

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144389.8 - phase: 0
(468 letters)
Database: nr
2,540,612 sequences; 863,360,394 total letters
Searching..................................................done
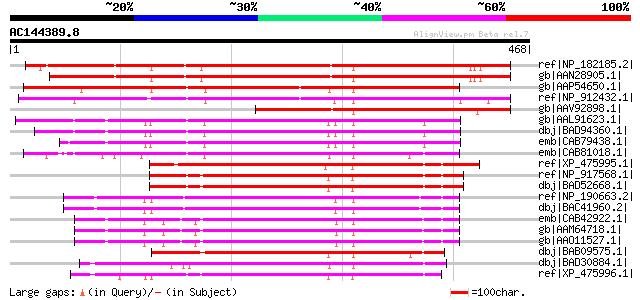
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_182185.2| AAA-type ATPase family protein [Arabidopsis tha... 506 e-142
gb|AAN28905.1| At2g46620/F13A10.15 [Arabidopsis thaliana] gi|441... 496 e-139
gb|AAP54650.1| hypothetical protein [Oryza sativa (japonica cult... 389 e-107
ref|NP_912432.1| Hypothetical protein [Oryza sativa (japonica cu... 379 e-104
gb|AAV92898.1| Avr9/Cf-9 rapidly elicited protein 102 [Nicotiana... 279 1e-73
gb|AAL91623.1| AT4g25830/F14M19_110 [Arabidopsis thaliana] gi|18... 262 2e-68
dbj|BAD94360.1| BCS1 like mitochondrial protein [Arabidopsis tha... 258 3e-67
emb|CAB79438.1| putative mitochondrial protein [Arabidopsis thal... 256 1e-66
emb|CAB81018.1| putative protein [Arabidopsis thaliana] gi|57301... 246 1e-63
ref|XP_475995.1| putative AAA-type ATPase [Oryza sativa (japonic... 235 2e-60
ref|NP_917568.1| P0681B11.25 [Oryza sativa (japonica cultivar-gr... 232 2e-59
dbj|BAD52668.1| BCS1 protein precursor-like [Oryza sativa (japon... 232 2e-59
ref|NP_190663.2| AAA-type ATPase family protein [Arabidopsis tha... 231 3e-59
dbj|BAC41960.2| putative BCS1 protein [Arabidopsis thaliana] 230 6e-59
emb|CAB42922.1| putative mitochondrial protein [Arabidopsis thal... 228 3e-58
gb|AAM64718.1| BCS1 protein-like protein [Arabidopsis thaliana] 228 3e-58
gb|AAO11527.1| At3g50930/F18B3_210 [Arabidopsis thaliana] gi|180... 228 3e-58
dbj|BAB09575.1| AAA-type ATPase-like protein [Arabidopsis thalia... 227 5e-58
dbj|BAD30884.1| AAA-type ATPase-like [Oryza sativa (japonica cul... 224 3e-57
ref|XP_475996.1| putative AAA-type ATPase [Oryza sativa (japonic... 222 2e-56
>ref|NP_182185.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 491
Score = 506 bits (1303), Expect = e-142
Identities = 275/476 (57%), Positives = 339/476 (70%), Gaps = 43/476 (9%)
Query: 15 ILLFSIYAIHMI----FETGLIHESTKLWRI-IEDWFHVYQVFHVPELNDNMQHNTLYRK 69
+LL S +A+ ++ F+TGLI+ KLWR I DWFHVYQ + VPE NDN+Q N LY+K
Sbjct: 10 LLLVSTFALFLVRILLFKTGLIY-MVKLWRRKIIDWFHVYQFYKVPEFNDNVQENHLYQK 68
Query: 70 LSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFNQTQPN--R 127
+ +Y +SL S++NS NL T + N+++L L NQ + D FLGA V W N + R
Sbjct: 69 VYMYLNSLSSIENSDFTNLFTGKKS-NEIILRLDRNQVVGDEFLGARVCWINGEDEDGAR 127
Query: 128 TFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNAS----------DFGPWR 177
F+L+IRK DK+RIL +Y+QHIH V DE+E Q N +L+ ++N G WR
Sbjct: 128 NFVLKIRKADKRRILGSYLQHIHTVSDELE-QRNTELKLFINVGIDDHLNKKKKKNGRWR 186
Query: 178 FVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSS 237
+PF HP TF+ I METDLKN+VKSDLESFLKGKQYY+RLGR+WKRS+LLYG SGTGKSS
Sbjct: 187 SIPFDHPCTFDNIAMETDLKNKVKSDLESFLKGKQYYNRLGRVWKRSYLLYGPSGTGKSS 246
Query: 238 FIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSS 297
F+AAMANFL YDVY IDLS++ DSDLK +LLQT KS+IV+EDLDR+L+ K ST V S
Sbjct: 247 FVAAMANFLDYDVYDIDLSKVVDDSDLKMLLLQTRGKSVIVIEDLDRHLSTK-STAVNLS 305
Query: 298 GILNFMDGIWS---GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASN 354
GILNF D I S +ER+MVFTM KE +DP +LRPGRVDVHIHFPLCDF++FKTLA+N
Sbjct: 306 GILNFTDSILSSCTADERIMVFTMTGKEQIDPAMLRPGRVDVHIHFPLCDFTAFKTLANN 365
Query: 355 YLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALKTDGDGRGCG 414
YLGVK+HKLF QV+ IF+NGASLSPAEIGELMIANRNSP+RA+K VI AL+TDGD RG G
Sbjct: 366 YLGVKEHKLFSQVEGIFQNGASLSPAEIGELMIANRNSPTRALKHVINALQTDGDRRGTG 425
Query: 415 ---FIE---RRIGNE-------------GDGVDEGARDTRKLYGFFRLKGPRKSSS 451
+E R+ +E G G ++ RKLYG R+K RKS S
Sbjct: 426 RRLLLENGSRKSTSEDVSDDMSGSLCGGGGGSSPAVKEFRKLYGLLRIKSSRKSGS 481
>gb|AAN28905.1| At2g46620/F13A10.15 [Arabidopsis thaliana]
gi|4415942|gb|AAD20172.1| hypothetical protein
[Arabidopsis thaliana] gi|18700084|gb|AAL77654.1|
At2g46620/F13A10.15 [Arabidopsis thaliana]
gi|25354579|pir||B84905 hypothetical protein At2g46620
[imported] - Arabidopsis thaliana
Length = 459
Score = 496 bits (1278), Expect = e-139
Identities = 266/450 (59%), Positives = 323/450 (71%), Gaps = 38/450 (8%)
Query: 37 KLWRI-IEDWFHVYQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQ 95
KLWR I DWFHVYQ + VPE NDN+Q N LY+K+ +Y +SL S++NS NL T +
Sbjct: 3 KLWRRKIIDWFHVYQFYKVPEFNDNVQENHLYQKVYMYLNSLSSIENSDFTNLFTGKKS- 61
Query: 96 NDVVLTLAPNQTIHDHFLGATVSWFNQTQPN--RTFILRIRKFDKQRILRAYIQHIHAVV 153
N+++L L NQ + D FLGA V W N + R F+L+IRK DK+RIL +Y+QHIH V
Sbjct: 62 NEIILRLDRNQVVGDEFLGARVCWINGEDEDGARNFVLKIRKADKRRILGSYLQHIHTVS 121
Query: 154 DEIEKQGNRDLRFYMNAS----------DFGPWRFVPFTHPSTFETITMETDLKNRVKSD 203
DE+E Q N +L+ ++N G WR +PF HP TF+ I METDLKN+VKSD
Sbjct: 122 DELE-QRNTELKLFINVGIDDHLNKKKKKNGRWRSIPFDHPCTFDNIAMETDLKNKVKSD 180
Query: 204 LESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSD 263
LESFLKGKQYY+RLGR+WKRS+LLYG SGTGKSSF+AAMANFL YDVY IDLS++ DSD
Sbjct: 181 LESFLKGKQYYNRLGRVWKRSYLLYGPSGTGKSSFVAAMANFLDYDVYDIDLSKVVDDSD 240
Query: 264 LKSILLQTAPKSIIVVEDLDRYLTEKSSTTVTSSGILNFMDGIWS---GEERVMVFTMNS 320
LK +LLQT KS+IV+EDLDR+L+ K ST V SGILNF D I S +ER+MVFTM
Sbjct: 241 LKMLLLQTRGKSVIVIEDLDRHLSTK-STAVNLSGILNFTDSILSSCTADERIMVFTMTG 299
Query: 321 KENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPA 380
KE +DP +LRPGRVDVHIHFPLCDF++FKTLA+NYLGVK+HKLF QV+ IF+NGASLSPA
Sbjct: 300 KEQIDPAMLRPGRVDVHIHFPLCDFTAFKTLANNYLGVKEHKLFSQVEGIFQNGASLSPA 359
Query: 381 EIGELMIANRNSPSRAIKTVITALKTDGDGRGCG---FIE---RRIGNE----------- 423
EIGELMIANRNSP+RA+K VI AL+TDGD RG G +E R+ +E
Sbjct: 360 EIGELMIANRNSPTRALKHVINALQTDGDRRGTGRRLLLENGSRKSTSEDVSDDMSGSLC 419
Query: 424 --GDGVDEGARDTRKLYGFFRLKGPRKSSS 451
G G ++ RKLYG R+K RKS S
Sbjct: 420 GGGGGSSPAVKEFRKLYGLLRIKSSRKSGS 449
>gb|AAP54650.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|37536122|ref|NP_922363.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
gi|10122056|gb|AAG13445.1| hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 532
Score = 389 bits (1000), Expect = e-107
Identities = 206/414 (49%), Positives = 279/414 (66%), Gaps = 23/414 (5%)
Query: 13 IAILLFSIYAIHMIFE-TGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQH---NTLYR 68
+A ++ A+ ++ + H ++WR ++W YQ + VP N L+R
Sbjct: 18 VAYAALAVVALRVVLSYKSVAHAVRRMWRWADEWAQAYQYYEVPRFGGGGGEGVENPLFR 77
Query: 69 KLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFNQTQPN-- 126
K + Y +LPSL+++ +++S ND L L P T HD FLGA ++W N
Sbjct: 78 KAAAYVAALPSLEDADAACVLSSACKTNDFSLQLGPGHTAHDAFLGARLAWTNAGPAGDG 137
Query: 127 ----RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGP----WRF 178
+LR+R+ D+ R+LR Y+QH+ +V DE+E + R+LR Y N G W
Sbjct: 138 GGGRERLVLRVRRHDRTRVLRPYLQHVESVADEMELR-RRELRLYANTGGDGAPSPKWTS 196
Query: 179 VPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSF 238
PFTHP+T ET+ M+ +LK RV++DLESFLKG+ YYHRLGR W+RS+LLYG SGTGKS+F
Sbjct: 197 APFTHPATLETVAMDPELKARVRADLESFLKGRAYYHRLGRAWRRSYLLYGPSGTGKSTF 256
Query: 239 IAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLT---EKSSTTVT 295
AAMA FL YDVY ID+SR D DL+++LL+T P+S+I+VEDLDRYL + ++
Sbjct: 257 AAAMARFLVYDVYDIDMSRGGCD-DLRALLLETTPRSLILVEDLDRYLRGGGDGETSAAR 315
Query: 296 SSGILNFMDGIWS--GEERVMVFTMN-SKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLA 352
+S +L+FMDG+ S GEERVMVFTM+ K+ VDP +LRPGR+DVHIHF +CDF FKTLA
Sbjct: 316 TSRMLSFMDGLSSCCGEERVMVFTMSGDKDGVDPAILRPGRLDVHIHFTMCDFEGFKTLA 375
Query: 353 SNYLGVKDHKLFPQVQEIFE-NGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
SNYLG+KDHKL+PQV+E F GA LSPAE+GE+M+ANR SPSRA++TVI AL+
Sbjct: 376 SNYLGLKDHKLYPQVEEGFHAAGARLSPAELGEIMLANRGSPSRALRTVINALQ 429
>ref|NP_912432.1| Hypothetical protein [Oryza sativa (japonica cultivar-group)]
gi|27476092|gb|AAO17023.1| Hypothetical protein [Oryza
sativa (japonica cultivar-group)]
Length = 523
Score = 379 bits (974), Expect = e-104
Identities = 211/491 (42%), Positives = 299/491 (59%), Gaps = 53/491 (10%)
Query: 9 ILLKIAILLFSIYAIHMIFE-TGLIHESTKLWRIIEDWFHVYQVFHVPEL--NDNMQHNT 65
+ + +A ++ A+ M ++ +LWR ++W YQ VP + N
Sbjct: 13 VAVAVAYAALAVVALRMALSYKSALYAVRRLWRWADEWAQAYQYHEVPRFACDGGGAENP 72
Query: 66 LYRKLSLYFHSLPSLQNSQLNNLVTSNTNQND-VVLTLAPNQTIHDHFLGATVSWFNQTQ 124
L+RK + Y LPSL+++ ++++S + N L L P T D FLGA ++W N+
Sbjct: 73 LFRKAAQYVAVLPSLEDADAASVLSSASRTNGGFSLQLGPGHTARDAFLGARLAWTNRGD 132
Query: 125 PNRTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGP-----WRFV 179
+LR+R+ D+ R+LR Y+QH+ +V DE+E + R+LR + N G W
Sbjct: 133 ---VLVLRVRRHDRTRVLRPYLQHVESVADEMELR-RRELRLFANTGVDGSTGTPRWASA 188
Query: 180 PFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFI 239
PFTHP+T +T+ M+ DLK RV++DLE+FLKG+ YYHRLGR+W+RS+LLYG GTGKS+F
Sbjct: 189 PFTHPATLDTVAMDPDLKARVRADLENFLKGRAYYHRLGRVWRRSYLLYGPLGTGKSTFA 248
Query: 240 AAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYLTEKSS--TTVTSS 297
AAMA FL YD+Y +DLSR +D DL+++LL T P+S+I+VEDLDR+L + ++
Sbjct: 249 AAMARFLGYDIYDVDLSRAGSD-DLRALLLHTTPRSLILVEDLDRFLQGGGAGDAEARAA 307
Query: 298 GILNFMDGIWS--GEERVMVFTMNS-KENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASN 354
+L+FMDG+ S GEERVMVFTM KE VD ++RPGR+DVHIHF LCDF +FK LASN
Sbjct: 308 RVLSFMDGVASCCGEERVMVFTMRGGKEGVDAAVVRPGRLDVHIHFTLCDFEAFKALASN 367
Query: 355 YLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALK--------- 405
YLG+KDHKL+PQV+E F GA LSPAE+GE+M+ANR+SPSRA++ VIT L+
Sbjct: 368 YLGLKDHKLYPQVEESFHGGARLSPAELGEIMLANRSSPSRALRNVITKLQHVSGAAAAP 427
Query: 406 -------TDGDGRGCGFIERRIGNEGDGVDEG------------------ARDTRKLYGF 440
T G G + E+ D D G R+ +KLYG
Sbjct: 428 RPPHRRNTSWSGAGGPWEEQAARASADAADGGEEAITATAACGVFAKDAPMREFKKLYGL 487
Query: 441 FRLKGPRKSSS 451
+++ ++ SS
Sbjct: 488 IKIRSRKEGSS 498
>gb|AAV92898.1| Avr9/Cf-9 rapidly elicited protein 102 [Nicotiana tabacum]
Length = 258
Score = 279 bits (714), Expect = 1e-73
Identities = 142/244 (58%), Positives = 186/244 (76%), Gaps = 15/244 (6%)
Query: 222 KRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVED 281
KRS+LLYG SGTGKS+FIA AN L YDVY +DLSR++ DSDLK +LLQT KS+IV+ED
Sbjct: 1 KRSYLLYGPSGTGKSTFIAGAANMLKYDVYDVDLSRVTDDSDLKLLLLQTTNKSLIVIED 60
Query: 282 LDRYLTEKSSTTVTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIH 339
LD YL KS T V+ SGILNF+DGI+S GEER+M+FT+N+K+ +DP +LRPGR+DVHIH
Sbjct: 61 LDSYLGNKS-TAVSLSGILNFLDGIFSCCGEERIMIFTVNNKDQIDPTVLRPGRIDVHIH 119
Query: 340 FPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKT 399
FPLCDF++FK+LA+++LG+KDHKLFPQV+EIF+ GA LSPAEI E+MI+NR+SP+RA+K+
Sbjct: 120 FPLCDFNAFKSLANSHLGLKDHKLFPQVEEIFQTGAVLSPAEISEIMISNRSSPTRALKS 179
Query: 400 VITALKTDGDGRGCGFIERRI------------GNEGDGVDEGARDTRKLYGFFRLKGPR 447
VI+AL + + R RR+ G+ G + R+ +KLYG R++ R
Sbjct: 180 VISALHINTESRAATRHARRLSESGSVRTVEETGDSGIFCKDSVREFKKLYGLLRIRSSR 239
Query: 448 KSSS 451
K SS
Sbjct: 240 KDSS 243
>gb|AAL91623.1| AT4g25830/F14M19_110 [Arabidopsis thaliana]
gi|18416629|ref|NP_567730.1| AAA-type ATPase family
protein [Arabidopsis thaliana]
Length = 506
Score = 262 bits (669), Expect = 2e-68
Identities = 164/453 (36%), Positives = 246/453 (54%), Gaps = 56/453 (12%)
Query: 6 YTPILLKIAILLFSIYAIHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNT 65
+T + + +L F ++ +F L +KL+ F + F + E+ D + N
Sbjct: 5 WTSLASLLGVLAFCQSLMNSVFPPELRFAISKLFNKFFKLFSTFCYFDITEI-DGVNTNE 63
Query: 66 LYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFN---- 121
LY + LY S S+ ++L+ +T N + V L+ N +I D F TV W +
Sbjct: 64 LYNAVQLYLSSSVSIAGNRLS--LTRAVNSSSVTFGLSNNDSIVDTFNSVTVVWEHIVTQ 121
Query: 122 -QTQP---------NRTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNAS 171
QTQ R F LRI+K DK IL +Y+ +I +EI + N+D Y N+
Sbjct: 122 RQTQTFAWRPMPEEKRGFTLRIKKKDKNLILDSYLDYIMEKANEIRRL-NQDRLLYTNSR 180
Query: 172 DFG------PWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSF 225
PW VPF HPSTF+T+ M+ K ++ DL+ F + + +Y R GR WKR +
Sbjct: 181 GGSLDSRGLPWESVPFKHPSTFDTLAMDPVKKQQIMEDLKDFAECQSFYERTGRAWKRGY 240
Query: 226 LLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRY 285
LLYG GTGKSS IAAMAN+L YD+Y ++L+ + ++S+L+ +L++T+ KSIIV+ED+D
Sbjct: 241 LLYGPPGTGKSSMIAAMANYLRYDIYDLELTEVKSNSELRKLLMKTSSKSIIVIEDIDCS 300
Query: 286 L-----TEKSST----------------------TVTSSGILNFMDGIWS--GEERVMVF 316
+ +K ST T+T SG+LNF DG+WS G ER+ VF
Sbjct: 301 INLTNRNKKQSTGSYNEPEMLTGSGLGDDLGDGNTITLSGLLNFTDGLWSCCGSERIFVF 360
Query: 317 TMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQV-QEIFE--N 373
T N E +DP LLR GR+D+HIH C FSS K L NYLG ++ L V +E+ E +
Sbjct: 361 TTNHIEKLDPALLRSGRMDMHIHMSYCTFSSVKILLRNYLGFEEGDLNDVVLKELAEVVD 420
Query: 374 GASLSPAEIGELMIANRNSPSRAIKTVITALKT 406
A ++PA++ E +I NR RA++ ++ L++
Sbjct: 421 RAEITPADVSEALIKNRRDKERAVRELLVDLRS 453
>dbj|BAD94360.1| BCS1 like mitochondrial protein [Arabidopsis thaliana]
Length = 485
Score = 258 bits (659), Expect = 3e-67
Identities = 161/436 (36%), Positives = 239/436 (53%), Gaps = 56/436 (12%)
Query: 23 IHMIFETGLIHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQN 82
++ +F L +KL+ F + F + E+ D + N LY + LY S S+
Sbjct: 1 MNSVFPPELRFAISKLFNKFFKLFSTFCYFDITEI-DGVNTNELYNAVQLYLSSSVSIAG 59
Query: 83 SQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVSWFN-----QTQP---------NRT 128
++L+ +T N + V L+ N +I D F TV W + QTQ R
Sbjct: 60 NRLS--LTRAVNSSSVTFGLSNNDSIVDTFNSVTVVWEHIVTQRQTQTFAWRPMPEEKRG 117
Query: 129 FILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFG------PWRFVPFT 182
F LRI+K DK IL +Y+ +I +EI + N+D Y N+ PW VPF
Sbjct: 118 FTLRIKKKDKNLILDSYLDYIMEKANEIRRL-NQDRLLYTNSRGGSLDSRGLPWESVPFK 176
Query: 183 HPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAM 242
HPSTF+T+ M+ K ++ DL+ F + + +Y R GR WKR +LLYG GTGKSS IAAM
Sbjct: 177 HPSTFDTLAMDPVKKQQIMEDLKDFAECQSFYERTGRAWKRGYLLYGPPGTGKSSMIAAM 236
Query: 243 ANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYL-----TEKSST----- 292
AN+L YD+Y ++L+ + ++S+L+ +L++T+ KSIIV+ED+D + +K ST
Sbjct: 237 ANYLRYDIYDLELTEVKSNSELRKLLMKTSSKSIIVIEDIDCSINLTNRNKKQSTGSYNE 296
Query: 293 -----------------TVTSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGR 333
T+T SG+LNF DG+WS G ER+ VFT N E +DP LLR GR
Sbjct: 297 PEMLTGSGLGDDLGDGNTITLSGLLNFTDGLWSCCGSERIFVFTTNHIEKLDPALLRSGR 356
Query: 334 VDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQV-QEIFE--NGASLSPAEIGELMIANR 390
+D+HIH C FSS K L NYLG ++ L V +E+ E + A ++PA++ E +I NR
Sbjct: 357 MDMHIHMSYCTFSSVKILLRNYLGFEEGDLNDVVLKELAEVVDRAEITPADVSEALIKNR 416
Query: 391 NSPSRAIKTVITALKT 406
RA++ ++ L++
Sbjct: 417 RDKERAVRELLVDLRS 432
>emb|CAB79438.1| putative mitochondrial protein [Arabidopsis thaliana]
gi|4539301|emb|CAB39604.1| putative mitochondrial
protein [Arabidopsis thaliana] gi|7485479|pir||T04237
hypothetical protein F14M19.110 - Arabidopsis thaliana
Length = 618
Score = 256 bits (654), Expect = 1e-66
Identities = 156/413 (37%), Positives = 233/413 (55%), Gaps = 56/413 (13%)
Query: 46 FHVYQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPN 105
F++++++ P+ D + N LY + LY S S+ ++L+ +T N + V L+ N
Sbjct: 157 FNLFRLYS-PKQIDGVNTNELYNAVQLYLSSSVSIAGNRLS--LTRAVNSSSVTFGLSNN 213
Query: 106 QTIHDHFLGATVSWFN-----QTQP---------NRTFILRIRKFDKQRILRAYIQHIHA 151
+I D F TV W + QTQ R F LRI+K DK IL +Y+ +I
Sbjct: 214 DSIVDTFNSVTVVWEHIVTQRQTQTFAWRPMPEEKRGFTLRIKKKDKNLILDSYLDYIME 273
Query: 152 VVDEIEKQGNRDLRFYMNASDFG------PWRFVPFTHPSTFETITMETDLKNRVKSDLE 205
+EI + N+D Y N+ PW VPF HPSTF+T+ M+ K ++ DL+
Sbjct: 274 KANEIRRL-NQDRLLYTNSRGGSLDSRGLPWESVPFKHPSTFDTLAMDPVKKQQIMEDLK 332
Query: 206 SFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLK 265
F + + +Y R GR WKR +LLYG GTGKSS IAAMAN+L YD+Y ++L+ + ++S+L+
Sbjct: 333 DFAECQSFYERTGRAWKRGYLLYGPPGTGKSSMIAAMANYLRYDIYDLELTEVKSNSELR 392
Query: 266 SILLQTAPKSIIVVEDLDRYL-----TEKSST----------------------TVTSSG 298
+L++T+ KSIIV+ED+D + +K ST T+T SG
Sbjct: 393 KLLMKTSSKSIIVIEDIDCSINLTNRNKKQSTGSYNEPEMLTGSGLGDDLGDGNTITLSG 452
Query: 299 ILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYL 356
+LNF DG+WS G ER+ VFT N E +DP LLR GR+D+HIH C FSS K L NYL
Sbjct: 453 LLNFTDGLWSCCGSERIFVFTTNHIEKLDPALLRSGRMDMHIHMSYCTFSSVKILLRNYL 512
Query: 357 GVKDHKLFPQV-QEIFE--NGASLSPAEIGELMIANRNSPSRAIKTVITALKT 406
G ++ L V +E+ E + A ++PA++ E +I NR RA++ ++ L++
Sbjct: 513 GFEEGDLNDVVLKELAEVVDRAEITPADVSEALIKNRRDKERAVRELLVDLRS 565
>emb|CAB81018.1| putative protein [Arabidopsis thaliana] gi|5730135|emb|CAB52469.1|
putative protein [Arabidopsis thaliana]
gi|15234674|ref|NP_194754.1| AAA-type ATPase family
protein [Arabidopsis thaliana] gi|7486760|pir||T14085
hypothetical protein F9N11.100 - Arabidopsis thaliana
Length = 512
Score = 246 bits (627), Expect = 1e-63
Identities = 151/455 (33%), Positives = 240/455 (52%), Gaps = 69/455 (15%)
Query: 13 IAILLFSIYAIHMIFETGL----IHESTKLWRIIEDWFHVYQVFHVPELNDNMQHNTLYR 68
+ +L F + ++F L +H T++ + H+Y F + E+ D + N LY
Sbjct: 5 LGMLAFCQTIVQLVFPPELRLAFLHFLTRIRHVFSS--HIY--FDITEI-DGVNTNELYN 59
Query: 69 KLSLYFHSLPSLQN---SQLNNLVTSNT---NQNDVVLTLAPNQTIHDHFLGATV----- 117
+ LY S ++ + S NN S T N + V L+ N I D F G T+
Sbjct: 60 AVQLYLSSSVTVNDAVSSSNNNTRLSLTRVPNSSSVTFGLSNNDRITDVFNGVTILWEHV 119
Query: 118 ---------SWFNQTQPNRTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYM 168
SW + R F L+I K DK +L +Y+ +I +EI ++ N + Y
Sbjct: 120 VVQRQVQSFSWRPMPEEKRGFTLQINKRDKALVLDSYLDYIVGKSEEIRRR-NEERLLYT 178
Query: 169 NASDFG------PWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWK 222
N+ PW V F HPSTF+T+ M+ + K R+ DL F G+ +Y + GR WK
Sbjct: 179 NSRGVSLDARSHPWDSVRFKHPSTFDTLAMDPEKKKRIMEDLREFANGQGFYQKTGRAWK 238
Query: 223 RSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDL 282
R +LLYG GTGKSS IAAMAN+L YD+Y ++L+ + +S+L+ +L++T+ KSIIV+ED+
Sbjct: 239 RGYLLYGPPGTGKSSLIAAMANYLGYDIYDLELTEVQNNSELRKLLMKTSSKSIIVIEDI 298
Query: 283 DRYLT--------------------------EKSSTTVTSSGILNFMDGIWS--GEERVM 314
D ++ E+ ++VT SG+LNF DG+WS G E++
Sbjct: 299 DCSISLTKRGKNKKKNGSYEYDPGLTNGSGLEEPGSSVTLSGLLNFTDGLWSCCGSEKIF 358
Query: 315 VFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVK----DHKLFPQVQEI 370
VFT N E +D L+R GR+D+H+H C F + K L NYL ++ D + +++E
Sbjct: 359 VFTTNHIEKLDSALMRSGRMDMHVHMGFCKFPALKILLKNYLRLEEEDMDSVVLKEMEEC 418
Query: 371 FENGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
E A ++PA++ E++I NR+ +A++ +++ LK
Sbjct: 419 VEE-AEITPADVSEVLIRNRSDAEKAVREIVSVLK 452
>ref|XP_475995.1| putative AAA-type ATPase [Oryza sativa (japonica cultivar-group)]
gi|47777363|gb|AAT37997.1| putative AAA-type ATPase
[Oryza sativa (japonica cultivar-group)]
Length = 479
Score = 235 bits (600), Expect = 2e-60
Identities = 123/317 (38%), Positives = 194/317 (60%), Gaps = 25/317 (7%)
Query: 127 RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPST 186
++F + K K++ LR+Y+ V+D + ++ M+ ++ W V HPST
Sbjct: 158 KSFEVSFHKKHKEKALRSYLPF---VIDTAKAMNDKHRNLKMHMIEYDAWTAVDLRHPST 214
Query: 187 FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFL 246
F+T+ M+ LK+ V DLE F+K K YY R+GR WKR +LLYG GTGKSS IAAMAN+L
Sbjct: 215 FDTLAMDHSLKHSVMYDLERFVKRKDYYRRIGRAWKRGYLLYGPPGTGKSSLIAAMANYL 274
Query: 247 SYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLD------------------RYLTE 288
+D+Y ++L+ + ++SDL+ +L+ + +SI+VVED+D Y E
Sbjct: 275 KFDIYDLELTEVKSNSDLRRLLVGMSNRSILVVEDIDCTIDLQQRDEGEIKRAKPTYSGE 334
Query: 289 KSSTTVTSSGILNFMDGIW--SGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFS 346
++ VT SG+LNF+DG+W SGEER++VFT N +E +DP LLRPGR+D+HIH C
Sbjct: 335 ENEDKVTLSGLLNFVDGLWSTSGEERIIVFTTNYRERLDPALLRPGRMDMHIHMGYCTRE 394
Query: 347 SFKTLASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALKT 406
+F+ LASNY V++H ++P+++++ E +PAE+ E+++ N + A++ + LK
Sbjct: 395 AFRVLASNYHNVENHAMYPEIEQLIEE-VLTTPAEVAEVLMRN-DDVDVALQVLAEFLKA 452
Query: 407 DGDGRGCGFIERRIGNE 423
+ G E + GN+
Sbjct: 453 KRNEPGETKAENKNGNQ 469
>ref|NP_917568.1| P0681B11.25 [Oryza sativa (japonica cultivar-group)]
Length = 486
Score = 232 bits (591), Expect = 2e-59
Identities = 123/298 (41%), Positives = 186/298 (62%), Gaps = 20/298 (6%)
Query: 127 RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPST 186
R+F + K K + L +Y+ HI A +I+ Q +R L+ YMN + W + HPST
Sbjct: 154 RSFEMSFHKKHKDKALNSYLPHILATAKKIKDQ-DRTLKIYMNEGE--SWFAIDLHHPST 210
Query: 187 FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFL 246
F T+ M+ K V DLE F+K K+YY ++G+ WKR +LLYG GTGKSS IAAMAN+L
Sbjct: 211 FTTLAMDHKQKQSVMDDLERFIKRKEYYKKIGKAWKRGYLLYGPPGTGKSSLIAAMANYL 270
Query: 247 SYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYL-------------TEKSSTT 293
+DVY ++L+ ++ +S L+ +L+ +SI+V+ED+D L + S
Sbjct: 271 KFDVYDLELTEVNWNSTLRRLLIGMTNRSILVIEDIDCTLELQQREEGQESSKSNPSEDK 330
Query: 294 VTSSGILNFMDGIW--SGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
VT SG+LNF+DG+W SGEER++VFT N KE +DP LLRPGR+D+H+H C SF+ L
Sbjct: 331 VTLSGLLNFVDGLWSTSGEERIIVFTTNYKERLDPALLRPGRMDMHVHMGYCCPESFRIL 390
Query: 352 ASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALKTDGD 409
ASNY + +H +P+++E+ + ++PAE+ E+++ N + A++ +I LK D
Sbjct: 391 ASNYHSIDNHATYPEIEELIKE-VMVTPAEVAEVLMRN-DDTDVALEGLIQFLKRKKD 446
>dbj|BAD52668.1| BCS1 protein precursor-like [Oryza sativa (japonica
cultivar-group)]
Length = 453
Score = 232 bits (591), Expect = 2e-59
Identities = 123/298 (41%), Positives = 186/298 (62%), Gaps = 20/298 (6%)
Query: 127 RTFILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPST 186
R+F + K K + L +Y+ HI A +I+ Q +R L+ YMN + W + HPST
Sbjct: 121 RSFEMSFHKKHKDKALNSYLPHILATAKKIKDQ-DRTLKIYMNEGE--SWFAIDLHHPST 177
Query: 187 FETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFL 246
F T+ M+ K V DLE F+K K+YY ++G+ WKR +LLYG GTGKSS IAAMAN+L
Sbjct: 178 FTTLAMDHKQKQSVMDDLERFIKRKEYYKKIGKAWKRGYLLYGPPGTGKSSLIAAMANYL 237
Query: 247 SYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYL-------------TEKSSTT 293
+DVY ++L+ ++ +S L+ +L+ +SI+V+ED+D L + S
Sbjct: 238 KFDVYDLELTEVNWNSTLRRLLIGMTNRSILVIEDIDCTLELQQREEGQESSKSNPSEDK 297
Query: 294 VTSSGILNFMDGIW--SGEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTL 351
VT SG+LNF+DG+W SGEER++VFT N KE +DP LLRPGR+D+H+H C SF+ L
Sbjct: 298 VTLSGLLNFVDGLWSTSGEERIIVFTTNYKERLDPALLRPGRMDMHVHMGYCCPESFRIL 357
Query: 352 ASNYLGVKDHKLFPQVQEIFENGASLSPAEIGELMIANRNSPSRAIKTVITALKTDGD 409
ASNY + +H +P+++E+ + ++PAE+ E+++ N + A++ +I LK D
Sbjct: 358 ASNYHSIDNHATYPEIEELIKE-VMVTPAEVAEVLMRN-DDTDVALEGLIQFLKRKKD 413
>ref|NP_190663.2| AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 451
Score = 231 bits (590), Expect = 3e-59
Identities = 137/389 (35%), Positives = 220/389 (56%), Gaps = 37/389 (9%)
Query: 49 YQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTI 108
YQ+ V E +HN ++ Y + + NS V Q++ +T+ ++ +
Sbjct: 58 YQMTAVIEEFGGFEHNQVFEAAEAYLST--KISNSTRRIKVNKLEKQSNYSVTVERDEEV 115
Query: 109 HDHFLGATVSWF-----------------NQTQPN--RTFILRIRKFDKQRILRAYIQHI 149
D F G +SW N T + R++ L RK K +L +Y+ +
Sbjct: 116 VDIFDGVKLSWILVCRHVDKKDFRNPRDLNSTLKSEVRSYELSFRKKFKNMVLESYLPFV 175
Query: 150 HAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLK 209
I KQ + L+ + S W V HPSTF T+ ++ ++K + DL+ F++
Sbjct: 176 VEQAASI-KQKFKTLKIFTVDSYSVEWTSVTLDHPSTFRTLALDPEVKKNLVEDLDRFVQ 234
Query: 210 GKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILL 269
K +Y R+G+ WKR +LLYG GTGKSS IAA+AN L++D+Y +DL+ ++ +++L+ +L+
Sbjct: 235 RKGFYGRVGKAWKRGYLLYGPPGTGKSSLIAAIANHLNFDIYDLDLTSLNNNAELRRLLM 294
Query: 270 QTAPKSIIVVEDLDRYLTEKSST-----------TVTSSGILNFMDGIWS--GEERVMVF 316
TA +SI+VVED+D + K + TVT SG+LNF+DG+WS G ER++VF
Sbjct: 295 STANRSILVVEDIDCSIELKDRSTDQENNDPLHKTVTLSGLLNFVDGLWSSCGNERIIVF 354
Query: 317 TMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGAS 376
T N +E +DP LLRPGR+D+HIH C ++FK LASNYL ++DH LF Q++E F
Sbjct: 355 TTNYREKLDPALLRPGRMDMHIHMSYCTPAAFKVLASNYLEIQDHILFEQIEE-FIREIE 413
Query: 377 LSPAEIGELMIANRNSPSRAIKTVITALK 405
++PAE+ E ++ + +S + ++ ++ LK
Sbjct: 414 VTPAEVAEQLMRS-DSVDKVLQGLVEFLK 441
>dbj|BAC41960.2| putative BCS1 protein [Arabidopsis thaliana]
Length = 451
Score = 230 bits (587), Expect = 6e-59
Identities = 136/389 (34%), Positives = 220/389 (55%), Gaps = 37/389 (9%)
Query: 49 YQVFHVPELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTI 108
YQ+ V E +HN ++ Y + + NS V Q++ +T+ ++ +
Sbjct: 58 YQMTAVIEEFGGFEHNQVFEAAEAYLST--KISNSTRRIKVNKLEKQSNYSVTVERDEEV 115
Query: 109 HDHFLGATVSWF-----------------NQTQPN--RTFILRIRKFDKQRILRAYIQHI 149
D F G +SW N T + R++ L RK K +L +Y+ +
Sbjct: 116 VDIFDGVKLSWILVCRHVDKKDFRNPRDLNSTLKSEVRSYELSFRKKFKNMVLESYLPFV 175
Query: 150 HAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLK 209
I KQ + L+ + S W V HPSTF T+ ++ ++K + DL+ F++
Sbjct: 176 VEQAASI-KQKFKTLKIFTVDSYSVEWTSVTLDHPSTFRTLALDPEVKKNLVEDLDRFVQ 234
Query: 210 GKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILL 269
K +Y R+G+ WKR +LLYG GTGKSS IAA+AN L++D+Y +DL+ ++ +++L+ +L+
Sbjct: 235 RKGFYGRVGKAWKRGYLLYGPPGTGKSSLIAAIANHLNFDIYDLDLTSLNNNAELRRLLM 294
Query: 270 QTAPKSIIVVEDLDRYLTEKSST-----------TVTSSGILNFMDGIWS--GEERVMVF 316
TA +SI+VVED+D + K + TVT SG+LNF+DG+WS G ER++VF
Sbjct: 295 STANRSILVVEDIDCSIELKDRSTDQENNDPLHKTVTLSGLLNFVDGLWSSCGNERIIVF 354
Query: 317 TMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGAS 376
T N +E +DP LLRPGR+D+HIH C ++FK LASNYL ++DH LF Q++E F
Sbjct: 355 TTNYREKLDPALLRPGRMDMHIHMSYCTPAAFKVLASNYLEIQDHILFEQIEE-FIREIE 413
Query: 377 LSPAEIGELMIANRNSPSRAIKTVITALK 405
++P+E+ E ++ + +S + ++ ++ LK
Sbjct: 414 VTPSEVAEQLMRS-DSVDKVLQGLVEFLK 441
>emb|CAB42922.1| putative mitochondrial protein [Arabidopsis thaliana]
gi|20856863|gb|AAM26687.1| AT3g50930/F18B3_210
[Arabidopsis thaliana] gi|13877697|gb|AAK43926.1|
putative mitochondrial protein [Arabidopsis thaliana]
gi|7485703|pir||T08414 hypothetical protein F18B3.210 -
Arabidopsis thaliana
Length = 534
Score = 228 bits (581), Expect = 3e-58
Identities = 136/393 (34%), Positives = 217/393 (54%), Gaps = 53/393 (13%)
Query: 59 DNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVS 118
+ HN ++ Y + S N ++ V+ + +N+ +T+ ++ + D + G
Sbjct: 72 EGFAHNEVFEAAEAYLATKISPSNKRIK--VSKHEKENNYNVTVERDEEVVDTYNGVKFQ 129
Query: 119 WF------------NQTQPNRTFILRIRKFD-------KQRILRAYIQHIHAVVDEIEKQ 159
W N N T +R F+ K L +Y+ + + KQ
Sbjct: 130 WILHCRHVESKHFHNPRDLNSTLRSEVRSFELNFHKKFKDVALESYLPFMVKRAT-LMKQ 188
Query: 160 GNRDLRF--------YMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGK 211
+ L+ Y N SD W V HPSTF+T+ M++D+K V DL+ F+K +
Sbjct: 189 EKKTLKIFTLSPENMYGNYSD--AWTSVTLDHPSTFKTLAMDSDVKTSVMEDLDKFVKRR 246
Query: 212 QYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQT 271
+Y R+G+ WKR +LLYG GTGKSS IAAMAN L++D+Y ++L+ ++ +S+L+ +L+ T
Sbjct: 247 DFYKRVGKAWKRGYLLYGPPGTGKSSLIAAMANHLNFDIYDLELTAVNNNSELRRLLIAT 306
Query: 272 APKSIIVVEDLDRYLTEKSSTT-----------------VTSSGILNFMDGIWS--GEER 312
A +SI++VED+D L K T+ VT SG+LNF+DG+WS G+ER
Sbjct: 307 ANRSILIVEDIDCSLELKDRTSDEPPRESDDIEDPRYKKVTLSGLLNFIDGLWSSCGDER 366
Query: 313 VMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFE 372
+++FT N KE +D LLRPGR+D+HIH C S+FK LA NYL +K+H+LF +++E E
Sbjct: 367 IIIFTTNYKEKLDAALLRPGRMDMHIHMSYCTPSTFKALALNYLEIKEHRLFSKIEEGIE 426
Query: 373 NGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
++PAE+ E ++ N +S + ++ +I LK
Sbjct: 427 -ATEVTPAEVAEQLMRN-DSVDKVLEGLIEFLK 457
>gb|AAM64718.1| BCS1 protein-like protein [Arabidopsis thaliana]
Length = 534
Score = 228 bits (581), Expect = 3e-58
Identities = 136/393 (34%), Positives = 217/393 (54%), Gaps = 53/393 (13%)
Query: 59 DNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVS 118
+ HN ++ Y + S N ++ V+ + +N+ +T+ ++ + D + G
Sbjct: 72 EGFAHNEVFEAAEAYLATKISPSNKRIK--VSKHEKENNYNVTVERDEEVVDTYNGVKFQ 129
Query: 119 WF------------NQTQPNRTFILRIRKFD-------KQRILRAYIQHIHAVVDEIEKQ 159
W N N T +R F+ K L +Y+ + + KQ
Sbjct: 130 WILHCRHVESKHFHNPRDLNSTLRSEVRSFELNFHKKFKDVALESYLPFMVKRAT-LMKQ 188
Query: 160 GNRDLRF--------YMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGK 211
+ L+ Y N SD W V HPSTF+T+ M++D+K V DL+ F+K +
Sbjct: 189 EKKTLKIFTLSPENMYGNYSD--AWTSVTLDHPSTFKTLAMDSDVKTSVMEDLDKFVKRR 246
Query: 212 QYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQT 271
+Y R+G+ WKR +LLYG GTGKSS IAAMAN L++D+Y ++L+ ++ +S+L+ +L+ T
Sbjct: 247 DFYKRVGKAWKRGYLLYGPPGTGKSSLIAAMANHLNFDIYDLELTAVNNNSELRRLLIAT 306
Query: 272 APKSIIVVEDLDRYLTEKSSTT-----------------VTSSGILNFMDGIWS--GEER 312
A +SI++VED+D L K T+ VT SG+LNF+DG+WS G+ER
Sbjct: 307 ANRSILIVEDIDCSLELKDRTSDEPPRESDDIEDPRYKKVTLSGLLNFIDGLWSSCGDER 366
Query: 313 VMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFE 372
+++FT N KE +D LLRPGR+D+HIH C S+FK LA NYL +K+H+LF +++E E
Sbjct: 367 IIIFTTNYKEKLDAALLRPGRMDMHIHMSYCTPSTFKALALNYLEIKEHRLFSKIEEGIE 426
Query: 373 NGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
++PAE+ E ++ N +S + ++ +I LK
Sbjct: 427 -ATEVTPAEVAEQLMRN-DSVDKVLEGLIEFLK 457
>gb|AAO11527.1| At3g50930/F18B3_210 [Arabidopsis thaliana]
gi|18086343|gb|AAL57634.1| AT3g50930/F18B3_210
[Arabidopsis thaliana] gi|30693378|ref|NP_190662.2|
AAA-type ATPase family protein [Arabidopsis thaliana]
Length = 576
Score = 228 bits (581), Expect = 3e-58
Identities = 136/393 (34%), Positives = 217/393 (54%), Gaps = 53/393 (13%)
Query: 59 DNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQNDVVLTLAPNQTIHDHFLGATVS 118
+ HN ++ Y + S N ++ V+ + +N+ +T+ ++ + D + G
Sbjct: 114 EGFAHNEVFEAAEAYLATKISPSNKRIK--VSKHEKENNYNVTVERDEEVVDTYNGVKFQ 171
Query: 119 WF------------NQTQPNRTFILRIRKFD-------KQRILRAYIQHIHAVVDEIEKQ 159
W N N T +R F+ K L +Y+ + + KQ
Sbjct: 172 WILHCRHVESKHFHNPRDLNSTLRSEVRSFELNFHKKFKDVALESYLPFMVKRAT-LMKQ 230
Query: 160 GNRDLRF--------YMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGK 211
+ L+ Y N SD W V HPSTF+T+ M++D+K V DL+ F+K +
Sbjct: 231 EKKTLKIFTLSPENMYGNYSD--AWTSVTLDHPSTFKTLAMDSDVKTSVMEDLDKFVKRR 288
Query: 212 QYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQT 271
+Y R+G+ WKR +LLYG GTGKSS IAAMAN L++D+Y ++L+ ++ +S+L+ +L+ T
Sbjct: 289 DFYKRVGKAWKRGYLLYGPPGTGKSSLIAAMANHLNFDIYDLELTAVNNNSELRRLLIAT 348
Query: 272 APKSIIVVEDLDRYLTEKSSTT-----------------VTSSGILNFMDGIWS--GEER 312
A +SI++VED+D L K T+ VT SG+LNF+DG+WS G+ER
Sbjct: 349 ANRSILIVEDIDCSLELKDRTSDEPPRESDDIEDPRYKKVTLSGLLNFIDGLWSSCGDER 408
Query: 313 VMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFE 372
+++FT N KE +D LLRPGR+D+HIH C S+FK LA NYL +K+H+LF +++E E
Sbjct: 409 IIIFTTNYKEKLDAALLRPGRMDMHIHMSYCTPSTFKALALNYLEIKEHRLFSKIEEGIE 468
Query: 373 NGASLSPAEIGELMIANRNSPSRAIKTVITALK 405
++PAE+ E ++ N +S + ++ +I LK
Sbjct: 469 -ATEVTPAEVAEQLMRN-DSVDKVLEGLIEFLK 499
>dbj|BAB09575.1| AAA-type ATPase-like protein [Arabidopsis thaliana]
gi|30686502|ref|NP_850841.1| AAA-type ATPase family
protein [Arabidopsis thaliana]
Length = 505
Score = 227 bits (579), Expect = 5e-58
Identities = 119/284 (41%), Positives = 179/284 (62%), Gaps = 24/284 (8%)
Query: 129 FILRIRKFDKQRILRAYIQHIHAVVDEIEKQGNRDLRFYMNASDFGPWRFVPFTHPSTFE 188
F L K K IL +Y+ +I + EI + + +N+ W V HPSTFE
Sbjct: 163 FELSFDKKHKDLILNSYVPYIESKAKEIRDERRILMLHSLNSLR---WESVILEHPSTFE 219
Query: 189 TITMETDLKNRVKSDLESFLKGKQYYHRLGRLWKRSFLLYGSSGTGKSSFIAAMANFLSY 248
T+ ME DLK V DL+ F++ K++Y R+G+ WKR +LLYG GTGKSS +AAMAN+L +
Sbjct: 220 TMAMEDDLKRDVIEDLDRFIRRKEFYKRVGKAWKRGYLLYGPPGTGKSSLVAAMANYLKF 279
Query: 249 DVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDLDRYL--------------TEKSSTTV 294
DVY + L+ + DSDL+ +LL T +SI+V+ED+D + +S +
Sbjct: 280 DVYDLQLASVMRDSDLRRLLLATRNRSILVIEDIDCAVDLPNRIEQPVEGKNRGESQGPL 339
Query: 295 TSSGILNFMDGIWS--GEERVMVFTMNSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLA 352
T SG+LNF+DG+WS G+ER+++FT N K+ +DP LLRPGR+D+HI+ C F FKTLA
Sbjct: 340 TLSGLLNFIDGLWSSCGDERIIIFTTNHKDRLDPALLRPGRMDMHIYMGHCSFQGFKTLA 399
Query: 353 SNYLGVKD----HKLFPQVQEIFENGASLSPAEIGELMIANRNS 392
SNYLG+ D H+LFP+++ + + G ++PA++ E ++ + ++
Sbjct: 400 SNYLGLSDAAMPHRLFPEIERLID-GEVMTPAQVAEELMKSEDA 442
>dbj|BAD30884.1| AAA-type ATPase-like [Oryza sativa (japonica cultivar-group)]
Length = 575
Score = 224 bits (572), Expect = 3e-57
Identities = 130/374 (34%), Positives = 208/374 (54%), Gaps = 47/374 (12%)
Query: 64 NTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQND-VVLTLAPNQTIHDHFLGATVSWFNQ 122
+T Y + Y L + +S+ L + D +V+++ Q + D F GAT+ W +
Sbjct: 97 DTTYDEAKAY---LSATCSSEARELHAEGAEEGDGLVISMRDGQDVADEFGGATMWWSSV 153
Query: 123 TQPNRTFILRIRKFDKQRILRA--YIQHIHAVVDE----IEKQG--------------NR 162
+ + ++R LR +++H VVDE + ++G N
Sbjct: 154 AAEQQAAPPPPQGAAERRCLRLTFHMRHRRLVVDEYLPHVRREGREVLFSSRRRRLYTNN 213
Query: 163 DLRFYMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLGRLWK 222
+ Y + SD W +V F HP+TFET+ ME K + DL++F + +++Y R G+ WK
Sbjct: 214 KMSEYASYSDEKAWSYVDFDHPTTFETLAMEPAKKKAIMDDLDAFRRSREFYRRTGKPWK 273
Query: 223 RSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIVVEDL 282
R +LL+G GTGKS+ +AAMAN+L YD+Y ++L+ + +++L+ +L++T KSIIV+ED+
Sbjct: 274 RGYLLHGPPGTGKSTMVAAMANYLDYDIYDVELTVVGNNNNLRKLLIETTSKSIIVIEDI 333
Query: 283 DRYLT--------------------EKSSTTVTSSGILNFMDGIWS--GEERVMVFTMNS 320
D L ++ S+ VT SG+LNF+DG+WS G ER++VFT N
Sbjct: 334 DCSLDITGDRAARRSRPPPSYRDGHDRRSSDVTLSGLLNFIDGLWSACGGERIVVFTTNH 393
Query: 321 KENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLSPA 380
+ +DP L+R GR+D+HI C F +FKTLA NYL V H LF V+E+ + +L+PA
Sbjct: 394 LDKLDPALIRRGRMDMHIEMSYCGFEAFKTLAKNYLDVDAHHLFDAVEELLRD-VNLTPA 452
Query: 381 EIGELMIANRNSPS 394
++ E ++ R S S
Sbjct: 453 DVAECLMTARRSGS 466
>ref|XP_475996.1| putative AAA-type ATPase [Oryza sativa (japonica cultivar-group)]
gi|47777364|gb|AAT37998.1| putative AAA-type ATPase
[Oryza sativa (japonica cultivar-group)]
Length = 484
Score = 222 bits (565), Expect = 2e-56
Identities = 129/371 (34%), Positives = 204/371 (54%), Gaps = 43/371 (11%)
Query: 56 ELNDNMQHNTLYRKLSLYFHSLPSLQNSQLNNLVTSNTNQND--VVLTLAPNQTIHDHFL 113
E + HN +Y + Y L + N+ + L S+ +++ +V+T+ + + D
Sbjct: 64 EETEGWSHNHVYNAVRAY---LATRINNNMQRLRVSSMDESSEKMVVTMEEGEELVDMHE 120
Query: 114 GATVSWFN-----QTQPN----------RTFILRIRKFDKQRILRAYIQHIHAVVDEIEK 158
G W PN R++ L + K++ L++Y+ I A I+
Sbjct: 121 GTEFKWCLISRSISADPNNGNGSGQREVRSYELSFHRKHKEKALKSYLPFIIATAKAIKD 180
Query: 159 QGNRDLRFYMNASDFGPWRFVPFTHPSTFETITMETDLKNRVKSDLESFLKGKQYYHRLG 218
Q R L+ YMN W + HPSTF+T+ M+ LK + DL+ F+K K YY R+G
Sbjct: 181 Q-ERILQIYMNEYS-DSWSPIDLHHPSTFDTLAMDQKLKQSIIDDLDRFIKRKDYYKRIG 238
Query: 219 RLWKRSFLLYGSSGTGKSSFIAAMANFLSYDVYYIDLSRISTDSDLKSILLQTAPKSIIV 278
+ WKR +LLYG GTGKSS IAAMAN L +D+Y ++L+ + ++S+L+ +L+ +SI+V
Sbjct: 239 KAWKRGYLLYGPPGTGKSSLIAAMANHLKFDIYDLELTGVHSNSELRRLLVGMTSRSILV 298
Query: 279 VEDLDRYL------------------TEKSSTTVTSSGILNFMDGIW--SGEERVMVFTM 318
VED+D + +K VT SG+LNF+DG+W SGEER++VFT
Sbjct: 299 VEDIDCSIELKQREAGEERTKSNSTEEDKGEDKVTLSGLLNFVDGLWSTSGEERIIVFTT 358
Query: 319 NSKENVDPNLLRPGRVDVHIHFPLCDFSSFKTLASNYLGVKDHKLFPQVQEIFENGASLS 378
N KE +D L+RPGR+D+HIH C +F+ LASNY + H +P+++E+ + ++
Sbjct: 359 NYKERLDQALMRPGRMDMHIHMGYCTPEAFRILASNYHSIDYHVTYPEIEELIKE-VMVT 417
Query: 379 PAEIGELMIAN 389
PAE+ E ++ N
Sbjct: 418 PAEVAEALMRN 428
Database: nr
Posted date: Jul 5, 2005 12:34 AM
Number of letters in database: 863,360,394
Number of sequences in database: 2,540,612
Lambda K H
0.322 0.138 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 797,217,016
Number of Sequences: 2540612
Number of extensions: 34313263
Number of successful extensions: 99547
Number of sequences better than 10.0: 1890
Number of HSP's better than 10.0 without gapping: 788
Number of HSP's successfully gapped in prelim test: 1102
Number of HSP's that attempted gapping in prelim test: 96488
Number of HSP's gapped (non-prelim): 2393
length of query: 468
length of database: 863,360,394
effective HSP length: 132
effective length of query: 336
effective length of database: 527,999,610
effective search space: 177407868960
effective search space used: 177407868960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC144389.8


