 6/152 (3%)
Query: 92 QWWGVRV-SRVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSKPLFP 150
+W+ V+V S + + A RS+ + ++ P K K GS + +FP
Sbjct: 20 RWYAVQVASSCEKKVKATLEQRSVTLGVNNRIIEIEIPQTPGIKLKKDGSRQTTEEKVFP 79
Query: 151 GCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQIN---KPRPVAEDDMEAIFKQAK 207
G + +R L++ +K V F+G++ G + KPRP++ ++ IFK+A
Sbjct: 80 GYVLVRMILDEDTMMAVKSTPNVINFVGAEDGRGSGRSRGHIKPRPLSRQEVNRIFKRAS 139
Query: 208 VEQENADKAFEEEEKKAAVNSGNPNKELESDV 239
E++ K EE+ + V SG P K+ + +V
Sbjct: 140 -EKKAVIKLDLEEKDRIIVTSG-PFKDFQGEV 169
>ref|YP_159171.1| transcription antitermination protein [Azoarcus sp. EbN1]
gi|56313625|emb|CAI08270.1| transcription
antitermination protein [Azoarcus sp. EbN1]
Length = 177
Score = 46.6 bits (109), Expect = 0.001
Identities = 26/86 (30%), Positives = 41/86 (47%), Gaps = 10/86 (11%)
Query: 130 IHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQIN 189
+ E +KSG S+ + FPG + + E+N +K V GF+G N
Sbjct: 42 VEEVVEMKSGQKSISERKFFPGYVLVEMEMNDESWHLVKSTPKVTGFVGGTA-------N 94
Query: 190 KPRPVAEDDMEAIFKQAKVEQENADK 215
KP P++E ++E I +Q QE +K
Sbjct: 95 KPSPISEKEVEKIMQQM---QEGVEK 117
Score = 37.7 bits (86), Expect = 0.47
Identities = 22/81 (27%), Positives = 39/81 (47%), Gaps = 2/81 (2%)
Query: 246 SKPKRGSRKTSNQLTITEEASSAKKKPKLV--TGSTVQIISGSFLGFAGTLKKLNSKTKM 303
+KP S K ++ + K +PK++ G V++ G F F G ++ +N
Sbjct: 94 NKPSPISEKEVEKIMQQMQEGVEKPRPKVLWELGEVVRVKEGPFTDFHGAVEDVNYDKSK 153
Query: 304 ATVHFTMFGKENIVDLDVSEI 324
V T+FG+ V+LD S++
Sbjct: 154 LRVSVTIFGRATPVELDFSQV 174
>emb|CAE08859.1| transcription antitermination protein, NusG [Synechococcus sp. WH
8102] gi|33866874|ref|NP_898433.1| transcription
antitermination protein, NusG [Synechococcus sp. WH
8102]
Length = 222
Score = 46.2 bits (108), Expect = 0.001
Identities = 41/157 (26%), Positives = 76/157 (48%), Gaps = 16/157 (10%)
Query: 92 QWWGVRVS-----RVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSK 146
+W+ V+V+ +VK A+ ++K +IE PA+ KK GS +
Sbjct: 39 RWYAVQVASSCEKKVKATLEQRAITLGVSKRILEIEIP-QTPAVKVKK---DGSRQSTEE 94
Query: 147 PLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSK----VGNTKKQINKPRPVAEDDMEAI 202
+FPG + +R L++ ++ V F+G++ G + I KPRP++ +++ I
Sbjct: 95 KVFPGYVLVRMVLDEDTMMAVRSTPNVINFVGAEDRRAAGKARGHI-KPRPLSRAEVDRI 153
Query: 203 FKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDV 239
FK+A E++ K E + V +G P K+ + +V
Sbjct: 154 FKRA-AEKKTVVKVDLTEGDQILVTAG-PFKDFQGEV 188
>ref|YP_067091.1| transcription antitermination protein NusG (N utilization substance
protein G) [Rickettsia typhi str. Wilmington]
gi|51459646|gb|AAU03609.1| transcription antitermination
protein NusG (N utilization substance protein G)
[Rickettsia typhi str. Wilmington]
Length = 192
Score = 46.2 bits (108), Expect = 0.001
Identities = 55/254 (21%), Positives = 93/254 (35%), Gaps = 77/254 (30%)
Query: 80 ELNLDNLMKLGP----QWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAI 130
E +DN++ QW+ V + + E + R +AK FF DI V+ +
Sbjct: 3 EQTIDNILPASKNNVKQWYVVHTASGAEKRIKEDILRKIAKQKMTDFFEDILIPVFG--V 60
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
E KR K+ + K L P I ++ + +K GV GF+GSK+
Sbjct: 61 SEVKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNISGVTGFLGSKI--------V 109
Query: 191 PRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKR 250
P+ + E +++ IF E E K A NS K E
Sbjct: 110 PKALTESEIQNIFNNL------------EAEAKVAKNS----KLYE-------------- 139
Query: 251 GSRKTSNQLTITEEASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSKTKMATVHFTM 310
G V + G F F GT+++++ + V ++
Sbjct: 140 -------------------------VGEVVTVTDGPFETFMGTVEEIDKERNRLKVSVSI 174
Query: 311 FGKENIVDLDVSEI 324
FGK ++L+ +++
Sbjct: 175 FGKATPIELNFNQV 188
>ref|ZP_00328344.1| COG0250: Transcription antiterminator [Trichodesmium erythraeum
IMS101]
Length = 224
Score = 45.8 bits (107), Expect = 0.002
Identities = 53/209 (25%), Positives = 92/209 (43%), Gaps = 27/209 (12%)
Query: 64 NKLIQKTKKVNKSWKDELNLDNLMKLGPQWWGVRVSRVKGQYTAEALARSLAKF-FPDIE 122
N +++T++ + + E K +W+ V+V+ + L++ + D
Sbjct: 2 NLSLEETQQTTQLDQGEEKTTEFAKGELRWYAVQVASGCEKRVKTNLSQRIQTLDITDKI 61
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYE----------- 171
FKV P I K K GS + +FPG + ++ +LN H+ KE+
Sbjct: 62 FKVEIPHIAAIKLRKDGSRQHSEEKVFPGYVLVQMKLN--WHEEQKEWRIDDEAWQIVKS 119
Query: 172 --GVGGFIGS----KVGNTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAA 225
V F+G+ + G + + KP P++ ++E IF++A+ EQE K E
Sbjct: 120 TPNVINFVGAEQKRRYGRGRGHV-KPLPLSSSEVERIFRRAQ-EQEPYLKVTVAEGDHIT 177
Query: 226 VNSGNPNKELESDVSKAIVDSKPKRGSRK 254
V SG P K+ E DV ++ P+R K
Sbjct: 178 VLSG-PFKDFEGDV----IEVSPERNKLK 201
>ref|NP_387982.1| transcription antitermination factor [Bacillus subtilis subsp.
subtilis str. 168] gi|2632368|emb|CAB11877.1|
transcription antitermination factor [Bacillus subtilis
subsp. subtilis str. 168] gi|541423|pir||S39859
transcription antitermination factor nusG - Bacillus
subtilis gi|548391|sp|Q06795|NUSG_BACSU Transcription
antitermination protein nusG gi|285628|dbj|BAA02560.1|
transcription antitermination factor NusG [Bacillus
subtilis]
Length = 177
Score = 45.8 bits (107), Expect = 0.002
Identities = 33/127 (25%), Positives = 57/127 (43%), Gaps = 11/127 (8%)
Query: 123 FKVYAPAIHEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVG 182
F+V P E+ +K+G V K +FPG + + + ++ GV GF+GS
Sbjct: 36 FRVVVPE-EEETDIKNGKKKVVKKKVFPGYVLVEIVMTDDSWYVVRNTPGVTGFVGSAGS 94
Query: 183 NTKKQINKPRPVAEDDMEAIFKQAKVEQENADKAFEEEEKKAAVNSGNPN-----KELES 237
+KP P+ + E I K+ +++ D FE +E ++ N +E++
Sbjct: 95 G-----SKPTPLLPGEAETILKRMGMDERKTDIDFELKETVKVIDGPFANFTGSIEEIDY 149
Query: 238 DVSKAIV 244
D SK V
Sbjct: 150 DKSKVKV 156
>emb|CAE22261.1| transcription antitermination protein, NusG [Prochlorococcus
marinus str. MIT 9313] gi|33864351|ref|NP_895911.1|
transcription antitermination protein, NusG
[Prochlorococcus marinus str. MIT 9313]
Length = 223
Score = 44.7 bits (104), Expect = 0.004
Identities = 40/157 (25%), Positives = 75/157 (47%), Gaps = 16/157 (10%)
Query: 92 QWWGVRVS-----RVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSK 146
+W+ ++V+ +VK A+ ++ +IE PA+ KK GS +
Sbjct: 40 RWYAIQVASSCEKKVKATLEQRAVTLGVSNRILEIEIP-QTPAVKLKK---DGSRQSTEE 95
Query: 147 PLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSK----VGNTKKQINKPRPVAEDDMEAI 202
+FPG + +R L++ ++ V F+G++ G + I KPRP++ +++ I
Sbjct: 96 KVFPGYVLVRMVLDEDTMMAVRSTPNVINFVGAEDRRATGKARGHI-KPRPLSRQEVDRI 154
Query: 203 FKQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDV 239
FK+A E++ K E + V SG P K+ + +V
Sbjct: 155 FKRA-AEKKTLVKVDLAEGDQILVTSG-PFKDFQGEV 189
>ref|NP_359813.1| transcription antitermination protein NusG [Rickettsia conorii str.
Malish 7] gi|15619223|gb|AAL02714.1| transcription
antitermination protein NusG [Rickettsia conorii str.
Malish 7] gi|25299615|pir||H97721 transcription
antitermination protein NusG nusG [imported] -
Rickettsia conorii (strain Malish 7)
gi|32129825|sp|Q92J91|NUSG_RICCN Transcription
antitermination protein nusG
Length = 192
Score = 44.7 bits (104), Expect = 0.004
Identities = 37/141 (26%), Positives = 62/141 (43%), Gaps = 22/141 (15%)
Query: 82 NLDNLM----KLGPQWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAIHE 132
++DN++ K QW+ V + + E + R +AK FF DI V+ + E
Sbjct: 5 SIDNILSSSEKNVKQWYVVHTASGAEKRIKEDMLRKIAKQNMTHFFEDILIPVFG--VSE 62
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
KR K+ + K L P I ++ + +K GV GF+GSK P+
Sbjct: 63 VKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNISGVTGFLGSK--------TTPK 111
Query: 193 PVAEDDMEAIFKQAKVEQENA 213
+ E +++ IF + E + A
Sbjct: 112 ALTESEIQNIFNNLEAEAKEA 132
Score = 36.6 bits (83), Expect = 1.1
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 10/84 (11%)
Query: 251 GSRKTSNQLTITE----------EASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSK 300
GS+ T LT +E EA AK G V + G F F GT+++++ +
Sbjct: 105 GSKTTPKALTESEIQNIFNNLEAEAKEAKNSKLYEVGEIVTVTDGPFETFMGTVEEIDQE 164
Query: 301 TKMATVHFTMFGKENIVDLDVSEI 324
V +FGK ++L+ +++
Sbjct: 165 KNRLKVSVAIFGKATPIELNFNQV 188
>dbj|BAB76998.1| transcription antitermination protein [Nostoc sp. PCC 7120]
gi|17232791|ref|NP_489339.1| transcription
antitermination protein [Nostoc sp. PCC 7120]
gi|25299626|pir||AC2468 transcription antitermination
protein nusG [imported] - Nostoc sp. (strain PCC 7120)
Length = 210
Score = 44.7 bits (104), Expect = 0.004
Identities = 47/171 (27%), Positives = 77/171 (44%), Gaps = 18/171 (10%)
Query: 92 QWWGVRVS-----RVKGQYTAEALARSLAKFFPDIEFKVYAPAIHEKKRLKSGSISVKSK 146
+W+ V+V+ RVK +A+ +E + PA+ K K GS +
Sbjct: 27 RWYAVQVASGCEKRVKTNLEQRIQTFDVAEKIVQVEIP-HTPAV---KIRKDGSRQHTEE 82
Query: 147 PLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGS--KVGNTKKQIN-KPRPVAEDDMEAIF 203
+FPG + +R ++ ++ V F+G+ K G K + + KP P++ ++E IF
Sbjct: 83 KVFPGYVLVRMVMDDDTWQVVRNTSHVINFVGAEQKRGTGKGRGHVKPLPLSHSEVERIF 142
Query: 204 KQAKVEQENADKAFEEEEKKAAVNSGNPNKELESDVSKAIVDSKPKRGSRK 254
KQ EQE K K V SG P K+ E +V ++ P+R K
Sbjct: 143 KQT-TEQEPVVKIDMASGDKIIVLSG-PFKDFEGEV----IEVSPERSKLK 187
>ref|ZP_00153237.1| COG0250: Transcription antiterminator [Rickettsia rickettsii]
Length = 192
Score = 44.7 bits (104), Expect = 0.004
Identities = 37/141 (26%), Positives = 62/141 (43%), Gaps = 22/141 (15%)
Query: 82 NLDNLM----KLGPQWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAIHE 132
++DN++ K QW+ V + + E + R +AK FF DI V+ + E
Sbjct: 5 SIDNILPSSEKNVKQWYVVHTASGAEKRIKEDMLRKIAKQNMTDFFEDILIPVFG--VSE 62
Query: 133 KKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINKPR 192
KR K+ + K L P I ++ + +K GV GF+GSK P+
Sbjct: 63 VKRGKNFKVE---KKLMPSYILIKMNMTDKSWHLVKNISGVTGFLGSK--------TTPK 111
Query: 193 PVAEDDMEAIFKQAKVEQENA 213
+ E +++ IF + E + A
Sbjct: 112 ALTESEIQNIFNNLEAEAKEA 132
Score = 35.4 bits (80), Expect = 2.3
Identities = 22/84 (26%), Positives = 37/84 (43%), Gaps = 10/84 (11%)
Query: 251 GSRKTSNQLTITE----------EASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSK 300
GS+ T LT +E EA AK G V + G F F GT++ ++ +
Sbjct: 105 GSKTTPKALTESEIQNIFNNLEAEAKEAKNSKLYEVGEIVTVTDGPFETFMGTVEGIDQE 164
Query: 301 TKMATVHFTMFGKENIVDLDVSEI 324
V +FGK ++L+ +++
Sbjct: 165 KNRLKVSVAIFGKATPIELNFNQV 188
>ref|NP_220526.1| TRANSCRIPTION ANTITERMINATION PROTEIN NUSG (nusG) [Rickettsia
prowazekii str. Madrid E] gi|987968|emb|CAA90886.1|
transcription antitermination factor NusG [Rickettsia
prowazekii] gi|3860702|emb|CAA14603.1| TRANSCRIPTION
ANTITERMINATION PROTEIN NUSG (nusG) [Rickettsia
prowazekii] gi|7443530|pir||D71723 transcription
antitermination protein nusg (nusG) RP135 - Rickettsia
prowazekii gi|1709419|sp|P50056|NUSG_RICPR Transcription
antitermination protein nusG
Length = 192
Score = 44.3 bits (103), Expect = 0.005
Identities = 37/143 (25%), Positives = 62/143 (42%), Gaps = 22/143 (15%)
Query: 80 ELNLDNLMKLGP----QWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAI 130
E +DN++ QW+ V + + E + R +AK FF DI V+ +
Sbjct: 3 EQTIDNILPASKNNVKQWYVVHTASGAEKRIKEDILRKIAKQKMTDFFEDILIPVFG--V 60
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
E KR K+ + K L P I ++ + +K GV GF+GSK+
Sbjct: 61 SEVKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNIPGVTGFLGSKI--------V 109
Query: 191 PRPVAEDDMEAIFKQAKVEQENA 213
P+ + E +++ IF + E + A
Sbjct: 110 PKALTESEIQNIFNNLEAEAKVA 132
>ref|YP_247167.1| Transcription antitermination protein NusG [Rickettsia felis
URRWXCal2] gi|67005076|gb|AAY62002.1| Transcription
antitermination protein NusG [Rickettsia felis
URRWXCal2]
Length = 192
Score = 44.3 bits (103), Expect = 0.005
Identities = 40/153 (26%), Positives = 68/153 (44%), Gaps = 24/153 (15%)
Query: 80 ELNLDNLM----KLGPQWWGVRVSRVKGQYTAEALARSLAK-----FFPDIEFKVYAPAI 130
E ++DN++ K QW+ V + + E + R +AK FF DI V+ +
Sbjct: 3 EQSIDNILPSSEKNVKQWYVVHTASGAEKRIKEDMLRRIAKQKMTDFFEDILIPVFG--V 60
Query: 131 HEKKRLKSGSISVKSKPLFPGCIFLRCELNKPLHDYIKEYEGVGGFIGSKVGNTKKQINK 190
E KR K+ + K L P I ++ + +K GV GF+GSK
Sbjct: 61 SEVKRGKNVKVE---KKLMPSYILIKMNMTDKSWHLVKNIPGVTGFLGSK--------TT 109
Query: 191 PRPVAEDDMEAIFK--QAKVEQENADKAFEEEE 221
P+ + E +++ IF +A+ ++ K +E E
Sbjct: 110 PKALTESEIQNIFNNLEAEAKETKNSKLYEVGE 142
Score = 35.0 bits (79), Expect = 3.1
Identities = 21/84 (25%), Positives = 38/84 (45%), Gaps = 10/84 (11%)
Query: 251 GSRKTSNQLTITE----------EASSAKKKPKLVTGSTVQIISGSFLGFAGTLKKLNSK 300
GS+ T LT +E EA K G V + G F F GT+++++ +
Sbjct: 105 GSKTTPKALTESEIQNIFNNLEAEAKETKNSKLYEVGEIVIVTDGPFETFTGTVEEIDQE 164
Query: 301 TKMATVHFTMFGKENIVDLDVSEI 324
V ++FGK ++L+ +++
Sbjct: 165 KNRLKVSVSIFGKATPIELNFNQV 188
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.311 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 545,452,251
Number of Sequences: 2540612
Number of extensions: 22472758
Number of successful extensions: 93293
Number of sequences better than 10.0: 482
Number of HSP's better than 10.0 without gapping: 112
Number of HSP's successfully gapped in prelim test: 385
Number of HSP's that attempted gapping in prelim test: 92347
Number of HSP's gapped (non-prelim): 1158
length of query: 329
length of database: 863,360,394
effective HSP length: 128
effective length of query: 201
effective length of database: 538,162,058
effective search space: 108170573658
effective search space used: 108170573658
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 75 (33.5 bits)
Medicago: description of AC144474.7


